Abstract
FEZ family zinc finger 1 (FEZF1) is an essential transcription factor during olfactory development. In gastrointestinal tumors, FEZF1 plays an oncogenic role through DNA demethylation. However, the role of FEZF1 in the prognosis of human glioma prognosis remains unclear. In this research, we discovered that FEZF1 was significantly increased in glioma tissues in contrast to normal brain tissues (NBTs; P < 0.05). Moreover, the expression of FEZF1 showed a significant correlation with Eastern Cooperative Oncology Group performance status, World Health Organization grade, isocitrate dehydrogenase 1 mutation, over-expression of glial fibrillary acidic protein and 1p19q co-deletion. Furthermore, a high level of FEZF1 in patients with glioma acted as an independent biomarker to predict reduced survival (P = 0.026). In an in vitro experiment, FEZF1 can promote the proliferation, migration, and invasion of glioma cells and inhibit cell apoptosis by activating Akt-ERK pathway. All these findings suggest that FEZF1 acts as a key oncogene and predicts a poor prognosis in glioma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As the most common malignant tumor in the brain, gliomas are grouped into two subtypes: low-grade gliomas (LGG, grades I and grade II) and high-grade gliomas (HGG, grades III and grade IV) (Xi et al. 2016). Due to rapid progression and resistance to chemoradiotherapy, the outcomes for patients with glioma are dismal, with an extremely low 5-year survival rate and a median survival of only 12–15 months after diagnosis (Lv et al. 2016; Marfia et al. 2016; Penas-Prado and Gilbert 2007). Therefore, identification of crucial regulator that control glioma risk stratification is critically important for developing more effective therapeutics.
Several markers that predict a good or poor treatment have been reported (Meng et al. 2017). Specifically, isocitrate dehydrogenase 1 (IDH1) mutation, glial fibrillary acidic protein (GFAP), and 1p19q co-deletion were written in the 2016 World Health Organization (WHO) classification of central nervous system (CNS) tumors. However, the clinical relevance of these clinicobiological markers has not yet been fully studied (Cancer Genome Atlas Research et al. 2015; Paulus 2009; Yan et al. 2009). Therefore, the identification of new biomarkers to develop patient-specific treatments as well as improve the prognosis of patients with glioma becomes imperative.
The forebrain embryonic zinc finger 1 (FEZF1), also known as FEZ or ZNF312B, is a transcription factor that encodes a protein which promotes the migration of GnRH neurons (Kotan et al. 2014). Moreover, FEZF1 was reported to involve in olfactory development (Eckler et al. 2011). Further study indicated that FEZF1 promoted the progression of gastric cancer through epigenetic modification, including DNA demethylation and histone acetylation (Song et al. 2011). Nevertheless, the relationship between FEZF1 and the outcomes of glioma remains ambiguous.
In this study, we detected the expression of FEZF1 in 166 clinical glioma samples and assessed its correlation with gender, age, WHO grades, Eastern Cooperative Oncology Group (ECOG) performance status, IDH1 mutation, GFAP over-expression and 1p19q co-deletion. Our results revealed that expression of FEZF1 was closely related to the prognosis of gliomas so that FEZF1 may be employed as an independent prognostic biomarker of glioma.
Materials and Methods
Oncomine Analysis
The mRNA levels of FEZF1 in the brain, brain glioblastomas, and brain astrocytomas were determined through analysis of oncomine database (www.oncomine.org). The dataset included 582 brain glioblastomas, 63 brain astrocytomas, and 37 normal brains. Students’ t test was used to calculate the p value for comparison between glioma and normal control datasets.
Glioma Specimens
We studied a total of 166 glioma tissue samples and 30 NBTs from patients with epilepsy and brain injury in the Department of Pathology and Neurosurgery, Shengjing Hospital of China Medical University, admitted from October 2007 to September 2016. After resection, brain tissues were immediately frozen in liquid nitrogen and clinical pathological data, such as pathological grade, ECOG score, sex, age, IDH1 mutation, expression of GFAP, and 1p19q co-deletions were collected according to the 2016 WHO classification of CNS tumors. Furthermore, the date of deaths directly related to glioma was recorded. The interval from the date of diagnosis to death was defined as the survival time. Permission was obtained from the patients, and the study was permitted by the Ethics Committee of Shengjing Hospital (No. 2016PS366K).
RNA Isolation and Quantitative RT–PCR
Total RNA was extracted from 17 NBTs and 34 gliomas with the use of TRIzol (Life Technologies Corporation, CA, USA). After measuring the concentration of RNA, we employed a One-Step SYBR PrimeScript RT–PCR Kit (Takara Bio, Inc., Kusatsu, Japan) to detect the expression of FEZF1 mRNA by qRT–PCR. GAPDH was employed as an endogenous control. The relative expression of FEZF1 mRNA was normalized to GAPDH by applying the ΔΔCt method. Primer sequences for FEZF1, forward 5′-GCCGAAAGGAATAAACTGGTGG-3′, reverse 5′-CGAGGTTTTGAACGCGATTTTT-3′; GAPDH forward 5′-TGCACCACCAACTGCTTAGC-3′, reverse 5′-GGCATGCACTGTGGTCATGAG-3′.
Western Blot Analysis
The lysates were extracted from NBTs and glioma tissues, U87 and U251 cells. Protein levels of FEZF1, p-Akt/Akt, and p-ERK/ERK were further analyzed by SDS–polyacrylamide gel electrophoresis (PAGE) and transferred for immunoblotting with antibodies against FEZF1 (1:200, Santa Cruz Biotechnology, Dallas, TX, USA), p-ERK (1:1000; Cell Signaling, Beverly, MA, USA), ERK (1:1000; Cell Signaling, Beverly, MA, USA), p-AKT (1:1000; Cell Signaling, Beverly, MA, USA), AKT (1:1000; Cell Signaling, Beverly, MA, USA), or GAPDH (1:1000, Santa Cruz Biotechnology) overnight at 4 °C. The protein bands were scanned by an ECL chemiluminescence detection system, and relative integrated density values (IDVs) were calculated by FLuor Chem2.0 software using GAPDH as a loading control for protein normalization.
Immunohistochemistry
One hundred and sixty-six glioma samples (WHO grade I 26 cases, grade II 42 cases, grade III 39 cases, and grade IV 59 cases) and 30 NBTs were paraffin-embedded and sectioned. The paraffin-embedded sections were then stained with an antibody directly against FEZF1 (1:200, Abcam, Cambridge, UK) at 4 °C overnight. Biotinylated goat anti-rabbit IgG was applied as secondary antibody. Finally, the sections were counterstained with 3,3′-diaminobenzidine solution. Images were recorded under a light microscope (Olympus, Japan) at ×200 and ×400 magnifications.
FEZF1 expression was scored by two pathologists based on the staining intensity as well as the proportion of positive cells. We defined the score for the percentage of FEZF1-positive stained cells as follows: 0 (less than 10% cells stained), 1 (10–50% cells stained), 2 (50–90% cells stained), and 3 (more than 90% cells stained). Staining intensity was rated in a range of 0–3. For every patient, three tissue section samples were used in the scoring. Based on the sum of percentage and intensity scores, low [0 (−) to 1 (+)] or high expressions [2 (++) to 3 (+++)] were defined.
Cell Culture
Human U87 and U251 glioma cell lines were purchased from the Chinese Academy of Medical Sciences (Beijing, China). Cells were cultured in DMEM/high glucose with added 10% fetal bovine serum in a humidified incubator (37 °C, 5% CO2).
Cell Transfection and Generation of Stable Transfected Cells
An FEZF1(−) plasmid and its negative control (NC) sequence were purchased from GenePharma (GenePharma, Suzhou, China). An FEZF1(+) plasmid and its NC sequence were purchased from GenScript (Piscataway, NJ, USA). U87 and U251 glioma cells in 24-well plates (Corning, New York, NY, USA) were transfected at approximately 60% confluence using Opti-MEM and Lipofectamine 3000 reagents (Invitrogen, CA, USA) according to the manufacturer’s instructions. Stable cell lines were selected by G418 (Geneticin; Invitrogen).
Cell Proliferation, Migration, Invasion, and Apoptosis Detection
Cell proliferation assay was detected using a CCK-8 assay. Cell migration and invasion assay was detected by transwell assay. We employed a dual-color flow cytometric method to measure apoptosis. Detailed methods have been described previously (Yu et al. 2017a).
Statistical Analyses
All data were expressed as means ± standard deviation (SD) from three independent experiments. We applied SPSS 17.0 software for statistical analysis. The Kaplan–Meier method with a log-rank test for comparisons was utilized for calculating survival curves. A χ2 test was employed to evaluate the relationship between FEZF1 and clinicopathological characteristics. Univariable and multivariate Cox analysis was applied to analyze the prognostic value of clinicopathological characteristics and independent prognostic factors, respectively. P < 0.05 was regarded as statistically significant.
Results
FEZF1 Expression Was Up-Regulated in Glioma Tissues
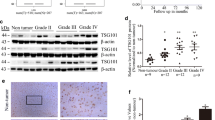
First, we analyzed the expression of FEZF1 in the Cancer Genome Atlas (TCGA) gene expression database (UCSC refGene) derived from patients with glioma. We found that it was significantly elevated in both glioblastoma (582 cases) and astrocytoma (63 cases) samples compared with normal tissue samples (37 cases; P < 0.01, Fig. 1a). We then determined the mRNA level and protein level of FEZF1 in NBTs, LGG, and HGG by qRT–PCR and Western blot. As expected, the mRNA and protein levels of FEZF1 were distinctly increased in gliomas compared with NBTs (P < 0.01; Fig. 1b, c). In addition, the levels of FEZF1 in HGG were significantly increased in contrast with LGG (P < 0.01) (Table 1).
FEZF1 was up–regulated in glioma tissues. a Expression of FEZF1 was up-regulated in 582 brain glioblastomas and 63 astrocytomas (tumor) compared with 37 adjacent normal brain tissues (NBTs) (adjacent) in the Oncomine database: TCGA brain 2 (RefSeq Genes (UCSC refGene, July 2009, hg18, NCBI 36.1, March 2006). b qRT–PCR analysis of FEZF1 mRNA expression in NBTs, low-grade gliomas (LGG) and high-grade gliomas (HGG), with GAPDH used as an endogenous control. c, d Expression of FEZF1 protein was detected by western blot in NBTs, LGG, and HGG, with GAPDH used as an endogenous control. Data are presented as the mean ± standard deviation (SD; n = 15, each group). **P < 0.01 vs. NBTs group; ##P < 0.01 vs. LGG. *P < 0.05 vs. NBTs group; #P < 0.05 vs. LGG
IHC revealed that FEZF1 was mainly located in the nucleus in glioma cells. In 166 patients with glioma, 36 patients were strongly positive (+++) for FEZF1 staining, 61 patients were moderately positive (++), 41 patients were weakly positive (+), and 28 patients showed negative staining (−). Furthermore, IHC revealed that the levels of FEZF1 were markedly increased in glioma tissues compared with NBTs (Fig. 2), a similar trend to that observed in western blot analysis. These results demonstrate the pivotal role of FEZF1 in the aggressive behavior of glioma.
Typical images of FEZF1 detected by immunohistochemistry in glioma and normal brain tissues. a1, a2 Normal brain tissues (NBTs). b1, b2 Grade I gliomas. c1, c2 Grade II gliomas. d1, d2 Grade III gliomas. e1, e2 Grade IV gliomas. The photographs were taken at ×200 magnification for a1–e1, and ×400 magnification for a2–e2
The Correlation between FEZF1 and Clinical Characteristics
After appraising the expression of FEZF1 in gliomas of various grades by western blot and IHC, we further assessed the relationship between FEZF1 expression and WHO grade. We considered moderate (++) and strong positive staining (+++) as high expression of FEZF1, while weak positive (+) and negative staining (−) as low expression of FEZF1. For the 26 patients with a glioma of WHO grade I and 42 patients with a glioma of grade II, 17 (65.4%) and 26 cases (61.9%), respectively, were defined as FEZF1 low expression, while the FEZF1 low expression percentages for patients with glioma WHO grades III and IV were 41.0% (16/39 cases) and 16.9% (10/59 cases), respectively. Our data showed that expression of FEZF1 was significantly correlated with the WHO grade (P < 0.001), the ECOG score standard (P = 0.044), age (P = 0.043), IDH1 mutation (P = 0.001), and GFAP over-expression (P = 0.021). However, FEZF1 was not associated with sex (P = 0.264) and 1p19q co-deletion (P = 0.065) (Table 2).
Over-Expressed FEZF1 Predicted a Poor Prognosis in Patients with Glioma
The correlation of FEZF1 with WHO grade suggested that FEZF1 may play a significant role in the prognosis of glioma. We, therefore, further explored the effects of FEZF1 on the prognosis of patients with glioma. Of the 166 patients, 155 patients died of glioma during the follow-up period in our study. Kaplan–Meier analysis with a log-rank test revealed that patients with glioma in the high FEZF1 expression group manifested a worse prognosis compared with those in the low FEZF1 expression group (P < 0.001; Fig. 3a). The median survival time was 33.86 months (95% CI 28.95–38.76) for patients with glioma in the low FEZF1 expression group, while it was 20.55 months (95% CI 17.57–23.52) in the high FEZF1 expression group. Thus, the high expression of FEZF1 was associated with decreased survival.
Kaplan–Meier survival curve for patients with glioma and different FEZF1 expression. a Kaplan–Meier survival curve for all patients with glioma (n = 166). Patients showing a low expression of FEZF1 manifested a significantly higher probability of survival. b Kaplan–Meier survival curve for patients with low-grade gliomas (LGG; n = 68). The probability of survival in patients with glioma and high FEZF1 expression was markedly lower than those with low FEZF1 expression. c Kaplan–Meier survival curve for patients with high-grade gliomas (HGG). The probability of survival in patients with glioma and high FEZF1 expression was lower than for those with low FEZF1 expression (log-rank test P < 0.001)
To further assess the effect of FEZF1 on the prognosis of glioma, we evaluated the correlation between the expression of FEZF1 and prognosis in LGG and HGG, separately. FEZF1 was shown to negatively correlate with overall survival. Moreover, FEZF1 was a significant prognostic biomarker in both LGG (P < 0.001) and HGG (P < 0.001; Fig. 3b, c). Among 68 cases of patients with LGG, the median survival time was 39.88 months (95% CI 34.06–45.71) for patients in the low FEZF1 expression group, while it was 32.84 months (95% CI 25.85–39.83) for those in the high FEZF1 expression group. Similarly, the median survival time was 23.88 months (95% CI 16.16–31.60) in the low FEZF1 expression group of HGG patients compared with 16.27 months (95% CI 13.64–18.91) in the high FEZF1 expression group. Thus, high FEZF1 expression was inversely correlated with poor median overall survival.
After univariable Cox analysis, the high expression of FEZF1 (P < 0.001) was found to closely correlate with survival time. The other correlating factors included age > 50 (P = 0.001), HGG (P < 0.001), ECOG score ≥ 2 (P < 0.001), IDH mutation (P < 0.001), GFAP over-expression (P < 0.001), and 1p19q co-deletion (P < 0.001) (Table 3). Multivariable Cox analysis indicated that the high expression of FEZF1 was an independent prognostic factor for shorter survival in patients with glioma (P = 0.026). Moreover, other data, including WHO grade (P = 0.004), ECOG (P = 0.021), IDH mutation (P = 0.009), and 1p19q (P = 0.016) co-deletion, were obviously associated with the survival of patients with glioma (Table 4).
Over-Expressed FEZF1 Accelerated Proliferation, Migration and Invasion While Restrained Glioma Cell Apoptosis by Regulating Akt/ERK Pathway
The expression of FEZF1 closely correlated with the prognosis of glioma, indicating the potential regulatory role of FEZF1 in glioma malignant biological properties. As a consequence, we constructed cells showing up-regulated and down-regulated FEZF1 expression and measured the effect of FEZF1 on U87 and U251 glioma cells. The results implied that in comparison with the FEZF1(+)-NC group, the proliferation, migration, and invasion of U87 and U251 glioma cells were enhanced in the FEZF1(+) group (Fig. 4b, c). Moreover, up-regulated FEZF1 significantly suppressed apoptosis in U87 and U251 cells (Fig. 4d). Thus, FEZF1 appeared to enhance proliferation migration and invasion, while it appeared to decrease apoptosis in glioma cells.
FEZF1 accelerated the cell proliferation, migration, and invasion while inhibited apoptosis of glioma cells. a Relative expression of FEZF1 after U87 and U251 glioma cells were transfected with FEZF1(+), or FEZF1(−) as well as their scrambled vectors (NC). b A CCK8 assay was performed to evaluate the effect of FEZF1 on the proliferation of glioma cells. c A transwell assay was performed to evaluate the effect of FEZF1 on the migration and invasion of glioma cells. Representative images and accompanying statistical plots are presented. d Flow cytometry analysis of the effects of FEZF1 on glioma cells. Data are presented as the mean ± standard deviation (SD; n = 3, each group). *P < 0.05 vs. FEZF1 (+)-NC group; #P < 0.05 vs. FEZF1 (−)-NC group. Scale bars 20 μm. The photographs were taken at ×200 magnification
To investigate the mechanism of FEZF1 promoting the malignant biological behaviors of glioma cells, we detected the pathways changed in FEZF1(+) glioma cells. As shown in Fig. 5, the ratio of p-AKT/t-AKT and p-ERK1/2/t-ERK1/2 was significantly raised in the FEZF1(+) group as compared to FEZF1(+)-NC group.
Discussion
As the most prevalent central nervous system tumors, glioma accounts for 50% of all the intracranial tumors (Chen et al. 2015). Because of its dismal prognosis and the difficulty of an early diagnosis, an urgent need exists to identify molecular biomarkers that can predict the prognosis for glioma. Our research revealed that FEZF1 mRNA and protein expression was elevated in glioma tissues. In addition, we appraised the clinical significance of FEZF1 in 166 patients with glioma. We found that the expression of FEZF1 negatively correlated with overall survival and FEZF1 acts as an independent prognostic factor for a low probability of survival in patients with glioma. In in vitro experiments, FEZF1 was shown to accelerate the proliferation, migration, and invasion of U87 and U251 glioma cells and to inhibit apoptosis in glioma cells.
FEZF1, also known as HH22 or ZNF312B, is a transcription factor belonging to the FEZ family. Our previous research indicated that FEZF1 was highly expressed in glioma stem cells (GSCs) in which it acted as an oncogene (Yu et al. 2017a, b). In this research, we evaluated the expression of FEZF1 in gliomas as well as NBTs. FEZF1 was found up-regulated in gliomas compared to NBTs. Furthermore, FEZF1 expression was elevated in HGG compared to LGG. Previous articles uncovered a similar oncogenic function of FEZF1. In gastric cancer, FEZF1 was activated by the K-ras oncogene and promoted cancer progression and metastasis (Song et al. 2009). Our results illustrated, for the first time, that the expression of FEZF1 was positively correlated with a progression in the pathological grade of the glioma; however, whether FEZF1 is a potential prognostic predictor of glioma needs to be further investigated.
Correlation analysis implied that the expression of FEZF1 was closely associated with glioma grade, ECOG standard score, age, IDH mutation, and GFAP over-expression. Patients with an ECOG ≥ 2, IDH mutation, and GFAP over-expression manifested a higher FEZF1 expression compared with patients with ECOG < 2. In addition, older patients with glioma (age > 50 years) showed higher FEZF1 expression. However, gender and 1p19q co-deletion were discovered to have no association with the expression of FEZF1. The data above highlight the potential role of FEZF1 as a biomarker of glioma.
Kaplan–Meier survival analysis implied that glioma patients with high FEZF1 expression exhibited a low probability of survival. Further analysis indicated that patients with low FEZF1 expression tend to have a high probability of survival with either LGG or HGG. In addition, we applied univariate and multivariate analyses to the survival of patients with glioma and confirmed that the expression of FEZF1 was an independent prognostic factor for the survival of patients with glioma. The other prognostic factors were WHO grade, ECOG, IDH1 mutation, and 1p19q co-deletion.
Furthermore, we appraised the regulatory role of FEZF1 in glioma and obtained stable glioma cell lines overexpressing FEZF1 or where FEZF1 was silenced by transfection. The CCK-8 assay and transwell assay suggested that glioma cell proliferation, migration, and invasion were elevated in the FEZF1(+) group compared to those of the FEZF1(+)-NC group. In addition, the over-expression of FEZF1 markedly hindered apoptosis in glioma cells. Taken together, FEZF1 acted as an oncogene in U87 and U251 glioma cells.
It was reported that Akt and ERK activation promoted glioma cell proliferation, migration, and invasion (Liu et al. 2018; Yu et al. 2017b). Our research certified that p-AKT/AKT and p-ERK1/2/ERK1/2 were significantly decreased in glioma cells after inhibition of FEZF1, which proved that FEZF1 regulated malignant biological behavior of glioma by activating AKT/ERK1/2 pathways.
In summary, our study found that FEZF1 was elevated in gliomas. Kaplan–Meier survival analysis implied that patients with glioma and a high FEZF1 level exhibited a low probability of survival; FEZF1 was also found to be an independent prognostic factor for survival in patients with glioma. In in vitro experiments, the knockdown of FEZF1 hindered cellular proliferation, migration, and invasion, but promoted apoptosis in glioma cells. Therefore, FEZF1 is a poor prognostic factor and may become a new therapeutic target in glioma.
References
Cancer Genome Atlas Research, N, Brat DJ, Verhaak RG et al (2015) Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372:2481–2498
Chen Y, Wu Y, Huang X, Qu P, Li G, Jin T, Xing J, He S (2015) Leukocyte telomere length: a novel biomarker to predict the prognosis of glioma patients. J Cancer Res Clin Oncol 141:1739–1747
Eckler MJ, McKenna WL, Taghvaei S, McConnell SK, Chen B (2011) Fezf1 and Fezf2 are required for olfactory development and sensory neuron identity. J Comp Neurol 519:1829–1846
Kotan LD, Hutchins BI, Ozkan Y, Demirel F, Stoner H, Cheng PJ, Esen I, Gurbuz F, Bicakci YK, Mengen E, Yuksel B, Wray S, Topaloglu AK (2014) Mutations in FEZF1 cause Kallmann syndrome. Am J Hum Genet 95:326–331
Liu YY, Chen MB, Cheng L, Zhang ZQ, Yu ZQ, Jiang Q, Chen G, Cao C (2018) microRNA-200a downregulation in human glioma leads to Galphai1 over-expression, Akt activation, and cell proliferation. Oncogene 37:2890–2902
Lv QL, Hu L, Chen SH, Sun B, Fu ML, Qin CZ, Qu Q, Wang GH, He CJ, Zhou HH (2016) A long noncoding RNA ZEB1-AS1 promotes tumorigenesis and predicts poor prognosis in glioma. Int J Mol Sci 17
Marfia G, Navone SE, Fanizzi C, Tabano S, Pesenti C, Abdel Hadi L, Franzini A, Caroli M, Miozzo M, Riboni L, Rampini P, Campanella R (2016) Prognostic value of preoperative von Willebrand factor plasma levels in patients with glioblastoma. Cancer Med 5:1783–1790
Meng L, Wang X, Liao W, Liu J, Liao Y, He Q (2017) BAF53a is a potential prognostic biomarker and promotes invasion and epithelial-mesenchymal transition of glioma cells. Oncol Rep 38:3327–3334
Paulus W (2009) GFAP, Ki67 and IDH1: perhaps the golden triad of glioma immunohistochemistry. Acta Neuropathol 118:603–604
Penas-Prado M, Gilbert MR (2007) Molecularly targeted therapies for malignant gliomas: advances and challenges. Expert Rev Anticancer Ther 7:641–661
Song IS, Ha GH, Kim JM, Jeong SY, Lee HC, Kim YS, Kim YJ, Kwon TK, Kim NS (2011) Human ZNF312b oncogene is regulated by Sp1 binding to its promoter region through DNA demethylation and histone acetylation in gastric cancer. Int J Cancer 129:2124–2133
Song IS, Oh NS, Kim HT, Ha GH, Jeong SY, Kim JM, Kim DI, Yoo HS, Kim CH, Kim NS (2009) Human ZNF312b promotes the progression of gastric cancer by transcriptional activation of the K-ras gene. Cancer Res 69:3131–3139
Xi Z, Xue Y, Zheng J, Liu X, Ma J, Liu Y (2016) WTAP expression predicts poor prognosis in malignant glioma patients. J Mol Neurosci 60:131–136
Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD (2009) IDH1 and IDH2 mutations in gliomas. N Engl J Med 360:765–773
Yu M, Xue Y, Zheng J, Liu X, Yu H, Liu L, Li Z, Liu Y (2017a) Linc00152 promotes malignant progression of glioma stem cells by regulating miR-103a-3p/FEZF1/CDC25A pathway. Mol Cancer 16:110
Yu Y, Fu X, Ran Q, Yang K, Wen Y, Li H, Wang F (2017b) Globularifolin exerts anticancer effects on glioma U87 cells through inhibition of Akt/mTOR and MEK/ERK signaling pathways in vitro and inhibits tumor growth in vivo. Biochimie 142:144–151
Funding
This work is supported by grants from the Natural Science Foundation of China (81672511, 81372484, and 81573010) and Scientific Fund of Shengjing hospital (MF43).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Patients gave their signed informed consent for publication. This study was permitted by the Ethics Committee of Shengjing Hospital.
Conflict of Interest
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Yu, M., Yu, S., Xue, Y. et al. Over-Expressed FEZF1 Predicts a Poor Prognosis in Glioma and Promotes Glioma Cell Malignant Biological Properties by Regulating Akt-ERK Pathway. J Mol Neurosci 65, 411–419 (2018). https://doi.org/10.1007/s12031-018-1108-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-018-1108-0