Abstract
Purpose
The correlation of IL-8 and IL-18 gene polymorphisms with colorectal cancer (CRC) was investigated by previous studies, though the results remained conflicting. Thus, the meta-analysis was performed to investigate the association of IL-8 -251T>A and IL-18 -607C>A polymorphisms with CRC risk.
Methods
A comprehensive search of the PubMed, Web of Science, CNKI, SciELO, and Wanfang databases was performed up to February 20, 2020. The strength of the associations was calculated with odds ratios (ORs) and their corresponding 95% of confidence intervals (CIs).
Results
A total of 16 case-control studies including 13 studies with 3908 cases and 5005 controls on IL-8 -251T>A polymorphism and three studies with 396 cases and 560 controls on IL-18 -607C>A polymorphism were selected. Pooled data revealed that the IL-8 -251T>A and IL-18 -607C>A polymorphisms were not significantly associated with an increased risk of CRC in global population. When stratified by ethnicity, source of controls, sample size, and Hardy-Weinberg equilibrium (HWE), there were still no significant association between IL-8 -251T>A polymorphism and risk of CRC.
Conclusions
Our results revealed that the IL-8 -251T>A and IL-18 -607C>A polymorphisms were not associated with an increased susceptibility to CRC. We strongly call for further studies with larger sample sizes and different ethnicities to confirm our findings.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is one of the leading causes of mortality and morbidity worldwide, accounting for an estimated 1.8 million new cases and 881,000 deaths in 2018 [1,2,3]. Globally, CRC represents the most common cancer of the digestive system, and about 1 in 10 cancer deaths is due to CRC [4, 5]. CRC is the third cause of cancer after prostate and lung cancer in men (10% of total) and the second most common diagnosed cancer type after breast cancer in women (9.4% of total) [6, 7]. Despite decades of investigations, the etiology, pathogenesis, and risk factors of CRC remain unknown [8]. CRC is a complex disease that develops as a complex interactions between genetic and environmental factors [9,10,11]. Hereditary predisposition plays an obvious part in development of CRC, although 80% of colorectal neoplasms occur in the absence of a family history of CRC [12].
Several prior studies have demonstrated that cytokine levels in plasma or serum varied significantly between CRC patients and healthy individuals, indicating that cytokine levels may be useful in screening or detecting CRC [13,14,15]. Interleukin 8 (IL-8), also named C-X-C motif chemokine ligand 8 (CXCL8), is located on chromosome 4q-13-21, contains 10 exons, and spans 5.2 kb in length [16, 17]. IL-8 plays important role in tumor formation processes such as angiogenesis, tumorigenesis, tissue invasion, and metastasis [18]. Moreover, the IL-18 was initially identified as a protein that induces interferon γ (IFNγ) production which is an ambiguous role of the immune system in cancer progression [19, 20]. The human IL-18 gene is located on chromosome 11q22.2-q22.3, contains six exons, and encompasses several polymorphisms within the promoter region [21].
Some epidemiologic studies evaluated the association of IL-8 -251T>A and IL-18 -607C>A polymorphisms with susceptibility to CRC. However, the results were inconsistent or even contradictory. Some reasons may be due to ethnic background of population, small sample size, inclusion or exclusion criteria, study design, and genotyping methods [22]. Moreover, the reason for this disagreement may be related to gene-gene and gene-environment interactions on development of CRC. Therefore, we performed a comprehensive meta-analysis to identify statistical evidence of the association between IL-8 -251T>A and IL-18 -607C>A polymorphisms and risk of CRC by retrieving all eligible studies.
Materials and Methods
Identification of Relevant Studies
A comprehensive literature search from PubMed, Goggle Scholar, EMBASE, Cochrane Library database, Springer Link, Chinese Biomedical Database (CBD), China National Knowledge Infrastructure (CNKI) platforms, Wanfang, and VIP database was conducted to identify all relevant studies on IL-8 -251T>A and IL-18 -607C>A polymorphisms with CRC risk up to February 20, 2020. We used the combination of the following search terms and keywords: (“Colorectal Cancer” OR “CRC” OR “Bowel Cancer” OR “Colon Cancer” OR “Rectal Cancer” OR “Tumor” OR “Cancer” OR “Neoplasm”) AND (“Interleukin-8” OR “IL-8” OR “chemokine (C-X-C motif) ligand 8” OR “CXCL8”) AND (“-251T>A” OR “rs4073”) AND (“Interleukin-18” OR “IL-18” OR “ Interferon-Gamma Inducing Factor”) AND (“-607C>A” OR “rs1946518”) AND (“Gene” OR “Polymorphism” OR “SNPs” OR “Mutation” OR “Variation” OR “Allele”). We have also manually screened the reference lists of eligible articles and reviews to retrieve additional articles. There was no language, ethnicity, or country restriction, and the literature search was limited to humans.
Inclusion and Exclusion Criteria
The following inclusion criteria were used to select literatures for the meta-analysis: (1) studies with case-control and cohort design, (2) studies evaluated the association of IL-8 -251T>A and IL-18 -607C>A polymorphisms with CRC risk, and (3) providing sufficient genotype data for both cases and controls to calculate the odds ratio (OR) with 95% confidence interval (CI). In addition, the following exclusion criteria were applied: (1) no usable data reported; (2) animal studies; (3) studies only involved a case population (without controls); (4) linkage studies and family-based studies; (5) repeated or overlapping studies; and (6) case report, reviews, commentaries, posters, abstracts, reviews, editorials, and conference papers.
Data Extraction
Data were carefully extracted independently and systematically from all eligible articles by two investigators. Any disagreements of the studies were resolved by third author. For each study, the following data were retrieved: first author’s name, year of publication, country or region, ethnic group of the study population, source of controls, genotyping methods, sample size, genotype and allele frequencies for the IL-8 -251T>A and IL-18 -607C>A polymorphisms in cases and healthy subjects, and minor allele frequency (MAFs) and Hardy-Weinberg equilibrium (HWE) in healthy subjects. Ethnicity was categorized as Asian, Caucasian, and mixed. For studies including subjects of different ethnic groups or for both IL-8 -251T>A and IL-18 -607C>A polymorphisms, the data were extracted separately.
Statistical Analysis
The strength of the association of the IL-8 -251T>A and IL-18 -607C>A polymorphisms with risk of CRC was assessed by odds ratios (ORs) with 95% confidence intervals (CIs). The Z test was applied to assess the significance of pooled ORs, in which P < 0.05 defined as the significance. The pooled ORs were calculated under five genetic models, i.e., allele (B vs. A), homozygote (BB vs. AA), heterozygote (BA vs. AA), dominant (BB + BA vs. AA), and the recessive (BB vs. BA + AA). A Cochran’s Q statistic was used to assess between-study heterogeneity, in which P ≤ 0.10 indicated significant heterogeneity was found. Moreover, we used the I2 statistic to qualify (degree of heterogeneity) the heterogeneity (range of 0 to 100%: I2 = 0–25%, no heterogeneity; I2 = 25–50%, moderate heterogeneity; I2 = 50–75%, large heterogeneity; I2 = 75–100%, extreme heterogeneity). When P < 0.1 (I2 > 50%), statistically significant heterogeneity was assumed, and a random effect (DerSimonian and Laird’s method) was used to calculate the pooled OR when heterogeneity was found; otherwise, a fixed-effect model (Mantel-Haenszel method) in absence of heterogeneity was applied. The agreement of genotype frequencies in healthy subjects with Hardy-Weinberg equilibrium (HWE) was tested using the Pearson’s χ2 test. A P value of < 0.05 indicated deviation from HWE. Subgroup analyses were performed based on ethnicity, genotyping methods, source of controls (hospital- and population-based), and sample size (< 250 and ≥ 250). Sensitivity analyses were performed to assess influence of each single study on pooled ORs and the stability of the meta-analysis results by sequential remove of individual studies. In addition, sensitivity analysis was performed by excluding HWE violating studies to examine the stability of the pooled data. Egger’s and Begg-Mazumdar’s tests were used to identify possible publication bias. A nonsymmetrical funnel plot or P less than 0.05 indicated that there was significant publication bias. If the publication bias tests indicated bias existed, the Duval and Tweedie “trim and fill” method was used to adjust the bias. All of the statistical calculations were performed using Comprehensive Meta-Analysis (CMA) software version 2.0 (Biostat, USA). Two-sided P values < 0.05 were considered statistically significant.
Results
Study Characteristics
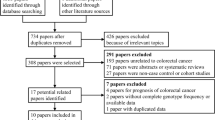
Figure 1 shows the flowchart of literature search and selection process. The initial literature searches retrieved 114 potentially relevant studies. After reading the titles and abstracts, 50 studies were excluded. Subsequently, 48 studies were excluded because not reporting useful data for the analysis, reviews, case only study, and not being case-control studies. Finally, 16 case-control studies including 13 case-control studies with 3908 CRC cases and 5005 controls on IL-8 -251T>A polymorphism [23,24,25,26,27,28,29,30,31,32,33,34,35] and three case-control studies with 396 CRC cases and 560 controls on IL-18 -607C>A polymorphism [14, 36, 37] were selected. The main characteristics of the studies were shown in Table 1. All included studies were published in English and Chinese between July 2003 and July 2015. The sample size of an individual cohort ranged from 191 to 1023 for CRC cases and 191 to 1121 for healthy controls. The subjects included Spanish, Grecian, US-American, Dane, Francis, Croatian, Dutch, Polish, Malaysian, Romanian, and Scottish individuals. As for ethnicity, eleven studies were conducted among Caucasians and one article among Asians. TaqMan, ARMS-PCR, PCR-RFLP, and AS-PCR methods were applied for genotyping. The genotype and minor allele frequency (MAF) distributions in the studies considered in the present meta-analysis are shown in Table 1. Moreover, the distribution of genotypes in the controls was in agreement with Hardy-Weinberg equilibrium (HWE) for all selected studies, except for four studies (Table1).
Quantitative Data Synthesis
IL-8 -251T>A Polymorphism
The summary of the meta-analysis of the association of IL-8 -251T>A polymorphism with CRC risk is shown in Table 2. Overall, pooled ORs showed that IL-8 -251T>A polymorphism was not significantly associated with an increased risk of CRC under all five genetic models, i.e., allele (T vs. A: OR = 1.024, 95% CI 0.898–1.168, P = 0.724, Fig. 2a), homozygote (TT vs. AA: OR = 1.173, 95% CI 0.941–1.461, P = 0.156), heterozygote (TA vs. AA: OR = 1.060, 95% CI 0.876–1.283, P = 0.549), dominant (TT + TA vs. AA: OR = 1.190, 95% CI 0.980–1.447, P = 0.752), and recessive (TT + TA vs. AA: OR = 1.119, 95% CI 0.954–1.311, P = 0.167, Fig. 2b). Moreover, stratified analysis by ethnicity, source of controls, and genotyping methods still showed that IL-8 -251T>A polymorphism was not associated with risk of CRC (Table 2).
IL-18 -607C>A Polymorphism
The summary of the meta-analysis of the association of IL-18 -607C>A polymorphism with CRC risk is shown in Table 2. Overall, pooled results revealed that IL-18 -607C>A polymorphism was not significantly associated with risk of CRC under all five genetic models, i.e., allele (A vs. C: OR = 1.066, 95% CI 0.883–1.288, P = 0.506), homozygote (AA vs. CC: OR = 1.086, 95% CI 0.732–1.611, P = 0.682), heterozygote (AC vs. CC: OR = 1.349, 95% CI 0.706–2.578, P = 0.365), dominant (AA + AC vs. CC: OR = 1.207, 95% CI 0.906–1.608, P = 0.198), and recessive (AA + AC vs. CC: OR = 0.898, 95% CI 0.642–1.255, P = 0.528, Table 2).
Between-Study Heterogeneity Test
There were statistically significant heterogeneity for IL-8 -251T>A polymorphism under all five genetic models, i.e., allele (I2 = 78.84; PH ≤ 0.001), homozygote (I2 = 63.29; PH = 0.002), heterozygote (I2 = 67.57; PH ≤ 0.001), dominant (I2 = 71.64; PH ≤ 0.001), and recessive (I2 = 54.65; PH = 0.012). Thus, subgroup analysis by ethnicity, genotyping methods, source of controls, and HWE was performed in order to determine the source of heterogeneity among the studies and to assess the effect of race on the association between IL-8 -251T>A polymorphism and CRC risk. As shown in Table 2, most of the heterogeneity disappeared in the subgroup analysis in group of studies used TaqMan approach, indicating that genotyping methods might be the major source of heterogeneity in this meta-analysis.
Sensitivity Analysis
Sensitivity analysis was performed by excluding one study at a time and subsequently recalculating the overall effect to assess the influence of each study on pooled results and robustness of the pooled ORs. The results showed that the significance of the OR was not affected by any single study. Then, sensitivity analysis was conducted by excluding those studies departure from the HWE. Therefore, the sensitivity analysis suggested that the current meta-analysis were relatively consistent even when a single study or some studies were excluded.
Publication Bias
Publication bias was assessed with Begg’s funnel plots and Egger’s test (Table 2). The shape of the funnel plots and Egger’s regression tests revealed evidence of publication bias for IL-8 -251T>A under allele model (A vs. T: PBeggs = 0.046; PEggers = 0.002, Fig. 3) and for IL-18 -607C>A under two genetic models, i.e., heterozygote (AC vs. CC: PBeggs = 0.296; PEggers = 0.020) and dominant (AA + AC vs. CC: PBeggs = 0.296; PEggers = 0.029). Thus, to adjust these biases, we have used a trim-and-fill method developed by Duval and Tweedie. However, after trimming we have yield similar results, indicating that the publication bias has little effect on the results of our study and the results of our meta-analysis are relatively stable.
Discussion
To date, some epidemiological association studies have been performed to evaluate the potential roles of IL-8 -251T>A and IL-18 -607C>A polymorphisms in development of CRC, but the results of these studies were inconsistent and the sample size. Meta-analysis is a statistical procedure for combining data from individual studies to increase the sample size and enhance statistical power. Therefore, in order to resolve this conflict, a meta-analysis was conducted to explore the association of IL-8 -251T>A and IL-18 -607C>A polymorphisms with CRC risk. When all the eligible studies were pooled into the meta-analysis of polymorphism, no significant association was found between IL-8 -251T>A and IL-18 -607C>A polymorphisms and an increased risk of CRC. In further stratified and sensitivity analyses, no significant association was found in any subgroup analysis.
Up to now, there are two previous meta-analyses which have been done on the association of IL-8 -251T>A polymorphism with CRC risk [38, 39]. However, the number of original studies regarding this issue was statistically less for the estimation of the association. In 2012, Hu et al., in a meta-analysis of nine case-control studies with 3019 CRC cases and 3984 controls, evaluated the association between IL-8 -251T>A polymorphism and CRC risk. Their result revealed that IL-8 -251T>A polymorphism was not associated with an increased risk of CRC. Moreover, their subgroup analysis by ethnicity (Caucasians) and source of controls showed that there was no significant association between the polymorphism and CRC risk [38]. In the same year, Wang et al., in another meta-analysis of five studies with 1242 CRC cases and 1880 healthy subjects, have evaluated the association of IL-8 -251T>A polymorphism with CRC risk in Europeans. Similarly, their results showed that IL-8 -251T>A polymorphism was not significantly associated risk of CRC. However, the results of our meta-analysis are in accordance with those reported the previous two meta-analyses on IL-8 -251T>A polymorphism [39]. Our meta-analysis included more studies than previous two meta-analyses; there are 3908 CRC cases and 5005 controls from 13 case-control studies. From a statistical perspective, the previous meta-analyses with relatively inadequate sample sizes have low power to identify genetic association. Moreover, we have performed subgroup analysis by sample size and by excluding the HWE-violating studies.
IL-18 is a pleiotropic cytokine which is involved in the regulation of innate and acquired immune responses and plays key role in autoimmune diseases by controlling Th1- and Th2-type immune response [19]. Moreover, it induces the productions of TNF-α, granulocyte/macrophage colony-stimulating factor, and IFN-γ and increases the cytotoxic effects of NK and T cells in some disease. It was shown that IL-18 -607C>A polymorphism disrupts a potential cAMP-responsive element-binding protein-binding site [40]. These meta-analysis results showed that IL-18 -607C>A polymorphism was not significantly associated with CRC risk. Our results are in consistence with the previous meta-analysis. In 2015, Yao et al., in a meta-analysis of five case-control studies with 1618 cases and 1155 healthy controls, evaluated the relationship of IL-18 -607C>A polymorphism with gastrointestinal cancer risk. Their results showed that the polymorphism did not significantly associate with gastric cancer and CRC risk. However, they have revealed that IL-18 -607C>A polymorphism may be associated with susceptibility to esophageal cancer [41]. Guo et al., in a case-control study, showed that the distributions of IL-18 -607C>A polymorphism did not differ between CRC cases and healthy subjects in Chinese population. However, they found that the IL-18 -137C/-607A haplotype was associated with increased risk of CRC [41].
The heterogeneity is a crucial issue when elucidating the outcomes of a meta-analysis, and finding the possible sources for the high heterogeneity is very important [42]. The studies included in the current meta-analysis probably have different ethnic backgrounds, environmental exposures, methodology, and sample size, thus causing inconsistent conclusions. Moreover, it is evident that other factors such as diversity in source of controls, genotyping , and Hardy-Weinberg equilibrium (WHE) might contribute to potential sources of heterogeneity. In this meta-analysis, there was a significant heterogeneity under all five genetic models in the overall population. Thus, we attempted to minimize this issue by developing strict criteria and subgroup analyses. In the subgroup analysis based on ethnicity, the subgroup results were consistent with the overall results. However, due to the limitation of small sample size of the study in Asians, African, and mixed population, further epidemiological studies should be conducted in different ethnicities to clarify this issue. Moreover, stratification analysis revealed that genotyping methods may be source of heterogeneity in this study. Both funnel plot and Egger’s test were used to assess the publication bias of our meta-analysis. The shape of funnel plot and statistical results revealed an obvious publication bias under the allele genetic model. Therefore, to adjust this bias, we have used a trim-and-fill method developed by Duval and Tweedie. However, after trimming we have found similar results. This indicates that the publication bias has little effect on the results of our study and the results of our meta-analysis are relatively stable.
Although the current study has collected available data on the association between IL-8 -251T>A polymorphism with CRC risk, several limitations could not be neglected. First, the sample size for IL-18 -607C>A polymorphism was considerably small; there are only three studies with a total of 396 cases and 560 controls were selected. Thus, more studies with large sample size and well-designed are needed to further identify the association more comprehensively. Second, studies included in this meta-analysis mainly provided data on Caucasian populations, and the subgroup analyses for other ethnicities were not applicable. Therefore, other ethnicities including Asians, Africans, and mixed populations should be evaluated in future studies. Third, only published studies in English were included in the current study, which might have led to publication and also potential language biases. Fourth, considering the diversity of CRC etiology, its pathogenesis is likely to be affected by factors such as age, gender, ethnicity, environmental factors, and other variables. However, due to lack of the mentioned confounding factors in the original articles, we could not perform the corresponding subgroup analysis. Finally, we were also unable to evaluate the interactions among gene-gene and gene-environment, and the lack of the original data of the included studies limited our further evaluation of potential interactions, which may be an important component of the association between the IL-8 -251T>A and IL-18 -607C>A polymorphisms and risk of CRC.
In summary, our pooled data revealed that the IL-8 -251T>A and IL-18 -607C>A polymorphisms were not associated with susceptibility to CRC. Moreover, stratified analysis by ethnicity, source of controls, and genotyping methods showed that the IL-8 -251T>A polymorphism was not associated with risk of CRC. Given the limited sample size and ethnicities included in the meta-analysis, we strongly call for f further larger scaled, well-designed studies in different ethnicities to confirm these findings.
References
Gandomani HS, Yousefi SM, Aghajani M, Mohammadian-Hafshejani A, Tarazoj AA, Pouyesh V, et al. Colorectal cancer in the world: incidence, mortality and risk factors. Biomed Res Ther. 2017;4:1656.
Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. Termedia Publishing House Ltd. 2019;14:89–103.
Khoram-Abadi KM, Forat-Yazdi M, Kheirandish S, Saeidi N, Zarezade Z, Mehrabi N, et al. DNMT3B -149 C>T and -579 G>T polymorphisms and risk of gastric and colorectal cancer: a meta-analysis. Asian Pac J Cancer Prev. 2016;17:3015–20.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2018; 68, 394–424. CA Cancer J Clin. 2018;68:394–424.
Xie YH, Chen YX, Fang JY. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduction and Targeted Therapy. Springer Nature. 2020;5:1–30.
Ferlay J, Parkin DM, Steliarova-Foucher E. Estimates of cancer incidence and mortality in Europe in 2008. Eur J Cancer (Oxford, England : 1990). 2010;46:765–81.
Ferlay J, Shin H-R, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917.
Marley AR, Nan H. Epidemiology of colorectal cancer. Int J Mol Epidemiol Genet. E-Century Publishing Corporation. 2016;7:105–14.
Raskov H, Pommergaard HC, Burcharth J, Rosenberg J. Colorectal carcinogenesis-update and perspectives. World J Gastroenterol WJG Press. 2014;20:18151–64.
Migliore L, Migheli F, Spisni R, Copped F. Genetics, cytogenetics, and epigenetics of colorectal cancer. J Biomed Biotechnol. 2011;2011:1–19.
Mármol I, Sánchez-de-Diego C, Dieste AP, Cerrada E, Yoldi MJR. Colorectal carcinoma: a general overview and future perspectives in colorectal cancer. Int J Mol Sci. MDPI AG. 2017:18(1):197.
Lynch HT, de la Chapelle A. Hereditary colorectal cancer. Guttmacher AE, Collins FS, editors. N Engl J Med. Massachusetts Medical Society. 2003;348:919–32.
Yamaguchi M, Okamura S, Yamaji T, Iwasaki M, Tsugane S, Shetty V, et al. Plasma cytokine levels and the presence of colorectal cancer. PLoS One. 2019;14:e0213602.
Haghshenas MR, Hosseini SV, Mahmoudi M, Saberi-Firozi M, Farjadian S, Ghaderi A. IL-18 serum level and IL-18 promoter gene polymorphism in Iranian patients with gastrointestinal cancers. J Gastroenterol Hepatol (Australia). 2009;24:1119–22.
Crivello A, Giacalone A, Vaglica M, Scola L, Forte GI, Macaluso MC, et al. Regulatory cytokine gene polymorphisms and risk of colorectal carcinoma. Ann N Y Acad Sci. 2006;1089:98–103.
Zhang M, Fang T, Wang K, Mei H, Lv Z, Wang F, et al. Association of polymorphisms in interleukin-8 gene with cancer risk: a meta-analysis of 22 case–control studies. OncoTargets Ther. 2016;9:3727–37.
Li Y, Bai J, He B, Wang N, Wang H, Liu D. Weak association between the interleukin-8 rs4073 polymorphism and acute pancreatitis: a cumulative meta-analysis. BMC Med Genet. 2019;20:129.
Zhao Z, Wang S, Lin Y, Miao Y, Zeng Y, Nie Y, et al. Epithelial-mesenchymal transition in cancer: Role of the IL-8/IL-8R axis. Oncol Lett. 2017;13(6):4577–4584.
Salimi E, Karimi-Zarchi M, Dastgheib SA, Abbasi H, Tabatabaiee RS, Hadadan A, et al. Association of promoter region polymorphisms of IL-6 and IL-18 genes with risk of recurrent pregnancy loss: a systematic review and meta-analysis. Fetal Pediatr Pathol Taylor and Francis Ltd. 2019;39:346–59.
Palma G, Barbieri A, Bimonte S, Palla M, Zappavigna S, Caraglia M, et al. Interleukin 18: friend or foe in cancer. Biochimica et Biophysica Acta. 2013;1836(2):296–303.
Zhang M-J, Zhou Y, Wang X, Chen X, Pi Y, Guo L, et al. Interleukin-18 gene promoter 607A polymorphism, but not 137C polymorphism, is a protective factor for ischemic stroke in the Chinese population: a meta-analysis. Meta Gene Elsevier. 2016;9:165–72.
Bahrami R, Shajari A, Aflatoonian M, Noorishadkam M, Akbarian-Bafghi MJ, Morovati-Sharifabad M, et al. Association of rearranged during transfection (RET) c.73 + 9277T > C and c.135G > a polymorphisms with susceptibility to Hirschsprung disease: a systematic review and meta-analysis. Fetal Pediatr Pathol. 2019:1–15. https://doi.org/10.1080/15513815.2019.1672225
Landi S, Moreno V, Gioia-Patricola L, Guino E, Navarro M, de Oca J, et al. Association of common polymorphisms in inflammatory genes interleukin (IL)6, IL8, tumor necrosis factor alpha, NFKB1, and peroxisome proliferator-activated receptor gamma with colorectal cancer. Cancer Res. 2003;63:3560–6.
Theodoropoulos G, Papaconstantinou I, Felekouras E, Nikiteas N, Karakitsos P, Panoussopoulos D, et al. Relation between common polymorphisms in genes related to inflammatory response and colorectal cancer. World J Gastroenterol. 2006;12:5037–43.
Gunter MJ, Canzian F, Landi S, Chanock SJ, Sinha R, Rothman N. Inflammation-related gene polymorphisms and colorectal adenoma. Cancer Epidemiol Biomark Prev. 2006;15:1126–31.
Vogel U, Christensen J, Dybdahl M, Friis S, Hansen RD, Wallin H, et al. Prospective study of interaction between alcohol, NSAID use and polymorphisms in genes involved in the inflammatory response in relation to risk of colorectal cancer. Mutat Res. 2007;624:88–100.
Küry S, Buecher B, Robiou-du-Pont S, Scoul C, Colman H, Le Neel T, et al. Low-penetrance alleles predisposing to sporadic colorectal cancers: a French case-controlled genetic association study. BMC Cancer. 2008;8:326.
Cacev T, Radosević S, Krizanac S, Kapitanović S. Influence of interleukin-8 and interleukin-10 on sporadic colon cancer development and progression. Carcinogenesis. 2008;29:1572–80.
Wilkening S, Tavelin B, Canzian F, Enquist K, Palmqvist R, Altieri A, et al. Interleukin promoter polymorphisms and prognosis in colorectal cancer. Carcinogenesis. 2008;29:1202–6.
Tsilidis KK, Helzlsouer KJ, Smith MW, Grinberg V, Hoffman-Bolton J, Clipp SL, et al. Association of common polymorphisms in IL10, and in other genes related to inflammatory response and obesity with colorectal cancer. Cancer Causes Control. 2009;20:1739–51.
Walczak A, Przybylowska K, Dziki L, Sygut A, Chojnacki C, Chojnacki J, et al. The lL-8 and IL-13 gene polymorphisms in inflammatory bowel disease and colorectal cancer. DNA Cell Biol. 2012;31:1431–8.
Mustapha MA, Shahpudin SNM, Aziz AAA, Ankathil R. Risk modification of colorectal cancer susceptibility by interleukin-8 -251T>A polymorphism in Malaysians. World J Gastroenterol. 2012;18:2668–73.
Burada F, Dumitrescu T, Nicoli R, Ciurea ME, Rogoveanu I, Ioana M. Cytokine promoter polymorphisms and risk of colorectal cancer. Clin Lab. 2013;59:773–9.
Basavaraju U, Shebl FM, Palmer AJ, Berry S, Hold GL, El-Omar EM, et al. Cytokine gene polymorphisms, cytokine levels and the risk of colorectal neoplasia in a screened population of Northeast Scotland. Eur J Cancer Prev. 2015;24:296–304.
Dumitrescu T, Nicoli R, Serban SS, Moraru E, Cimpoeru A, Ivanov P, et al. Polymorphism is not correlated with colorectal cancer. Ann RSCB. 2012;12:197–201.
Nikiteas N, Yannopoulos A, Chatzitheofylaktou A, Tsigris C. Heterozygosity for interleukin-18 -607 A/C polymorphism is associated with risk for colorectal cancer. Anticancer Res. 2007;27:3849–53.
Guo J, Qin A, Li R, Yang C, Huang F, Huang Z, et al. [Association of the IL-18 gene polymorphism with susceptibility to colorectal cancer]. Zhonghua wei chang wai ke za zhi =. Chin J Gastrointest Surg. 2012;15:400–3.
Hu L-X, Du Y-Y, Zhang Y, Pan Y-Y. Lack of association between interleukin-8-251 T > A polymorphism and colorectal cancer risk: a meta-analysis based on 3,019 cases and 3,984 controls. Asian Pac J Cancer Prev. 2012;13:5075–9.
Wang N, Zhou R, Wang C, Guo X, Chen Z, Yang S, et al. 251 T/A polymorphism of the interleukin-8 gene and cancer risk: a HuGE review and meta-analysis based on 42 case-control studies. Mol Biol Rep. 2012;39:2831–41.
Song GG, Choi SJ, Ji JD, Lee YH. Association between interleukin-18 polymorphisms and systemic lupus erythematosus: a meta-analysis. Mol Biol Rep. 2013;40:2581–7.
Yao J, Li ZH, Li YX, Zhang R, Zhang DG, Xu ZL, et al. Association between the -607 C > A polymorphism in interleukin-18 gene promoter with gastrointestinal cancer risk: a meta-analysis. Genet Mol Res. 2015;14:16880–7.
Glasziou PP, Sanders SL. Investigating causes of heterogeneity in systematic reviews. Stat Med. 2002;21:1503–11.
Acknowledgments
We would like to express our sincere gratitude to Professor Seyed Mehdi Kalantar for his motivation, knowledge, and support during the course of this research.
Author information
Authors and Affiliations
Contributions
M.H.A and F.A are responsible as the guarantor of integrity of the entire study, study design and concepts, definition of intellectual content, and literature research. H.N, Y.G, and S.K. are responsible for the clinical studies, experimental studies, data acquisition, and manuscript preparation. S.A.D and H.N are responsible for the data analysis, statistical analysis, and manuscript review. J.S.Y is responsible for the manuscript editing. All authors have read and agreed with the final version of this manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Antikchi, M.H., Asadian, F., Dastgheib, S.A. et al. Cumulative Evidence for Association Between IL-8 -251T>A and IL-18 -607C>A Polymorphisms and Colorectal Cancer Susceptibility: a Systematic Review and Meta-analysis. J Gastrointest Canc 52, 31–40 (2021). https://doi.org/10.1007/s12029-020-00521-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12029-020-00521-w