Abstract
Background/Objective
Preclinical evidence suggests that iron homeostasis is an important biological mechanism following aneurysmal subarachnoid hemorrhage (aSAH); however, this concept is underexplored in humans. This study examined the relationship between patient outcomes following aSAH and genetic variants and DNA methylation in the hepcidin gene (HAMP), a key regulator of iron homeostasis.
Methods
In this exploratory, longitudinal observational study, participants with verified aSAH were monitored for acute outcomes including cerebral vasospasm (CV) and delayed cerebral ischemia (DCI) and evaluated post-discharge at 3 and 12 months for long-term outcomes of death and functional status using the Modified Rankin Scale (mRS; poor = 3–6) and Glasgow Outcome Scale (GOS; poor = 1–3). Participants were genotyped for two genetic variants, and DNA methylation data were collected from serial cerebrospinal fluid over 14 days post-aSAH at eight methylation sites within HAMP. Participants were grouped based on their site-specific DNA methylation trajectory, with and without correcting for cell-type heterogeneity (CTH), and the associations between genetic variants and inferred DNA methylation trajectory groups and patient outcomes were tested. To correct for multiple testing, an empirical significance threshold was computed using permutation testing.
Results
Genotype data for rs10421768 and rs7251432 were available for 241 and 371 participants, respectively, and serial DNA methylation data were available for 260 participants. Acute outcome prevalence included CV in 45% and DCI in 37.1% of the overall sample. Long-term outcome prevalence at 3 and 12 months included poor GOS in 23% and 21%, poor mRS in 31.6% and 27.3%, and mortality in 15.1% and 18.2%, respectively, in the overall sample. Being homozygous for the rs7251432 variant allele was significantly associated with death at 3 months (p = 0.003) and was the only association identified that passed adjustment for multiple testing mentioned above. Suggestive associations (defined as trending toward significance, p value < 0.05, but not meeting empirical significance thresholds) were identified between the homozygous variant allele for rs7251432 and poor GOS and mRS at 3 months (both p = 0.04) and death at 12 months (p = 0.02). For methylation trajectory groups, no associations remained significant after correction for multiple testing. However, for methylation trajectory groups not adjusted for CTH, suggestive associations were identified between cg18149657 and poor GOS and mRS at 3 months (p = 0.003 and p = 0.04, respectively) and death at 3 months (p = 0.04), and between cg26283059 and DCI (p = 0.01). For methylation trajectory groups adjusted for CTH, suggestive associations were identified between cg02131995 and good mRS at 12 months (p = 0.02), and between cg26283059 and DCI (p = 0.01).
Conclusions
This exploratory pilot study offers preliminary evidence that HAMP may play a role in patient outcomes after aSAH. Replication of this study and mechanistic investigation of the role of HAMP in patient outcomes after aSAH are needed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Aneurysmal subarachnoid hemorrhage (aSAH) is a substantial public health problem affecting approximately 30,000 people in the USA annually, who are on average 55 years of age [1], and is a leading contributor to loss of productive life-years [2]. There are functional, emotional, and psychological strains for survivors and their families, as well as a profound impact on personal and public healthcare expenditures [1,2,3]. Among survivors, recovery is variable. Unfortunately, healthcare providers are often unable to identify patients at risk of poor outcomes after aSAH [4].
Evidence suggests that iron homeostasis may be an important predictor of aSAH outcomes [5]. In aSAH, blood accumulates in the subarachnoid space and is catabolized by heme-oxygenase (inducted in response to cellular stress) into carbon monoxide, biliverdin, and labile free iron [6]. Under normal physiologic conditions, iron is tightly bound to carrier proteins as ferric iron and is recycled within the body [7]. However, because of physiologic changes associated with aSAH, including acidic brain tissue pH, hypoxia, and an influx of catecholamines in the extracellular fluid, iron is liberated to the less stable, non-protein-bound ferrous form [8]. This non-protein-bound iron has the potential to damage nearby tissues. The hepcidin antimicrobial peptide gene (HAMP) provides instructions for the production of a peptide called hepcidin [7], which is often referred to as the “master” of iron homeostasis. Although primarily synthesized and secreted by hepatocytes, hepcidin mRNA has been detected in normal brain tissue [9] and systematically regulates plasma levels of iron posttranslationally via interaction with its receptor, ferroportin [10].
Hepcidin levels have recently emerged as an important regulator in brain iron homeostasis [11], and there is some evidence that increased hepcidin expression is associated with poor health outcomes after neurologic injury [12, 13]. In preclinical models, increased hepcidin expression in the brain secondary to aSAH has been associated with lower neurological outcome scores [12]. Likewise, in humans, increased serum hepcidin levels have been associated with poor outcomes following intracranial hemorrhage [13]. We hypothesize that patients with aSAH may not uniformly respond to iron overload or potential therapeutics to mitigate the effects of iron overload potentially due to genetic variability in HAMP.
Moreover, because DNA methylation is extensively involved in oxidative stress responses [14], a common pathological complication associated with aSAH, variability of HAMP DNA methylation could impact a patient’s ability to handle a large influx of iron following aSAH. Notably, DNA methylation of HAMP within the cerebrospinal fluid (CSF) after aSAH has not been studied previously. Examining DNA methylation within the CSF, which is a more proximal tissue to aneurysm rupture (compared with traditionally studied blood), may offer a unique perspective into the pathophysiology of aSAH.
If a relationship between genetic or epigenetic variability of HAMP and outcomes in the aSAH population can be proven, it may be possible to predict who is at risk of poorer outcomes and to develop interventions that target HAMP to improve outcomes. It would also be helpful to know whether variability in HAMP impacts long-term outcomes directly or indirectly via acute aSAH outcomes such as cerebral vasospasm (CV) and delayed cerebral ischemia (DCI). To our knowledge, no studies to date have examined genetic and epigenetic variability of HAMP and their relationships to the development of acute outcomes of CV and DCI, and long-term outcomes of death and functional status following aSAH in humans. The purpose of this exploratory pilot study is to addresses this knowledge gap by investigating these relationships.
Methods
Study Design
This study was a longitudinal, observational, candidate gene polymorphism and DNA methylation association study that capitalized on existing genomic, clinical, and outcome data collected from a large cohort of aSAH participants. This study examined outcomes in the acute phase (inpatient stay from day of insult up to 14 days post-injury) and long-term outcome phase (interview at 3 and 12 months).
Setting and Sample
Participants were prospectively recruited between 2000 and 2013 from UPMC Presbyterian neurovascular intensive care unit in Pittsburgh, PA. Participants were eligible if they were diagnosed via cerebral angiogram with aSAH, between 18 and 75 years of age, and able to read and speak English (to facilitate functional status survey). Participants were excluded if the cause of their SAH was trauma, arteriovenous malformation, mycotic aneurysm, or unknown. Informed consent was obtained from all individual participants included in this study.
All participants were managed according to aSAH practice guidelines which included prophylactic nimodipine; blood pressure management to maintain adequate systolic pressure (< 140 mmHg before aneurysm securing and > 140 mmHg after aneurysm repair); invasive intracranial pressure monitoring as clinically indicated; and normovolemia guided by fluid balance. Sample sizes for each portion of the study vary as described below.
Genotype Data Collection
Single nucleotide polymorphisms (SNPs) tagging the HAMP gene were selected using the National Institute of Environmental Health Sciences Linkage Disequilibrium Tag SNP Selection database [15]. A linkage disequilibrium threshold of r2 = 0.8 was designated in addition to a 5-prime and 3-prime flanking region of 2000 base pairs. Two tagging SNPs (rs10421768 and rs7251432) were identified as representing the genetic variability in HAMP [16].
DNA was extracted from peripheral leukocytes using a simple salting out procedure [17]. Genome-wide data were collected for a subset of study participants with Affymetrix Gene Chip Assay SNP 6.0 (Affymetrix, Santa Clara, CA, USA) as previously described [18]. Genotype data for SNP rs10421768 were extracted from these data. Because data for SNP rs7251432 were not available in the genome-wide data, this SNP was genotyped using an ABI Prism® Sequence Detection System (Applied Bioscience, Carlsbad, CA, USA) to conduct allelic discrimination using a TaqMan® assay. Duplicate controls on each plate were included to assess consistency and integrity of genotyping data, and data were double-called by two individuals blinded to phenotype data. Genotyping success rates for rs10421768 and rs7251432 were 93.1% and 96.3%, respectively. Given the low amount of ancestral diversity in our sample and that allelic frequencies in HAMP differ based on ancestry, non-Caucasian participants were removed from SNP analyses in an attempt to control for population substructure [19].
DNA Methylation Data Collection
Genome-wide DNA methylation data were collected on a subset of study participants using bisulfite converted DNA extracted from biosamples of bagged CSF collected from a ventricular drain changed every 12 h on days 0 to 14 following aSAH. Unlike protein biomarkers, DNA methylation is a chemical modification that is highly stable after biospecimen collection and methods of biosample collection, time to processing, and storage of DNA confer insignificant effects on DNA methylation [20]. The CSF samples were centrifuged, and the cellular component separated from the supernatant fluid and stored in a − 80° freezer until DNA extraction from the cellular component. DNA extraction was performed with the Qiagen Midi kit (Qiagen, Valencia, CA, USA).
DNA methylation data were collected with the Infinium Human Methylation450 Beadchip and scanned using an Illumina iSCAN (Illumina, Incorporated, San Diego, CA, USA) by the Center for Inherited Disease Research. All samples from the same individual were on the same chip and technical replicates were included to assess data reliability. The raw data were analyzed using Genome Studio Software (Illumina, Incorporated, San Diego, CA, USA), and data cleaning and quality control were performed. Our data-cleaning pipeline was implemented in R using functions from several packages, including Minfi [21] and ENmix [22], and included identification and exclusion of low-quality probes and poorly performing samples and functional normalization. For each sample, cell-type make-up, as a percentage of five cell types, was estimated using Houseman’s reference-free method; [23] these estimates were later used in the analyses adjusted for cell-type heterogeneity.
DNA methylation data within the HAMP transcript region ± 2000 kb upstream and downstream (GRCh37/hg19, chr19:35771410:35778045) were extracted from the genome-wide data. A total of eight CpG sites within HAMP(cg04668516, cg02131995, cg18149657, cg23677000, cg04085447, cg17907567, cg26283059, and cg27273033) passed quality control procedures and were included in the current analyses. Participants with genome-wide methylation data were included in the current study if they had between two and four CSF DNA methylation measurements (to allow for group-based trajectory analysis) over 14 days following aSAH.
Outcome Phenotyping
Demographic data, including age, sex, and self-reported race, and clinical data, including severity of hemorrhage as measured by Fisher grade, were obtained from the medical record. Clinical data throughout the inpatient stay (through 14 days post-aSAH) were collected by a trained study nurse. Following discharge, a study staff member trained in neuropsychological testing collected long-term outcome measures during face-to-face interviews or a telephone call with the patient or proxy at 3 and 12 months following aSAH. Interviewers were initially trained and routinely observed for ongoing quality assurance.
A total of eight (two acute and six long-term) outcomes were examined in the current study. The primary acute outcome measures in this study were CV and DCI. CV was defined as the presence of cerebral vessel narrowing of ≥ 25% on cerebral angiogram as evaluated by a neurosurgeon. DCI was defined as the presence of neurological deterioration, after exclusion of non-ischemic causes, which was accompanied by evidence of abnormal cerebral blood flow. In evaluating DCI, neurological deterioration measures were abstracted from the medical record and included any of the following: a decline in level of consciousness as measured by an increase ≥ 2 points on the National Institutes of Health Stroke Scale, a drop in the Glasgow Coma Scale, and/or new and persistent (> 1 h) focal neurological deficit. Cerebral blood flow measures used to diagnose DCI included either cerebral angiography performed by the treating physician, or transcranial Doppler performed daily for 14 days by a trained study staff member.
Long-term outcome measures of death and functional status were measured at 3 and 12 months after aSAH. Mortality data were obtained from medical records, caregiver report, or the Social Security Death Index. Global functional outcomes were measured using the Glasgow Outcome Scale (GOS) [24] and the Modified Rankin Scale (mRS) [25]. The GOS rates level of functioning on a scale of 1 (death) to 5 (good recovery) and has a well-established validity in patients with neurological insult [24]. The mRS measures mental and functional deficits using a scale from 0 (no symptoms) to 6 (death) and has a established validity in stroke patients [26, 27]. All study staff involved in data collection were blinded to participant genotype and DNA methylation status.
Statistical Analysis
Demographics and Clinical Characteristics
Statistical analysis was conducted using R statistical software (version 3.4.1, Vienna, Austria) and SAS (version 9.4, SAS Institute Incorporated, Cary, NC, USA). CV, DCI, and death were treated as binary outcomes (occurrence vs. no occurrence), and the mRS and GOS scores were dichotomized as good (mRS 0 to 2; GOS 4 to 5) or poor (mRS 3 to 6; GOS 1 to 3). Standard descriptive statistics were computed. A more detailed statistical methodological narrative is available in the Supplemental Material.
HAMP Single Nucleotide Polymorphisms
All analyses exploring the associations between SNPs and outcomes were conducted using R. Allele frequencies and Hardy–Weinberg Equilibrium (HWE) were evaluated for each SNP. All statistical models used binary logistic regression while adjusting for age, sex, and Fisher grade. The relationship between genetic variability in HAMP and patient outcomes was evaluated based on presence (vs. absence) of the minor allele for both SNPs. Given the heavily skewed distribution of genotypes for rs10421768, no further analyses were conducted for this SNP. For rs7251432, genotypic analysis (two-degree-of-freedom test of genetic association) was conducted. Finally, based on results from the genotypic analysis, a model testing a recessive association for rs7251432 was computed.
Given the exploratory nature of this pilot study and correlation between outcomes of interest, correction for multiple testing was made by computing empirical significance thresholds using permutation analysis (10,000 permutations). Associations with p values < 0.05 that did not meet the empirical significance thresholds were considered to be “suggestive” (i.e., trending) associations.
For the genotypic model, a likelihood ratio test (LRT) was used to produce a global p value of the overall model fit by comparing the full model (including the SNP) with a restricted model (omitting the SNP). As described above, a global empirical significance threshold of 0.008 was computed for this global p value to correct for testing eight correlated outcomes.
HAMP DNA Methylation
Group-based Trajectory Analysis
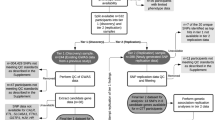
Using group-based trajectory analysis (GBTA) as implemented in the PROC TRAJ macro in SAS, the change pattern over time was used to infer distinct trajectory groups for DNA methylation at each CpG site using a censored normal model [28, 29]. DNA methylation data were available on days 0 to 14 following aSAH. Due to limited available information on days 0 and 14 (only 2 and 17 observations were available, respectively), we removed these days from our trajectory analysis. M-values were used for all methylation analyses. GBTA was largely automated for 39 possible models (Supplemental Table 1) with a maximum of three groups and comprehensive combinations of polynomial orders of 0 (intercept-only), 1 (linear), and 2 (quadratic). A secondary evaluation of model adequacy was performed to ensure models moving forward for binary logistic regression analysis met quality control standards. An expanded methods section includes a more detailed statistical methodological narrative (Supplemental Material) in addition to explanatory flowcharts of DNA methylation analysis workflow (Supplemental Figs. 1a, 1b, and 1c).
Cell-Type Heterogeneity
Cell-type heterogeneity (CTH) is an important consideration in methylation studies as differential methylation between cell types may impact overall methylation levels and confound results. In cases where genome-wide data are collected, there are methods to control for CTH [30]. However, if only CpG site-specific DNA methylation data are collected, as would be the case with a biomarker used clinically, controlling for CTH is not possible. In an effort to explore the potential clinical utility of methylation values unadjusted for CTH (as they would likely be in the hospital setting) as well as adjusted for CTH, the aforementioned procedures were conducted without adjustment for CTH (“unadjusted models”) and with adjustment for CTH (“CTH-adjusted models”) for all eight CpG sites. As described above, using previously collected genome-wide data, CTH data were computed using the R statistical software and Houseman’s reference-free method [23].
Trajectory Group—Patient Outcome Associations
The relationship between site-specific DNA methylation trajectory groups and patient outcomes were examined in R using binary logistic regression while controlling for age, sex, race, and Fisher grade. As stated above, p values < 0.05 were considered suggestive; however, permutation analysis was used to compute an empirical significance threshold of 0.002, which corrects for testing eight correlated outcomes for trajectory group–patient outcome associations. Again, models were evaluated using the LRT to produce a global p value to determine the overall effect of the predicted trajectory group membership within the model. A global empirical significance threshold of 0.001 was computed for this global p value to correct for testing eight correlated outcomes.
Results
Demographics and Clinical Characteristics
Because this study capitalized on existing data as previously described, sample sizes for each portion of the study varied. In our sample of 591 participants, genotype data for rs10421768 and rs7251432 were available for 241 and 371 Caucasian participants, respectively, and DNA methylation data were available for 260 participants (Table 1). The final sample sizes differed between analyses based on outcome data availability and analysis-specific sample sizes are presented (Tables 2, 3, 4 and 5). The overall sample was 93.4% Caucasian and 71.6% female with a mean(± SD) age of 53.4 ± 11.4 years. Fisher grades of 2, 3, and 4 comprised 40.8%, 43.0% and 16.2% of the overall sample, respectively. World Federation Neurosurgical Societies (WFNS) grading system scores of 1, 2, 3, 4, and 5 comprised 51.6%, 17.3%, 7.1%, 13.5%, and 10.5% of the overall sample, respectively. Of the overall sample, 37.7% had surgical intervention (vs. coil embolization). Older age was associated with poor GOS at 3 and 12 months (p = 0.04 and p = 0.03, respectively) and death at 3 months (p = 0.02) in the overall sample. No significant associations between sex or intervention (surgical vs. coil embolization) and outcome measures were found; however, sex was included in all logistic regression models as a covariate given the presence of an estrogen response element governing hepcidin expression [31]. Non-Caucasian race was associated with poor mRS at 3 months (p = 0.04) and poor GOS at 12 months (p = 0.02). Higher Fisher grade was associated with poor acute outcomes of CV (p = 0.006) and DCI (p = 0.04) as well as with poor long-term outcomes of GOS, mRS, and death measured at 3 and 12 months (p < 0.001 for all outcome measures and time points).
HAMP Single Nucleotide Polymorphisms
Allelic Associations
The association between genetic variability in HAMP and patient outcomes was first evaluated based on presence (vs. absence) of the minor allele (Supplemental Table 3). For tagging SNPs rs10421768 (minor allele, G) and rs7251432 (minor allele, A), the minor allele frequencies were 23.0% and 43.1%, respectively. No significant associations were found between the presence of the minor allele and patient outcomes for either SNP after controlling for age, sex, and Fisher grade. A figure depicting the locations of these SNPs is presented (Supplemental Fig. 2).
Genotypic Associations
rs10421768
The distribution of rs10421768 genotypes showed AA as the most common (56.8%), followed by AG (40.2%), and GG (2.9%). In our sample, HWE was violated for rs10421768 (p = 0.036). Given the sparse sample size for the GG genotype, we were not able to perform statistically meaningful subgroup analyses based on genotype for rs10421768.
rs7251432
The distribution of rs7251432 genotypes showed AG as the most common (41.5%), followed by GG (36.1%), and AA (22.4%). In our sample, HWE was violated (p = 0.003). Next, we tested associations between rs7251432 genotype and patient outcomes in a genotypic model (Table 2). Compared with the GG genotype, suggestive associations (defined as trending toward significance with a p value < 0.05 but not meeting empirical significance thresholds) were identified between the AA genotype and mRS at 3 months (p = 0.05) and death at 3 months (p = 0.02) after adjusting for age, sex, and Fisher grade (with odds ratios consistent with a recessive model). However, these p values did not meet the empirical significance threshold of 0.005. No differences in patient outcomes were identified between the AG genotype and the GG genotype.
The global test of SNP association within the models based on the LRT identified a suggestive overall effect for only mortality at 3 months (global p = 0.01). However, this global p value did not meet the global empirical significance threshold of p = 0.008 calculated in permutation testing.
Recessive Associations
rs7251432
Next, we tested for association between rs7251432 and patient outcomes under a recessive model (Table 3). Compared with combined GG and AG genotype groups, the AA genotype group was significantly associated with death at 3 months (p = 0.003) and suggestively associated with poor GOS at 3 months (p = 0.04), poor mRS at 3 months (p = 0.04), and death at 12 months (p = 0.02) after adjusting for age, sex, and Fisher grade. Only the p value for death at 3 months met the empirical significance threshold of 0.01.
HAMP DNA Methylation
Group-based Trajectory Analysis
Based on the GBTA procedures outlined (Supplemental Fig. 1), distinct methylation trajectory groups passing model adequacy assessment were inferred at six CpG sites for unadjusted models (cg02131995, cg17907567, cg18149657, cg27273033, cg04668516, and cg26283059) and at all eight CpG sites for CTH-adjusted models (Supplemental Table 2). Of the 14 candidate models presented, three could not be tested for association with outcomes because only one trajectory group was inferred at those CpG sites.
Trajectory Group—Patient Outcome Associations
Out of the 11 candidate models eligible for binary logistic regression (five unadjusted models and six CTH-adjusted models), we found suggestive associations between trajectory groups and patient outcomes in four models (two unadjusted models and two CTH-adjusted models). Given the breadth of this study, we have only discussed the CpG sites with suggestive patient outcome associations below. A figure depicting the locations of these CpG sites is presented (Supplemental Fig. 2).
Unadjusted models
Binary logistic regression analysis exploring associations of unadjusted methylation trajectory groups with patient outcomes was performed (Table 4). Trajectory groups at two CpG sites (cg18149657 and cg26283059) had suggestive associations (unadjusted p < 0.05) with patient outcomes after aSAH. For both cg18149657 and cg26283059, three distinct trajectory groups were identified, and trajectory plots are presented (Fig. 1). At cg18149657, predicted membership assignment to Group 3 (vs. reference Group 1) was suggestively associated with poor GOS and mRS at 3 months (p = 0.003 and p = 0.04 respectively) and death at 3 months (p = 0.04) after controlling for age, sex, race, and Fisher grade, while at cg26283059, predicted membership assignment to Group 3 (vs. reference Group 1) was suggestively associated with DCI (p = 0.01). However, these results did not meet the empirical significance threshold for unadjusted models of 0.002 calculated in permutation testing. No other associations were identified between unadjusted methylation trajectory groups and patient outcomes after aSAH (data not shown).
Methylation Trajectory Group Plots for Unadjusted Models. a Methylation Trajectory Group Plot at cg18149657 (Unadjusted Model with Polynomial Order 222a), b Methylation Trajectory Group Plot at cg26283059 (Unadjusted Model with Polynomial Order 000a), a Postulated model indicates the number of groups and polynomial orders (trajectory group shapes) of 0 (intercept-only), 1 (linear), or 2 (quadratic). Note: Solid lines represent group-specific average trajectories; dashed lines represent 95% Confidence Interval for group-specific trajectories; group number displayed on group-specific solid fitted lines
Based on the global test of trajectory group significance within the models using the LRT, a suggestive overall effect was identified for only GOS at 3 months at cg18149657 (global p = 0.004). At cg26283059, a suggestive overall effect was identified for not only DCI (global p = 0.003), but also CV (global p = 0.02). However, these global p values did not meet the global empirical significance threshold for unadjusted models of p = 0.001 calculated in permutation testing.
CTH-adjusted models
Binary logistic regression analysis exploring associations of CTH-adjusted methylation trajectory groups with patient outcomes was performed (Table 5). Trajectory groups at two CpG sites (cg02131995 and cg26283059) had suggestive associations with patient outcomes after aSAH. For both cg02131995 and cg26283059, three distinct trajectory groups were identified, and trajectory plots are presented (Fig. 2). At cg02131995, predicted membership assignment to Group 2 (vs. reference Group 1) was suggestively associated with good mRS at 12 months (p = 0.02), while at cg26283059, predicted membership assignment to Group 3 (vs. reference Group 1) was suggestively associated with DCI (p = 0.01). However, these results did not meet the empirical significance threshold for CTH-adjusted models of 0.002 calculated in permutation testing. No other associations were identified between CTH-adjusted methylation trajectory groups and patient outcomes after aSAH (data not shown).
Methylation Trajectory Group Plots for CTH-Adjusted Models. a Methylation Trajectory Group Plot at cg02131995 (CTH-adjusted Model with Polynomial Order 000a), b Methylation Trajectory Group Plot at cg26283059 (CTH-adjusted Model with Polynomial Order 000a). aPostulated model indicates the number of groups and polynomial orders (trajectory group shapes) of 0 (intercept-only), 1 (linear), or 2 (quadratic). CTH cell-type heterogeneity; solid lines represent group-specific average trajectories; dashed lines represent 95% Confidence Interval for group-specific trajectories; group number displayed on group-specific solid fitted lines; intercept-only polynomials appear jagged due to the inclusion of CTH data as time-varying covariates
Based on the global test of trajectory group significance within the models using the LRT, a suggestive overall effect was identified for mRS at 12 months at cg02131995 (global p = 0.05), whereas a suggestive overall effect was identified for DCI at cg26283059 (global p = 0.01). However, these global p values did not meet the global empirical significance threshold for unadjusted models of p = 0.001 calculated in permutation testing.
Discussion
HAMP Single Nucleotide Polymorphisms
To our knowledge, this is the first demonstration that variability in HAMP genotype may be associated with patient outcomes following aSAH in humans. In this exploratory pilot study, individuals with the AA genotype had between two and three times higher odds of death at 3 and 12 months following aSAH, even after accounting for age and injury severity, as well as two times higher odds of poor GOS and mRS at 3 months. Although the associations observed for this SNP were relatively consistent across time points and outcomes, it is important to note that only the association between the AA genotype and death at 3 months remained significant after correction for multiple testing described above. While little is known about rs7251432, this SNP (located in an intronic region of HAMP) has been associated with Kawasaki disease and nominal increased risk of abdominal aortic aneurysm (AAA), which are characterized by inflammation of blood vessels [32, 33]. Interestingly, Kawasaki and aSAH have been linked in three case reports of young children [34,35,36]. Similarly, cerebral aneurysm and AAA have been shown to share biological pathways in their hemodynamic pathogenesis [37].
Additionally, one study showed that the AA genotype of rs7251432 was associated with higher hemoglobin levels in a simulated hypoxic environment compared with GG or AG genotypes [38,39,40]. While the literature surrounding the effects of hemoglobin concentrations after aSAH is mixed, one theory suggests that lower hemoglobin levels may offer protective effects, including up-regulation of nitric oxide and decreased blood viscosity which improves brain perfusion [41]. In aSAH, brain tissue oxygen pressure and pH can be altered leading to ischemic changes and secondary injury [42]. This is important because if patients with the rs7251432 AA genotype have higher hemoglobin levels in the hypoxic environment created by aSAH, this could be a potential explanation for the increased odds of death in this subset because they do not have the potential protective response described above. Although this study did not evaluate hemoglobin levels, this is an important future direction of this work.
For rs10421768, we identified modest positive odds ratios of around 1.2 or higher (Supplemental Table 3). Although these modest associations were not significant in our small sample size, there could be a real effect that could be detected in larger sample sizes. Notably, this SNP has been previously identified to play a role in iron homeostasis in cases of iron overload [43] warranting future investigation in a larger sample.
In this sample, both rs10421768 and rs7251432 violated HWE. After a third party reviewed the double-called raw genotype data, we determined that genotyping error was unlikely. A potential explanation for this violation, however, could be that our sample of aSAH cases is enriched for associated variants and may not be representative of the general population, further supporting the potential role of genetic variability of HAMP with aSAH.
HAMP DNA Methylation
This study suggests variability in HAMP DNA methylation trajectories may be associated with patient outcomes following aSAH, although none of the associations identified survived correction for multiple testing. For the unadjusted model at cg18149657, Group 3 had between two and eight times increased odds of unfavorable 3-month outcomes. This group had the highest methylation levels across groups peaking between days 4 and 10 (Fig. 1a), a key window in recovery after aSAH in which DCI often occurs. However, the global test of trajectory group significance identified only a suggestive association for GOS at 3 months. An interesting finding in this study is that at cg26283059, suggestive associations were found in both the unadjusted model and CTH-adjusted model between Group 3 and DCI. At this site and similar to above, participants in Group 3 had the highest overall methylation levels and had increased odds of DCI for both the unadjusted model and CTH-adjusted model (Figs. 1b and 2b). Based on the global p values for unadjusted methylation trajectory groups at cg26283059, post hoc analyses revealed that by changing the reference group from Group 1 to Group 3, we achieved suggestively significant results for not only DCI, but also CV (data not shown). Again, we observe consistent results that hypermethylation is associated with both acute outcomes supporting our findings further. These results are inconsistent with our expectations that hypomethylation (leading to increased gene expression) versus the observed hypermethylation would be associated with unfavorable outcomes, suggesting other regulatory mechanisms might be at play.
Beyond this study, few studies to date have specifically examined DNA methylation of HAMP and of those that have, none have been in aSAH. However, similar to rs7251432 discussed above, hypomethylation of CpG sites within the HAMP promoter region (cg23677000 and cg04085447) has been associated with cases of Kawasaki disease [44]. Recently, it has become more clear that complex signaling pathways, plasma and tissue iron levels, inflammatory cytokines, erythropoiesis, and estrogen all play an important role in hepcidin expression by inducing or inhibiting hepcidin synthesis [31, 45, 46]. For example, endothelins are potent vasoconstrictors released during inflammatory responses that have been shown to be associated with poor outcomes following aSAH [45, 47]. Specific to this study, endothelin-1 has been shown to lead to increases in mRNA levels of iron homeostasis genes (including HAMP) in the brain [45]. Also, estrogen has been implicated as a regulator of hepcidin expression via transcriptional inhibition through a functional estrogen response element in the promoter region of the HAMP gene [31]. These potential confounders will be important to consider in future work of a larger sample size.
It is important to interpret these results with caution as none of our methylation findings remained significant after correction for multiple testing described above. It is also important to note that methylation was measured in the CSF as opposed to the major site of hepcidin production (the liver) or more commonly examined tissue of blood which makes it difficult to compare the results of this study with existing studies of HAMP. Finally, it should be acknowledged that Housman’s reference-free method for correcting for CTH [23] has not been validated in CSF and cell count data for associated DNA methylation data were unavailable. Nevertheless, given the proximal location of CSF within the central nervous system, CSF DNA methylation of HAMP could have important clinical implications and warrants more robust validation of the potential prognostic performance of these loci.
Limitations
This exploratory pilot study is significant because it suggests that HAMP may influence patient outcomes after aSAH; however, several important limitations should be acknowledged. First, by selecting specific DNA methylation targets located within the relatively narrow window of 2000 base pairs upstream and downstream of the HAMP gene region, we did not exhaust all potential regulatory regions of HAMP. Expanding this region to capture more CpG sites should be an area of future research in a larger study. Similarly, this study focused on genetic and epigenetic variability of HAMP and did not examine other loci relevant to the iron homeostasis pathway. While HAMP was carefully chosen based on the existing literature and its importance in iron homeostasis, interpretability of the results is limited without considering the intricate genetic network potentially moderating patient outcomes. Similarly, this study did not measure hepcidin or iron levels. Future investigation of hepcidin and iron levels, as well as additional genetic loci and their interactions within the iron homeostasis pathway, is warranted.
Furthermore, by capitalizing on data and samples from an existing cohort, we were limited to existing outcome and covariate data previously collected. Specifically, there is a large amount of missing data due to loss to follow-up. Because aSAH outcomes vary between and within patients over time, we did not fill in missing data with imputation or by carrying the last observation forward to avoid biasing the results. Similarly, the available CV data used in this study captured only the cases verified with angiogram (vs. ultrasound). Therefore, it is likely that we did not capture a portion of the overall sample who developed CV identified with transcranial Doppler. Similarly, severity of injury is an important predictor of outcomes after aSAH. In the available data, in addition to the Fisher grade, we had access to the WFNS grading system which is a score based on the patient’s presenting clinical condition and combines measures from the Glasgow Coma Scale and focal neurological deficits. Both WFNS and Fisher grade have been shown to be important predictors of outcomes after aSAH and are highly correlated in our data. In post hoc analyses, we repeated the analyses while controlling for WFNS (instead Fisher grade) and found that the results were consistent across outcomes and timepoints. In an effort to avoid multicollinearity with our models, we ultimately chose to control for Fisher grade (as opposed to WFNS) because it offers greater insight into the amount of blood within the subarachnoid space and is more scientifically meaningful to the conceptual foundation of this study.
An additional limitation is the relatively small sample size of this study. Specifically, there was an insufficient number of participants with the rs10421768 GG genotype for reliable analysis. Given this underrepresentation, we were unable to perform genotypic association analysis in this sample size. Next, race was self-reported by study participants and use of this variable limits the ability to stringently control for population substructure. Similarly, given the underrepresentation of non-Caucasian populations, we were unable to perform subgroup analyses for the SNP portion of this study in more diverse ancestries, which limits the generalizability of the findings. Future efforts are needed to replicate these findings in larger, more diverse samples. Finally, studies have demonstrated that smoking and body mass index (BMI) impact DNA methylation [48, 49] and that treatment strategies (e.g., induced hypertension, administration of a calcium channel blocker) impact the incidence of acute complications after aSAH. Unfortunately, our small sample size and amount of missing data prevented us from controlling for these factors.
Lastly and of particular notability, the interpretability of the results of this study are limited because of the large number of tests conducted. A simple Bonferroni correction for multiple testing would be too conservative due to the high correlation between outcomes in this cohort. Specifically, CV and DCI have a correlation of > 0.75. Likewise, mRS and GOS have a correlation of > 0.70 and were assessed at both 3 months and 12 months. Moreover, death is imbedded in both mRS and GOS scores. Despite these correlations, it was ultimately decided to retain both mRS and GOS in this study to allow for between study comparisons in the future. In an effort to carefully adjust for the multiple testing while properly taking the correlation between tests into account, we performed permutation testing to compute an empirical significance threshold within each model examined. While many of our results are consistent across correlated outcomes and time points suggesting possible associations of genetic and epigenetic variability of HAMP with patient outcomes after aSAH, it is imperative that this study be replicated in a larger sample to confirm these findings.
Conclusion
Outcomes following aSAH are variable creating an unyielding need for reliable biomarkers of poor outcomes. Hepcidin has surfaced as an important preclinical predictor of outcomes following experimental brain injury [12]. The results of this pilot study offer support to this theory in humans; however, due to the exploratory nature of this study, more research is needed before definitive conclusions can be made about the role of HAMP in outcomes after aSAH. In our sample, we identified an association between the AA genotype of rs7251432 and death at 3 months that remained significant after correction for multiple testing. If these findings can be replicated in an independent sample, this approach has the potential to provide a clinical tool for prediction of those at high-risk of poor outcomes following aSAH. We also believe that our exploratory study found interesting trends that may warrant further investigation into the role of DNA methylation of HAMP in patient outcomes after aSAH.
References
Zacharia BE, Hickman ZL, Grobelny BT, et al. Epidemiology of aneurysmal subarachnoid hemorrhage. Neurosurg Clin N Am. 2010;21(2):221–33. https://doi.org/10.1016/j.nec.2009.10.002.
Rosenbaum BP, Kelly ML, Kshettry VR, Weil RJ. Neurologic disorders, in-hospital deaths, and years of potential life lost in the USA, 1988–2011. J Clin Neurosci. 2014;21(11):1874–80. https://doi.org/10.1016/j.jocn.2014.05.006.
le Roux AA, Wallace MC. Outcome and cost of aneurysmal subarachnoid hemorrhage. Neurosurg Clin N Am. 2010;21(2):235–46. https://doi.org/10.1016/j.nec.2009.10.014.
Lantigua H, Ortega-Gutierrez S, Schmidt JM, et al. Subarachnoid hemorrhage: who dies, and why? Crit Care. 2015;19(1):309. https://doi.org/10.1186/s13054-015-1036-0.
Gomes JA, Selim M, Cotleur A, et al. Brain iron metabolism and brain injury following subarachnoid hemorrhage: iCeFISH-pilot (CSF iron in SAH). Neurocrit Care. 2014;21(2):285–93. https://doi.org/10.1007/s12028-014-9977-8.
Wagner KR, Sharp FR, Ardizzone TD, Lu A, Clark JF. Heme and iron metabolism: role in cerebral hemorrhage. J Cereb Blood Flow Metab. 2003;23(6):629–52. https://doi.org/10.1097/01.WCB.0000073905.87928.6D.
Ganz T. Hepcidin and iron regulation, 10 years later. Blood. 2011;117(17):4425–33. https://doi.org/10.1182/blood-2011-01-258467.
Bishop GM, Robinson SR. Quantitative analysis of cell death and ferritin expression in response to cortical iron: implications for hypoxia-ischemia and stroke. Brain Res. 2001;907(1–2):175–87. https://doi.org/10.1016/S0006-8993(01)02303-4.
Hänninen MM, Haapasalo J, Haapasalo H, et al. Expression of iron-related genes in human brain and brain tumors. BMC Neurosci. 2009;10(1):36. https://doi.org/10.1186/1471-2202-10-36.
Nemeth E, Tuttle MS, Powelson J, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306(5704):2090–3. https://doi.org/10.1126/science.1104742.
Vela D. Hepcidin, an emerging and important player in brain iron homeostasis. J Transl Med. 2018;16(1):25. https://doi.org/10.1186/s12967-018-1399-5.
Tan G, Liu L, He Z, Sun J, Xing W, Sun X. Role of hepcidin and its downstream proteins in early brain injury after experimental subarachnoid hemorrhage in rats. Mol Cell Biochem. 2016;418(1–2):31–8. https://doi.org/10.1007/s11010-016-2730-1.
Xiong X-YY, Chen J, Zhu W-YY, et al. Serum hepcidin concentrations correlate with serum iron level and outcome in patients with intracerebral hemorrhage. Neurol Sci. 2015;36(10):1843–9. https://doi.org/10.1007/s10072-015-2266-2.
Zhao H, Han Z, Ji X, Luo Y. Epigenetic regulation of oxidative stress in ischemic stroke. Aging Dis. 2016;7(3):295–306. https://doi.org/10.14336/AD.2015.1009.
Sciences NI of EH. LD TAG SNP Selection (TagSNP). https://snpinfo.niehs.nih.gov/snpinfo/snptag.html.
LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. https://analysistools.nci.nih.gov/LDlink/?var1=rs7251432&var2=rs10421768&pop=CEU&tab=ldpair. Accessed 2 Apr 2018.
Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. https://doi.org/10.1093/nar/16.3.1215.
Kim H, Crago E, Kim M, et al. Cerebral vasospasm after sub-arachnoid hemorrhage as a clinical predictor and phenotype for genetic association study. Int J Stroke. 2013;8(8):620–5. https://doi.org/10.1111/j.1747-4949.2012.00823.x.
Little J, Higgins JPT, Ioannidis JPA, et al. STrengthening the REporting of Genetic Association studies (STREGA)–an extension of the STROBE statement. Eur J Clin Invest. 2009;39(4):247-266. http://www.ncbi.nlm.nih.gov/pubmed/19297801. Accessed 12 Sep 2017.
Groen K, Lea RA, Maltby VE, Scott RJ, Lechner-Scott J. Letter to the editor: blood processing and sample storage have negligible effects on methylation. Clin Epigenet. 2018;10(1):22. https://doi.org/10.1186/s13148-018-0455-6.
Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–9. https://doi.org/10.1093/bioinformatics/btu049.
Xu Z, Niu L, Li L, Taylor JA. ENmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. Nucleic Acids Res. 2016;44(3):e20. https://doi.org/10.1093/nar/gkv907.
Houseman EA, Molitor J, Marsit CJ. Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics. 2014;30(10):1431–9. https://doi.org/10.1093/bioinformatics/btu029.
Jennett B, Bond M. Assessment of outcome after severe brain damage. Lancet (London, England). 1975;1(7905):480-484. http://www.ncbi.nlm.nih.gov/pubmed/46957. Accessed 3 Jan 2017.
van Swieten JC, Koudstaal PJ, Visser MC, Schouten HJ, van Gijn J. Interobserver agreement for the assessment of handicap in stroke patients. Stroke. 1988;19(5):604-607. http://www.ncbi.nlm.nih.gov/pubmed/3363593. Accessed 15 May 2017.
Banks JL, Marotta CA. Outcomes validity and reliability of the modified rankin scale: implications for stroke clinical trials - A literature review and synthesis. Stroke. 2007;38(3):1091–6. https://doi.org/10.1161/01.STR.0000258355.23810.c6.
Van Swieten JC, Koudstaal PJ, Visser MC, Schouten H, Van Gijn J. Interobserver agreement for the assessment of handicap in stroke patients. Stroke. 1988;19(5):604–7. https://doi.org/10.1161/01.STR.19.5.604.
Jones BL, Nagin DS, Roeder K. A SAS procedure based on mixture models for estimating developmental trajectories. Sociol Methods Res. 2001;29(3):374–93. https://doi.org/10.1177/0049124101029003005.
Jones BL, Nagin DS. Advances in group-based trajectory modeling and an SAS procedure for estimating them. Sociol Methods Res. 2007;35(4):542–71. https://doi.org/10.1177/0049124106292364.
Titus AJ, Gallimore RM, Salas LA, Christensen BC. Cell-type deconvolution from DNA methylation: a review of recent applications. Hum Mol Genet. 2017;26(R2):R216–24. https://doi.org/10.1093/hmg/ddx275.
Hou Y, Zhang S, Wang L, et al. Estrogen regulates iron homeostasis through governing hepatic hepcidin expression via an estrogen response element. Gene. 2012;511(2):398–403. https://doi.org/10.1016/j.gene.2012.09.060.
Lillvis JH, Kyo Y, Tromp G, et al. Analysis of positional candidate genes in the AAA1 susceptibility locus for abdominal aortic aneurysms on chromosome 19. BMC Med Genet. 2011;12(1):14. https://doi.org/10.1186/1471-2350-12-14.
Huang Y-H, Yang KD, Hsu Y-W, et al. Correlation of HAMP gene polymorphisms and expression with the susceptibility and length of hospital stays in Taiwanese children with Kawasaki disease. Oncotarget. July 2017. https://doi.org/10.18632/oncotarget.17700.
Ishida A, Matsuo S, Kawamura S, Nishikawa T. Subarachnoid hemorrhage due to nonbranching aneurysm of the middle cerebral artery in a young adult with a history of Kawasaki disease. Surg Neurol Int. 2014;5:5. https://doi.org/10.4103/2152-7806.125285.
Ahn JH, Phi JH, Kang H-S, et al. A ruptured middle cerebral artery aneurysm in a 13-month-old boy with Kawasaki disease. J Neurosurg Pediatr. 2010;6(2):150–3. https://doi.org/10.3171/2010.5.PEDS1012.
Tanaka S, Sagiuchi T, Kobayashi I. Ruptured pediatric posterior cerebral artery aneurysm 9 years after the onset of Kawasaki disease: a case report. Child’s Nerv Syst. 2007;23(6):701–6. https://doi.org/10.1007/s00381-006-0263-8.
Tanweer O, Wilson TA, Metaxa E, Riina HA, Meng H. A comparative review of the hemodynamics and pathogenesis of cerebral and abdominal aortic aneurysms: lessons to learn from each other. J Cerebrovasc Endovasc Neurosurg. 2014;16(4):335–49. https://doi.org/10.7461/jcen.2014.16.4.335.
Gerile W, Yang H, Longyan Y, Jing N, Jingling W, Hongshu J. Association between rs7251432 Polymorphism in hepcidin gene and change in hemogram after the HiHiLo training in men of Han Nationality. Chin J Sport Med. 2010;29(4):386-390. http://caod.oriprobe.com/articles/25996307/Association_between_rs7251432_Polymorphism_in_Hepcidin_Gene_and_Change.htm. Accessed June 6, 2017.
Chen Y, Jiang C, Luo Y, Liu F, Gao Y. Interaction of CARD14, SENP1 and VEGFA polymorphisms on susceptibility to high altitude polycythemia in the Han Chinese population at the Qinghai-Tibetan Plateau. Blood Cells Mol Dis. 2016;57:13–22. https://doi.org/10.1016/j.bcmd.2015.11.005.
Festic E, Rabinstein AA, Freeman WD, et al. Blood transfusion is an important predictor of hospital mortality among patients with aneurysmal subarachnoid hemorrhage. Neurocrit Care. 2013;18(2):209–15. https://doi.org/10.1007/s12028-012-9777-y.
Rebel A, Ulatowski JA, Kwansa H, Bucci E, Koehler RC. Cerebrovascular response to decreased hematocrit: effect of cell-free hemoglobin, plasma viscosity, and CO2. Am J Physiol Heart Circ Physiol. 2003;285(4):H1600–8. https://doi.org/10.1152/ajpheart.00077.2003.
Hoffman WE, Wheeler P, Edelman G, Charbel FT, Torres NJ, Ausman JI. Hypoxic brain tissue following subarachnoid hemorrhage. Anesthesiology. 2000;92(2):442–446. http://www.ncbi.nlm.nih.gov/pubmed/10691231. Accessed 3 April 2018.
Andreani M, Radio FC, Testi M, et al. Association of hepcidin promoter c.-582 A > G variant and iron overload in thalassemia major. Haematologica. 2009;94(9):1293–1296. https://doi.org/10.3324/haematol.2009.006270.
Huang YH, Kuo HC, Li SC, Cai XY, Liu SF, Kuo HC. HAMP promoter hypomethylation and increased hepcidin levels as biomarkers for Kawasaki disease. J Mol Cell Cardiol. 2018;117:82–7. https://doi.org/10.1016/j.yjmcc.2018.02.017.
Bickford JS, Ali NF, Nick JA, et al. Endothelin-1-mediated vasoconstriction alters cerebral gene expression in iron homeostasis and eicosanoid metabolism. Brain Res. 2014;1588:25–36. https://doi.org/10.1016/j.brainres.2014.09.022.
Liu J, Sun B, Yin H, Liu S. Hepcidin: a promising therapeutic target for iron disorders: a systematic review. Medicine (Baltimore). 2016;95(14):e3150. https://doi.org/10.1097/MD.0000000000003150.
Thampatty BP, Sherwood PR, Gallek MJ, et al. Role of endothelin-1 in human aneurysmal subarachnoid hemorrhage: associations with vasospasm and delayed cerebral ischemia. Neurocrit Care. 2011;15(1):19–27. https://doi.org/10.1007/s12028-011-9508-9.
Li S, Wong EM, Bui M, et al. Inference about causation between body mass index and DNA methylation in blood from a twin family study. International Journal of Obesity. https://www.biorxiv.org/content/early/2017/11/21/223040. Published November 21, 2018. Accessed 26 Aug 2018.
Li S, Wong EM, Bui M, et al. Causal effect of smoking on DNA methylation in peripheral blood: a twin and family study. Clin Epigenetics. 2018;10(1):18. https://doi.org/10.1186/s13148-018-0452-9.
Acknowledgements
We would also like to acknowledge the anonymous reviewers who took the time to critically evaluate this paper. Their detailed feedback greatly improved the clarity and quality of this work for the scientific community.
Funding
Research reported in this publication was supported by the National Instiute of Nursing Research of the National Institutes of Health under Award Numbers F31NR017311, R01NR004339, R01NR013610, and T32NR009759 and with additional support from the International Society of Nurses in Genetics, University of Pittsburgh Leslie A. Hoffman Endowed Research Award, Nightingale Awards of Pennsylvania, and Eta Chapter, Sigma Theta Tau, Inc. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or supporting foundations.
Author information
Authors and Affiliations
Contributions
LWH contributed to the study conception and design, analysis and interpretation of data, and drafted, critically revised, and gave final approval for the manuscript. AIA contributed to the analysis of data and critically revised and gave final approval for the manuscript. EAC contributed to the acquisition and interpretation of data and critically revised and gave final approval for the manuscript. DR contributed to the analysis and interpretation of data and critically revised and gave final approval for the manuscript. JRS contributed to analysis and interpretation of the data and critically revised and gave final approval for the manuscript. PRS contributed to the acquisition of data and critically revised and gave final approval for the manuscript. SMS contributed to the analysis and interpretation of data and critically revised and gave final approval for the manuscript. DEW contributed to the study design, analysis and interpretation of data, and critically revised and gave final approval for the manuscript. YPC contributed to the study conception and design, interpretation of data, and critically revised and gave final approval for the manuscript. All authors agree to be accountable for all aspects of the work in ensuring that questions relating to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All authors meet authorship criteria, have read and approved the published work, and certify that they have participated sufficiently in the work to take responsibility for the content including the concept, design, analysis, writing, or revision (see author contributions above).
Corresponding author
Ethics declarations
Conflict of interest
LW Heinsberg reports grants from the National Institute of Nursing Research, University of Pittsburgh, Eta Chapter, Sigma Theta Tau, Inc., and International Society of Nurses in Genetics during the conduct of this study. YP Conley and PR Sherwood reports grants from the National Institute of Nursing Research. EA Crago reports grants from the University of Pittsburgh. The remaining authors report no conflicts of interest to disclose.
Ethical Approval
Institutional review board approval at the University of Pittsburgh has been obtained (IRB Approval Number 021039), and we have adhered to ethical considerations in the protection of all human subjects involved.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Heinsberg, L.W., Arockiaraj, A.I., Crago, E.A. et al. Genetic Variability and Trajectories of DNA Methylation May Support a Role for HAMP in Patient Outcomes After Aneurysmal Subarachnoid Hemorrhage. Neurocrit Care 32, 550–563 (2020). https://doi.org/10.1007/s12028-019-00787-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12028-019-00787-4