Abstract
This study aims to investigate the role of circCBFB in hepatocellular carcinoma (HCC) cell proliferation and autophagy. qRT-PCR and Western blotting analyses quantified the expression levels of circCBFB, miR-424-5p, and ATG14 in HCC tissues and/or HCC cell lines. After transfection with pcDNA3.1-CircCBFB, sh-CircCBFB, miR-424-5p mimic, miR-424-5p inhibitor, pcDNA3.1-ATG14, sh-ATG14, sh-CircCBFB + miR-424-5p inhibitor, pcDNA3.1-CircCBFB + miR-424-5p mimic, sh-CircCBFB + pcDNA3.1-ATG14, or pcDNA3.1-CircCBFB + sh-ATG14, the proliferation, cell cycle, and apoptosis of Huh-7 and HCCLM3 cells were detected, respectively, through MTT assay and flow cytometry. Western blotting measured the expression levels of ATG14 and autophagy-related proteins (LC3-ΙΙ/LC3-Ι, Beclin1, and p62). The interactions among circCBFB, miR-424-5p, and ATG14 were identified through RNA fluorescence in situ hybridization and RNA immunoprecipitation. In HCC tissues, circCBFB and ATG14 were highly expressed, and miR-424-5p expression was downregulated. Transfection of pcDNA3.1-CircCBFB, miR-424-5p inhibitor, or pcDNA3.1-ATG14 into HCC cells facilitated HCC cell proliferation and autophagy, while suppressing cell apoptosis, evidenced by elevated cell viability, increased protein levels of autophagosome markers (LC3-ΙΙ/LC3-Ι and Beclin1), repressed apoptosis rate, and suppressed protein level of autophagy receptor p62. miR-424-5p was a target gene of circCBFB, and miR-424-5p negatively mediated ATG14. CircCBFB inhibits miR-424-5p and upregulates ATG14, thus promoting HCC cell proliferation and autophagy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatocellular carcinoma (HCC) is a dominating histological form of primary liver cancer [1], the prevalence of which occupies 70 ~ 90% of liver cancer [2]. Up to 2018, the incidence of liver cancer ranks the sixth, and the number of liver cancer-related deaths is approximately 781,000 [3]. The high risk of metastasis and recurrence for patients with HCC causes dismal 5-year survival rate of HCC even after the treatment of surgery, radiation, chemotherapy, and immunotherapy [4]. Hence, more studies are needed to enhance the understanding of mechanisms underlying HCC progression.
As a highly conserved catabolic process, autophagy is responsible for cellular homeostasis through degenerating damaged organelles and proteins [5]. Autophagy exerts a vital role in anti-HCC [6]. Induction of autophagy in HCC cells has been reported to promote liver cancer progression both in mice and human [7]. Autophagy-related genes (ATGs) are pivotal for the initiation of autophagy due to their implication in the production of autophagosome and the transferring of autophagic cargos to the lysosome [8]. Manipulations that trigger autophagy through ATGs have been regarded as promising targets for HCC treatment [9]. As a specific subunit for PI3K complex, ATG14 is regarded as a key modulator of autophagy in HCC, blockade of which can inhibit autophagy in HCC [10]. However, the molecular mechanisms modulating ATG14 in HCC remain to be fully illuminated.
Circular RNAs (circRNAs) are a peculiar class of endogenous non-coding RNAs that possess a covalent bond linking the 3′ and 5′ ends and originate from back splicing events of precursor mRNAs (pre-mRNAs) [11, 12]. Recently, circRNAs have been demonstrated to function as competing endogenous RNAs (ceRNAs) that sponge cytoplasmic microRNAs (miRNAs) or RNA binding proteins, thus regulating splicing or transcription [13]. In HCC, circRNAs have been reported to exert oncogenic effects through interacting with miRNAs [14, 15]. CircCBFB is capable of regulating cell proliferation, cell cycle, and apoptosis, acting through ceRNA network in diseases, such as chronic lymphocytic leukemia and abdominal aortic aneurysm [16, 17]. Since circCBFB has been mentioned for its promoting effect on acetaminophen-induced liver injury [18], circCBFB was also hypothesized to play a role in HCC.
In the current study, the effects of circCBFB on HCC prognosis and HCC cell autophagy were elucidated. To further illustrate the underlying molecular mechanism, the interactions between circCBFB and miR-424-5p were identified. More importantly, this study demonstrated the ceRNA mechanism through validating the co-expression network of circCBFB, miR-424-5p, and ATG14.
Materials and methods
Tissue samples
HCC and matched adjacent tissues were obtained from 43 patients with HCC who received hepatectomy at the Third Xiangya Hospital, Central South University. Patients who underwent chemoradiotherapy, radiofrequency ablation, or biotherapy were excluded from this study. A total of 43 patients (28 males and 15 females, 28 ~ 78 years old, average age of 55) were enrolled in this study. Collection of tissues and design of the following experiments were approved by the ethical committee of the Third Xiangya Hospital, Central South University, and informed consents were obtained from the patients or their family members. All samples were maintained in liquid nitrogen after collection.
Cell culture
Human HCC cell lines (HepG2, SMMC-7721, Huh-7, MHCC97H, and HCCLM3) and normal hepatocytes (THLE2) were purchased from Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. All cell lines were incubated in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37°C with 5% CO2.
qRT-PCR
Total RNA was extracted using TRIZOL (Invitrogen, Carlsbad, CA, USA) and reversely transcribed using a reverse transcriptase kit (TaKaRa, Tokyo, Japan) according to the directions of the manufacturer. PCR reactions were performed according to the instructions of the fluorescence quantitative PCR kit (SYBR Premix EX Taq™ ii, TaKaRa). The PCR was conducted with the following thermal parameters: 95 °C for 10 s and 45 cycles of 95 °C for 5 s, 60 °C for 10 s, and 72 °C for 10 s, followed by extension at 72 °C for 5 min. Each PCR was performed in triplicate. U6 and GAPDH were used to normalize miRNA and mRNA expression levels, respectively. The expression levels of circCBFB and miR-424-5p and the mRNA expression of ATG14 were quantified on a LightCycler 480 instrument (Roche, Indianapolis, IN, USA) by using 2−ΔΔCt method: ΔΔCt = (Cttarget gene – Ctinternal control)experimental group – (Cttarget gene – Ctinternal control)control group. Primer sequences used in this study are presented in Table 1.
Western blotting
Proteins were obtained after the cells were lysed by RIPA lysis buffer (Beyotime), and the concentration of extracted protein was determined using a BCA kit (Beyotime). The proteins were mixed with an equal volume of loading buffer (Beyotime) and heated in boiling water for 3 min to denature the proteins. Electrophoresis was initially run at 80 V for 30 min to separate the proteins and switched to 120 V for 1 ~ 2 h after the bromophenol blue entered the separation gel. Membrane transferring was conducted in an ice bath at 300 mA for 60 min, after which the membranes were rinsed in washing buffer for 1 ~ 2 min and blocked in blocking buffer for 60 min at room temperature or at 4 °C overnight. The membranes were then immunoblotted with the following antibodies for 1 h on a shaking table: anti-GAPDH (#5174, 1:1000), anti-LC3-I/LC3-II (#12741, 1:1000), anti-Beclin1 (#3495, 1:1000), p62 (#88588, 1:1000), and anti-ATG14 (#5504, 1:1000) (Cell Signaling Technology, Massachusetts, USA). After the membranes were washed with washing buffer for 3 × 10 min, secondary antibody (horseradish peroxidase-conjugated goat anti-rabbit IgG, 1:5000, Beijing ComWin Biotech Co., Ltd., Beijing, China) was added for incubation at room temperature for 1 h. After being washed for 3 × 10 min, the membranes were added with developing solution and quantified using chemiluminiscence imaging system (Bio-Rad).
Cell transfection
pcDNA3.1-CircCBFB (2 μg), short hairpin RNA (sh)-CircCBFB (2 μg), miR-424-5p mimic (50 nM), miR-424-5p inhibitor (100 nM), pcDNA3.1-ATG14 (2 μg), and their negative controls (NCs) were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). Cells were transfected with pcDNA3.1-CircCBFB, pcDNA3.1, sh-CircCBFB, sh-NC, miR-424-5p inhibitor, inhibitor NC, miR-424-5p mimic, mimic NC, and/or pcDNA3.1-ATG14 using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) and were assigned accordingly to CircCBFB group, pcDNA3.1 group, sh-CircCBFB group, sh-NC group, in-miR-424-5p group, in-NC group, mi-miR-424-5p group, mi-NC group, ATG14 group, NC group, CircCBFB + mi-miR-424-5p group, sh-CircCBFB + in-miR-424-5p group, CircCBFB + sh-ATG14 group, and sh-CircCBFB + ATG14 group.
MTT
After the transfections, cells were cultured for 24 h, 48 h, and 72 h, and then 20 μL of MTT solution (5 mg/mL, Merck KGaA, Darmstadt, Germany) was added into each well for incubation in a 37°C incubator with 5% CO2 for 4 h. After that, the culture solution was removed. DMSO (50 μL/well) was added, and the plates were slightly shaken to dissolve the crystal. The optical density value (OD value) was evaluated by a microplate reader at 490 nm for 3 times and then averaged. MTT curves were drawn using the OD value as the vertical coordinates and the time as the horizontal coordinates.
Flow cytometry (FCM)
Single cell suspension was prepared and then centrifuged at 2000 r/min before the cells were rinsed twice with PBS. Then, the cells were re-suspended in binding buffer, and 195 μL of cell suspension (about 105 cells) was homogenized with 5 μL of Annexin-V-FITC and PI solution for 10 min of incubation in dark. Cell cycle and apoptotic rate were determined using a FCM (BD Biosciences, Suzhou, China).
Dual luciferase reporter assay
The binding site between circCBFB and miR-424-5p or between miR-424-5p and ATG14 was predicted by starBase (http://starbase.sysu.edu.cn/). Accordingly, the wild-type (wt) sequences of circCBFB and the 3′untranslated region (3′UTR) of ATG14, as well as the mutant-type (mut) sequences of the independent binding sites of miR-424-5p (wt-CircCBFB, mut-CircCBFB, wt-ATG14, and mut-ATG14), were synthesized and then cloned into pGL3-Basic vectors, after which the vectors were separately transfected with miR-424-5p mimic (50 nM) and mimic NC into HEK293T cells. Finally, the firefly luciferase activity and renilla luciferase activity were determined by using a luminometer (Promega GLOMAX, Promega Corporation). The firefly luciferase activity was measured with the renilla luciferase activity as its internal control, and the ratio between the two was the relative luciferase activity.
RNA fluorescence in situ hybridization (FISH)
Isolation of cytoplasmic and nuclear RNA was performed using a PARIS Kit (Invitrogen). Then, the RNA expression of circCBFB, β-actin (a marker for cytoplasmic protein), or U6 (a marker for nuclear protein) was detected in cytoplasmic and protein RNA. The distribution of circCBFB and miR-424-5p was examined using an RNA FISH kit. CircCBFB and miR-424-5p probes (GenePharma) were used for hybridization overnight. DAPI was used for nuclear staining. The RNA samples were observed by a Nikon inverted fluorescence microscope.
RNA immunoprecipitation (RIP)
The binding between miR-424-5p and circCBFB or ATG14 was determined by RIP. The cells were collected and then washed with pre-cooled PBS twice before centrifugation at 1500 rpm for 5 min. An equal volume of RIP lysis buffer was added and homogenized. RIP wash buffer (100 μL) was applied to resuspend magnetic beads, and then 5 μg of anti-AGO2 (ab186733, 1:50, Abcam, Cambridge, UK) or NC IgG was added for incubation at 4°C on a rotator overnight. Prepared cell lysis buffer was rapidly dissolved and centrifuged at 14,000 rpm and 4°C for 10 min. The supernatant (100 μL) was mixed and incubated with bead-antibody complexes at 4°C overnight. After a transient centrifugation, the centrifuge tube containing the mixture was placed on a magnetic separation rack to discard the supernatant. RIP wash buffer (500 μL) was added, and the centrifuge tube was shaken by vortexing to wash complex. Then, the tube was maintained on the magnetic separation rack and the supernatant was removed. The complex was washed for 6 times as mentioned. The samples were then incubated with proteinase K buffer (150 μL) to re-suspend bead-antibody complexes at 55°C for 30 min, after which the tubes were placed on the magnetic separation rack to pipette off the supernatant. After RNAs were isolated, the expression levels of circCBFB and miR-424-5p were determined by qRT-PCR.
Statistical analysis
GraphPad prism7 software was applied for statistical analysis. We reported all data as average ± standard deviation and applied T-test to assess the difference between two groups. One-way analysis of variance was used to measure the differences among multiple groups. Correlations between circCBFB expression level and other indices were determined through Pearson correlation analysis, and survival analysis was performed using Kaplan–Meier. P value less than 0.05 was considered statistically significant.
Results
Highly expressed circCBFB in HCC tissues involves HCC prognosis
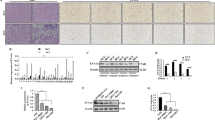
Firstly, we performed qRT-PCR to measure the expression of circCBFB in 43 human HCC tissues. Higher circCBFB level was presented in the HCC tissues compared with the adjacent tissues (Fig. 1A, P < 0.01).
Aberrant expression of circCBFB is associated with HCC prognosis. qRT-PCR detected the expression of circCBFB in HCC tissues and adjacent tissues (A); Kaplan–Meier analysis showed that the expression of circCBFB was of predictive value for overall survival in HCC patients (B); qRT-PCR revealed the upregulated expression of circCBFB in HCC cell lines (C). *P < 0.05, **P < 0.01, compared with peritumor group or THLE2 group. HCC, hepatocellular carcinoma
Next, patients were assigned into CircCBFB high group and CircCBFB low group with the median circCBFB expression in HCC tissues as the cutoff value, and then prognostic analysis was performed to evaluate the differences in the survival time for patients with high or low expression of circCBFB. Five-year survival rate of patients with higher circCBFB expression was lower than that in patients with lower circCBFB expression (Fig. 1B, P < 0.05). Overall, our findings indicated that circCBFB was upregulated in HCC patients and was associated with poor prognosis and survival rate, suggesting a potential role of circCBFB in HCC progression.
Additionally, circCBFB expression in HCC cell lines, including HepG2, SMMC-7721, Huh-7, MHCC97H, and HCCLM3, was significantly higher than that in THLE2 cells (Fig. 1C, P < 0.05).
CircCBFB promotes the proliferation and autophagy of HCC cells
Huh-7 cells were transfected with pcDNA3.1-CircCBFB (the CircCBFB group), and HCCLM3 cells were transfected with sh-CircCBFB (the sh-CircCBFB group) to artificially regulate circCBFB expression in these cells. The expression of cricCBFB in Huh-7 cells of the pcDNA3.1-CircCBFB group was markedly increased compared with that in the pcDNA3.1 group (Fig. 2A, P < 0.01). Consistently, circCBFB expression in HCCLM3 cells was much lower in the sh-CircCBFB group than in the sh-NC group (Fig. 2B, P < 0.01).
Promotive effects of circCBFB on HCC cell proliferation and autophagy. Huh-7 cells were transfected with pcDNA3.1-CircCBFB, and HCCLM3 cells were transfected with sh-CircCBFB, and then circCBFB expression was examined by qRT-PCR (A, B). MTT assay and FCM, respectively, showed the proliferation (C, D, E), cell cycle (F, G), and apoptosis (H) of Huh-7 and HCCLM3 cells; the levels of LC3II/LC3I, Beclin1, and p62 were detected through Western blotting (I, J). All data are presented as average ± standard deviation, *P < 0.05, **P < 0.01, compared with sh-NC group or pcDNA3.1 group. HCC, hepatocellular carcinoma; FCM, flow cytometry; NC, negative control
We performed MTT assay to detect HCC cell proliferation and found enhanced proliferative ability of Huh-7 cells after the transfection of pcDNA3.1-CircCBFB (Fig. 2C, P < 0.01). The proliferative ability of HCCLM3 cells transfected with sh-CircCBFB was noticeably inhibited (Fig. 2D, P < 0.01). Moreover, the proliferative ability of human normal hepatocytes (THLE2) transfected with pcDNA3.1-CircCBFB or sh-CircCBFB was detected. It was observed that overexpression or inhibition of circCBFB exerted no effect on the proliferative ability of THLE2 cells (Fig. 2E, P > 0.05). FCM displayed a higher proportion of Huh-7 cells in S phase in the CircCBFB group compared with the pcDNA3.1 group (Fig. 2F, P < 0.05). CircCBFB silencing caused a higher proportion of HCCLM3 cells in G1 phase (Fig. 2G, P < 0.05). In addition, FCM examined a decrease in Huh-7 cell apoptosis rate in the CircCBFB group compared with that in the pcDNA3.1 group and an increase in HCCLM3 cell apoptosis rate in the sh-CircCBFB group when compared with the sh-NC group (Fig. 2H, P < 0.01).
The effects of circCBFB on HCC cell autophagy were evaluated through detecting the levels of autophagy-related proteins, including LC3II/LC3I, Beclin1, and p62. The results of Western blotting manifested that the levels of LC3II/LC3I and Beclin1 were elevated in, and the expression of autophagy receptor p62 was inhibited in Huh-7 cells of the CircCBFB group, compared with the pcDNA3.1 group (Fig. 2I, J, P < 0.01). In the sh-CircCBFB group, the expression levels of LC3II/LC3I and Beclin1 were suppressed, and p62 expression was upregulated when compared with the sh-NC group. These results indicated that circCBFB enhanced HCC cell proliferation and autophagy.
CircCBFB promotes cancer cell development by enhancing autophagy
The aforementioned results indicated that circCBFB exacerbated HCC cell autophagy. To further validate the regulatory effect of circCBFB on cancer cell autophagy, we stimulated Huh-7 and HCCLM3 cells transfected with pcDNA3.1 or pcDNA3.1-CircCBFB with autophagy inhibitor chloroquine (CQ) (20 μM) for 48 h. Next, CCK-8 assay was used for detection of cell viability. The results showed a massive decrease in the CQ + pcDNA3.1 group (vs the control group) and a significant increase in the CQ + pcDNA3.1-CircCBFB group (vs the CQ + pcDNA3.1 group) (Fig. 3A, P < 0.01). Then, the detection of cell cycle by FCM further identified these results (Fig. 3B). Meanwhile, FCM detected the apoptosis rate of Huh-7 and HCCLM3 cells and demonstrated that the apoptosis rate was enhanced in the CQ + pcDNA3.1 group when compared with that in the control group, while it was reduced in the CQ + pcDNA3.1-CircCBFB group compared with that in the CQ + pcDNA3.1 group (Fig. 3C, P < 0.05). Overall, circCBFB facilitated HCC cell growth through exacerbating cell autophagy.
CircCBFB enhances cell autophagy to exacerbate HCC progression. MTT assay examined the proliferation rate of Huh-7 and HCCLM3 cells (A); FCM detected the cell cycle (B) and the apoptosis rate (C) of Huh-7 and HCCLM3 cells. Data are presented as average ± standard deviation. *P < 0.05, **P < 0.01, compared with the control group, #P < 0.05, ##P < 0.01 compared with the CQ + pcDNA3.1 group. HCC, hepatocellular carcinoma; CQ, chloroquine; FCM, flow cytometry
CircCBFB targets miR-424-5p in HCC
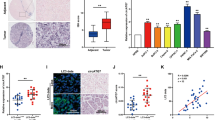
The binding site between circCBFB and miR-424-5p was predicted by the online software starBase (http://starbase.sysu.edu.cn), and the mutated sequence was designed (Fig. 4A). Then, we detected miR-424-5p expression in 43 HCC tissues and their adjacent tissues, and the results revealed the downregulated expression of miR-424-5p (Fig. 4B, P < 0.01). Moreover, correlation analysis confirmed that circCBFB level was negatively correlated with miR-424-5p level (Fig. 4C). Next, the enrolled patients were divided into miR-424-5p high or low group according to the expression of miR-424-5p, and the follow-up study compared the difference in clinical survival between these two groups by analyzing the prognosis. The results showed that patients with higher miR-424-5p expression had higher survival rate than those with lower miR-424-5p expression (Fig. 4D, P < 0.05).
CircCBFB directly binds to miR-424-5p in HCC. StarBase showed the binding site between circCBFB and miR-424-5p (A); qRT-PCR analysis of miR-424-5p expression in HCC tissues and the adjacent tissues (B); Pearson correlation analysis confirmed the negative correlation between circCBFB and miR-424-5p (C); Kaplan–Meier survival analysis (D); miR-424-5p expression in HCC cells was determined by qRT-PCR (E, F); FISH analysis of the co-localization of circCBFB and miR-424-5p in the cytoplasm, scale bar = 50 μm (G); anti-AGO2 RIP was analyzed by RT-PCR to detect the enrichment of circCBFB and miR-424-5p (H); Dual luciferase reporter assay examined the interactions between circCBFB and miR-424-5p (I). Data are presented as average ± standard deviation. **P < 0.01, ***P < 0.001, compared with sh-NC group, pcDNA3.1 group, mi-NC group, or in-NC group. HCC, hepatocellular carcinoma; RIP, RNA immunoprecipitation; FISH, fluorescence in situ hybridization
Thereafter, we examined miR-424-5p expression in Huh-7 and HCCLM3 cells after transfection with pcDNA3.1-CircCBFB and sh-CircCBFB. In Huh-7 cells transfected with pcDNA3.1-CircCBFB, miR-424-5p expression was downregulated compared with that in Huh cells transfected with pcDNA3.1 (Fig. 4E, P < 0.01). miR-424-5p expression was upregulated in HCCLM3 cells of the sh-CircCBFB group compared with that in the sh-NC group (Fig. 4F, P < 0.01). In addition, FISH analysis revealed that circCBFB bound to and co-expressed with miR-424-5p in the cytoplasm of HCC cells (Fig. 4G).
To identify whether circCBFB could directly bind to miR-424-5p, AGO2 antibody was used in HCCLM3 cells to perform RIP. The results displayed that AGO2 antibody enriched circCBFB significantly (Fig. 4H, P < 0.001), while IgG antibody could not enrich it. Additionally, dual luciferase reporter assay manifested that co-transfection of mut-CircCBFB and miR-424-5p mimic/inhibitor did not alter the luciferase activity in HEK293T cells; in HEK293T cells transfected with wt-CircCBFB, miR-424-5p mimic inhibited the luciferase activity, and miR-424-5p inhibitor remarkably elevated the luciferase activity (Fig. 4I, P < 0.05). These results supported that miR-424-5p was a target gene of circCBFB.
CircCBFB promotes HCC cell proliferation and autophagy through targeting miR-424-5p
To explore whether circCBFB could act as an oncogenic gene through regulating miR-424-5p, Huh-7 cells were transfected with pcDNA3.1-CircCBFB (the CircCBFB group) or co-transfected with pcDNA3.1-CircCBFB and miR-424-5p mimic (the CircCBFB + mi-miR-424-5p group), and HCCLM3 cells were transfected with sh-CircCBFB (the sh-CircCBFB group) or co-transfected with sh-CircCBFB and miR-424-5p inhibitor (the sh-CircCBFB + in-miR-424-5p group).
MTT assay showed that Huh-7 cell proliferation in the CircCBFB group was enhanced than that in the pcDNA3.1 group, while the cell proliferation in the CircCBFB + mi-miR-424-5p group was inhibited than that in the CircCBFB group (Fig. 5A, B, P < 0.05). HCCLM3 cell proliferation was suppressed in the sh-CircCBFB group but prompted in the sh-CircCBFB + in-miR-424-5p group when separately compared with the sh-NC group and sh-CircCBFB group (Fig. 5A, B, P < 0.05).
CircCBFB modulates HCC cell proliferation and autophagy through sponging miR-424-5p. HCC cell proliferation was assessed by MTT assay (A, B); FCM determined HCC cell cycle (C, D) and apoptosis (E, F); Western blotting of LC3II/LC3I, Beclin1, and p62 levels (G, H). Data are presented as average ± standard deviation. **P < 0.01, compared with sh-NC group or pcDNA3.1 group, ##P < 0.01, compared with sh-CircCBFB group or CircCBFB group. HCC, hepatocellular carcinoma; FCM, flow cytometry; NC, negative control
In the CircCBFB group, the proportion of Huh-7 cells in S phase was increased compared with that in the pcDNA3.1 group, while the CircCBFB + mi-miR-424-5p group had increased proportion of Huh-7 cells in G1 phase compared with that in the CircCBFB group (Fig. 5C, P < 0.05). HCCLM3 cell cycle was arrested in G1 phase in the sh-CircCBFB group in comparison with the sh-NC group, while the sh-CircCBFB + in-miR-424-5p showed elevated proportion of HCCLM3 cells in S phase compared with that in the sh-CircCBFB group (Fig. 5D, P < 0.05).
FCM displayed lower apoptosis rate of Huh-7 cells in the CircCBFB group (vs. the pcDNA3.1 group) and higher apoptosis rate in the CircCBFB + mi-miR-424-5p group (vs. the CircCBFB group) (Fig. 5E, P < 0.01). In HCCLM3 cells, the apoptosis rate of HCCLM3 cells in the sh-CircCBFB group was increased compared with the sh-NC group, and the sh-CircCBFB + in-miR-424-5p group showed decreased apoptosis rate of HCCLM3 cells when compared with the sh-CircCBFB group (Fig. 5F, P < 0.01).
Western blotting quantified the expression levels of LC3II/LC3I, Beclin1, and p62 in Huh-7 and HCCLM3 cells. Compared with the pcDNA3.1 group, the CircCBFB group exhibited higher levels of LC3II/LC3I and Beclin1 and lower expression of p62 in Huh-7 cells (Fig. 5G, P < 0.01). Compared with the CircCBFB group, the decreased levels of LC3II/LC3I and Beclin1 and increased level of p62 were found in Huh-7 cells of the CircCBFB + mi-miR-424-5p group (Fig. 5G, P < 0.01). In the sh-CircCBFB group, LC3II/LC3I and Beclin1 expression levels were suppressed, and p62 expression was elevated in HCCLM3 cells in comparison with the sh-NC group (Fig. 5H, P < 0.01). Reverse results were found in HCCLM3 cells of the sh-CircCBFB + in-miR-424-5p group when compared with the sh-CircCBFB group (Fig. 5H, P < 0.01). Taken together, circCBFB prompted autophagy and progression of HCC cells through acting as a sponge of miR-424-5p.
ATG14 is a downstream target gene of miR-424-5p in HCC
We had predicted the binding site between ATG14 and miR-424-5p through starBase (http://starbase.sysu.edu.cn) and designed the mutated sequence (Fig. 6A). The upregulated ATG14 expression in the 43 tissue samples was assessed (Fig. 6B, P < 0.001). Further analysis showed the negative association between ATG14 and miR-424-5p expression levels (Fig. 6C). Thereafter, we found that Huh-7 cells transfected with miR-424-5p mimic had lower ATG14 expression (Fig. 6D, P < 0.001), and HCCLM3 cells in the in-miR-424-5p group had higher ATG14 level (Fig. 6E, P < 0.001). Dual luciferase reporter assay exhibited no noticeable change in luciferase activity after HEK293T cell was transfected with mut-ATG14. Co-transfection of wt-ATG14 and miR-424-5p mimic into HEK293T cells reduced the luciferase activity, and co-transfection of wt-ATG14 and miR-424-5p inhibitor enhanced the luciferase activity (Fig. 6F, P < 0.05). These results implied that ATG14 was a target gene of miR-424-5p.
miR-424-5p directly targets ATG14. The binding site between miR-424-5p and ATG14 (A); ATG14 expression in HCC tissues and adjacent tissues was measured by RT-PCR (B); Pearson correlation analysis of the relationship between ATG14 and miR-424-5p (C); Western blotting quantified ATG14 expression after miR-424-5p expression was overexpressed or silenced in HCC cells (D, E); Dual luciferase reporter assay of luciferase activity (F). Data are presented as average ± standard deviation. * P < 0.05, **P < 0.01, compared with mi-NC group or in-NC group. HCC, hepatocellular carcinoma; NC, negative control
CircCBFB promotes HCC cell autophagy through ATG14
To identify whether ATG14 could be a downstream effector of circCBFB, sh-ATG14/pcDNA3.1-ATG14 and pcDNA3.1-CircCBFB/sh-CircCBFB were co-transfected into Huh-7 cells and HCCLM3 cells. ATG14 expression in these cells is shown in Fig. 7A and B. MTT assay manifested that the proliferation of Huh-7 cells in the CircCBFB + sh-ATG14 group was suppressed than that in the CircCBFB + sh-NC group (Fig. 7C), evidencing that ATG14 knockdown could inhibit the enhancement of HCC cell proliferation induced by circCBFB overexpression. HCCLM3 cell proliferation in the sh-CircCBFB + ATG14 group was raised compared with that in the sh-CircCBFB + pcDNA3.1 group (Fig. 7D), suggesting that ATG14 overexpression offset, in part, CircCBFB-silencing induced suppression of HCC cell proliferation.
CircCBFB regulates ATG14 to regulate HCC cell proliferation and autophagy. Huh-7 cells were separately transfected with pcDNA3.1-CircCBFB, pcDNA3.1-CircCBFB + sh-ATG14, and sh-ATG14, and HCCLM3 cells were treated with sh-CircCBFB, sh-CircCBFB + pcDNA3.1-ATG14, and pcDNA3.1-ATG14. Then, Western blotting examined ATG14 expression (A, B); MTT assay and FCM respectively detected HCC cell proliferation (C, D), cell cycle (E, F), and apoptosis (G, H); Western blotting of LC3II/LC3I, Beclin1, and p62 levels (I, J). Data are presented as average ± standard deviation. *P < 0.05, **P < 0.01, compared with sh-NC group or pcDNA3.1 group, #P < 0.05, ##P < 0.01, compared with sh-CircCBFB group or CircCBFB group. HCC, hepatocellular carcinoma; FCM, flow cytometry; NC, negative control
Compared with the CircCBFB + sh-NC group, the proportion of Huh-7 cells in G1 phase was significantly increased in the CircCBFB + sh-ATG14 group (Fig. 7E, P < 0.05). HCCLM3 cells in G1 phase were reduced in the sh-CircCBFB + -ATG14 group in comparison with cells in the sh-CircCBFB + pcDNA3.1 group (Fig. 7F, P < 0.05). Moreover, the apoptosis rate of Huh-7 cells in the CircCBFB + sh-ATG14 group was increased than that in the CircCBFB + sh-NC group (Fig. 7G, P < 0.01), and in the sh-CircCBFB + ATG14 group, the apoptosis rate of HCCLM3 cells was declined than that in sh-CircCBFB + pcDNA3.1 group (Fig. 7H, P < 0.01).
Western blotting of LC3II/LC3I, Beclin1, and p62 levels showed that Huh-7 cell autophagy in the CircCBFB + sh-ATG14 group was repressed than that in the CircCBFB + sh-NC group (Fig. 7I, P < 0.05), and HCCLM3 cell autophagy in the sh-CircCBFB + ATG14 group was increased than that in the sh-CircCBFB + pcDNA3.1 group (Fig. 7J, P < 0.05). These results identified that circCBFB regulated HCC cell autophagy through ATG14. To sum up, circCBFB regulated the miR-424-5p/ATG14 axis and then promoted HCC cell autophagy and proliferation.
Discussion
Although a number of studies have demonstrated abnormal alteration in circRNA expression in HCC [19,20,21], the regulation of circRNAs on autophagy in HCC development is largely unknown. Herein, we uncovered that circCBFB was upregulated in HCC tissues and promoted autophagy through sponging miR-424-5p and regulating ATG14 (Fig. 8). Increased circCBFB expression in HCC also displayed a correlation with poor prognosis and decreased survival rate, which indicated its therapeutic significance in disease management.
In the current study, the expression of circCBFB in HCC tissues and the adjacent tissues were identified. We confirmed that circCBFB was upregulated in HCC tissues and highly expressed circCBFB predicted dismal 5-year survival rate and poor prognosis, implying the significant involvement of circCBFB in HCC progression. Consistently, circCBFB overexpression was proven to prompt cell proliferation and discourage apoptosis in HCC cells. Considering the involvement of autophagy in HCC [22, 23], we speculated that circCBFB might also affect HCC cell autophagy. LC3-ΙΙ/LC3-Ι, Beclin1, and p62 are frequently used markers of autophagy. Cytosolic LC3-Ι is conjugated to phosphatidylethanolamine to become phagophore- or autophagosome-associated LC3-ΙΙ, which promotes autophagosome formation [24, 25]. Cytoplasmic Beclin1 is dysregulated and tightly correlated to autophagy in HCC tissues, which affects the clinicopathologic characteristics and survival time of HCC patients [26]. P62, a selective substrate of macroautophagy, functions as an adaptor protein that links ubiquitinated proteins to LC3, and inhibition of macroautophagy results in the accumulation of p62 [27]. In our study, LC3-ΙΙ/LC3-Ι and Beclin1 levels were positively associated with circCBFB expression in HCC cells, while p62 expression was decreased as an increase in circCBFB expression. More importantly, overexpression of circCBFB was found to offset the inhibition of CQ on cell autophagy and exacerbate cancer cell proliferation as well as suppress cell apoptosis. Therefore, it can be concluded that circCBFB aggravated HCC progression through facilitating cell autophagy.
A circRNA exerts biological functions through sponging miRNA and therefore regulates a downstream target gene of the miRNA [28]. CircCBFB has also been validated to act on cell progression through ceRNA mechanisms [16, 17], and circCBFB knockdown could alleviate mitochondrial dynamics in injured hepatocytes through sponging a certain miRNA [18]. Through Pearson correlation analysis, FISH, and RIP assays, we found that circCBFB directly bound to and co-localized with miR-424-5p in the cytoplasm, and then the interaction between circCBFB and miR-424-5p was further identified by dual luciferase reporter assay. miR-424-5p is downregulated both in vivo and in vitro and is related to intrahepatic metastasis and drug resistance [29, 30]. Consistently, miR-424-5p expression was significantly suppressed in HCC tissues. In addition, circCBFB overexpression suppressed miR-424-5p level in HCC cells. In the present study, circCBFB overexpression could increase proliferation and suppress apoptosis of HCC cells, which could be counteracted by upregulated miR-424-5p in HCC cells. Moreover, circCBFB- triggered autophagy was inhibited by miR-424-5p overexpression, supported by the declined levels of LC3-ΙΙ/LC3-Ι and Beclin1 and enhanced expression of p62. Hence, we confirmed that circCBFB regulated HCC cell proliferation, apoptosis, and autophagy through acting as a sponge of miR-424-5p.
ATG14 has been reported to implicate in the autophagy of both liver samples and hepatic cells [31, 32]. Herein, ATG14 expression was promoted in HCC tissues, and ATG14 was identified to be a target gene of miR-424-5p, suggesting an important role of ATG14 in HCC. Besides, ATG14 expression in HCC cells was negatively associated with miR-424-5p expression. Interestingly, the prompted proliferation of HCC cells induced by circCBFB overexpression was partly repressed by ATG14 silencing. CircCBFB had an inhibitory effect on the apoptosis of HCC cells, while this effect was counteracted by ATG14 silencing. Moreover, increased autophagy of HCC cells elicited by circCBFB overexpression was attenuated by ATG14 silencing. A protein complex, which consists of ATG14, Beclin1, vacuolar sorting protein 15 (Vps15), and Vps34 (the catalytic subunit of the class III PI3K), is crucial for autophagy initiation [33]. The binding of human ATG14 to Beclin1, through its coiled-coil domain, is required for autophagy, and ATG14 recruits a subset of class III PI3-kinase to the endoplasmic reticulum, which contributes to autophagosome formation [34]. Fogel et al. have reported human ATG14 as a critical regulator of autophagy-dependent phosphorylation of Beclin1, and the phosphorylation of Beclin1 at serines 90 and 93 is necessary for autophagy [35]. Also, the inhibitory effect of blocking Beclin1/Vps34/ATG14 complex activity on autophagy in HCC has been stated in an AKT-dependent manner [36].
In general, based on the results obtained from HCC tissues and cells, our study highlighted an important role of circCBFB in HCC progression. Herein, circCBFB was demonstrated to affect HCC progression by triggering cell autophagy through the miR-424-5p/ATG14 axis. Given the oncogenic role of circCBFB in HCC development, our study unravels a pharmacological target for HCC treatment.
References
Yang W, Sui J, Ma Y, Simon TG, Chong D, Meyerhardt JA, et al. A prospective study of dairy product intake and the risk of hepatocellular carcinoma in U. S. men and women. Int J Cancer. 2020;146(5):1241–9. https://doi.org/10.1002/ijc.32423.
Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep learning-based multi-omics integration robustly predicts survival in liver cancer. Clin Cancer Res. 2018;24(6):1248–59. https://doi.org/10.1158/1078-0432.CCR-17-0853.
Reynaert H, Colle I. Treatment of advanced hepatocellular carcinoma with somatostatin analogues: a review of the literature. Int J Mol Sci. 2019;20(19). doi:https://doi.org/10.3390/ijms20194811.
Ding H, Liu J, Zou R, Cheng P, Su Y. Long non-coding RNA TPTEP1 inhibits hepatocellular carcinoma progression by suppressing STAT3 phosphorylation. J Exp Clin Cancer Res. 2019;38(1):189. https://doi.org/10.1186/s13046-019-1193-0.
Zhou J, Zhang X, Tang H, Yu J, Zu X, Xie Z, et al. Nuclear factor erythroid 2 (NF-E2)-related factor 2 (Nrf2) in autophagy-induced hepatocellular carcinoma. Clin Chim Acta. 2020;506:1–8. https://doi.org/10.1016/j.cca.2020.02.028.
Kiruthiga C, Devi KP, Nabavi SM, Bishayee A. Autophagy: a potential therapeutic target of polyphenols in hepatocellular carcinoma. Cancers (Basel). 2020;12(3). doi:https://doi.org/10.3390/cancers12030562.
Niture S, Gyamfi MA, Lin M, Chimeh U, Dong X, Zheng W, et al. TNFAIP8 regulates autophagy, cell steatosis, and promotes hepatocellular carcinoma cell proliferation. Cell Death Dis. 2020;11(3):178. https://doi.org/10.1038/s41419-020-2369-4.
Wang CZ, Yan GX, Dong DS, Xin H, Liu ZY. LncRNA-ATB promotes autophagy by activating yes-associated protein and inducing autophagy-related protein 5 expression in hepatocellular carcinoma. World J Gastroenterol. 2019;25(35):5310–22. https://doi.org/10.3748/wjg.v25.i35.5310.
Xu WP, Liu JP, Feng JF, Zhu CP, Yang Y, Zhou WP, et al. miR-541 potentiates the response of human hepatocellular carcinoma to sorafenib treatment by inhibiting autophagy. Gut. 2019. https://doi.org/10.1136/gutjnl-2019-318830.
Huang KT, Kuo IY, Tsai MC, Wu CH, Hsu LW, Chen LY, et al. Factor VII-induced microRNA-135a inhibits autophagy and is associated with poor prognosis in hepatocellular carcinoma. Mol Ther Nucleic Acids. 2017;9:274–83. https://doi.org/10.1016/j.omtn.2017.10.002.
Qiu L, Huang Y, Li Z, Dong X, Chen G, Xu H, et al. Circular RNA profiling identifies circADAMTS13 as a miR-484 sponge which suppresses cell proliferation in hepatocellular carcinoma. Mol Oncol. 2019;13(2):441–55. https://doi.org/10.1002/1878-0261.12424.
Wang S, Cui S, Zhao W, Qian Z, Liu H, Chen Y, et al. Screening and bioinformatics analysis of circular RNA expression profiles in hepatitis B-related hepatocellular carcinoma. Cancer Biomark. 2018;22(4):631–40. https://doi.org/10.3233/CBM-170910.
Qu S, Liu Z, Yang X, Zhou J, Yu H, Zhang R, et al. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. 2018;414:301–9. https://doi.org/10.1016/j.canlet.2017.11.022.
Guan Z, Tan J, Gao W, Li X, Yang Y, Li X, et al. Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR-486/CDK4 pathway. J Cell Physiol. 2018;234(1):500–8. https://doi.org/10.1002/jcp.26612.
Huang XY, Huang ZL, Xu YH, Zheng Q, Chen Z, Song W, et al. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-100338/miR-141-3p pathway in hepatitis B-related hepatocellular carcinoma. Sci Rep. 2017;7(1):5428. https://doi.org/10.1038/s41598-017-05432-8.
Xia L, Wu L, Bao J, Li Q, Chen X, Xia H, et al. Circular RNA circ-CBFB promotes proliferation and inhibits apoptosis in chronic lymphocytic leukemia through regulating miR-607/FZD3/Wnt/beta-catenin pathway. Biochem Biophys Res Commun. 2018;503(1):385–90. https://doi.org/10.1016/j.bbrc.2018.06.045.
Yue J, Zhu T, Yang J, Si Y, Xu X, Fang Y, et al. CircCBFB-mediated miR-28-5p facilitates abdominal aortic aneurysm via LYPD3 and GRIA4. Life Sci. 2020;253: 117533. https://doi.org/10.1016/j.lfs.2020.117533.
Wang Z, Zhao Y, Sun R, Sun Y, Liu D, Lin M, et al. circ-CBFB upregulates p66Shc to perturb mitochondrial dynamics in APAP-induced liver injury. Cell Death Dis. 2020;11(11):953. https://doi.org/10.1038/s41419-020-03160-y.
Ding Z, Guo L, Deng Z, Li P. Circ-PRMT5 enhances the proliferation, migration and glycolysis of hepatoma cells by targeting miR-188-5p/HK2 axis. Ann Hepatol. 2020;19(3):269–79. https://doi.org/10.1016/j.aohep.2020.01.002.
Pu J, Wang J, Li W, Lu Y, Wu X, Long X, et al. hsa_circ_0000092 promotes hepatocellular carcinoma progression through up-regulating HN1 expression by binding to microRNA-338-3p. J Cell Mol Med. 2020. https://doi.org/10.1111/jcmm.15010.
Wei X, Zheng W, Tian P, He Y, Liu H, Peng M, et al. Oncogenic hsa_circ_0091581 promotes the malignancy of HCC cell through blocking miR-526b from degrading c-MYC mRNA. Cell Cycle. 2020;19(7):817–24. https://doi.org/10.1080/15384101.2020.1731945.
Chen DP, Ning WR, Li XF, Wei Y, Lao XM, Wang JC, et al. Peritumoral monocytes induce cancer cell autophagy to facilitate the progression of human hepatocellular carcinoma. Autophagy. 2018;14(8):1335–46. https://doi.org/10.1080/15548627.2018.1474994.
Xiong H, Ni Z, He J, Jiang S, Li X, He J, et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36(25):3528–40. https://doi.org/10.1038/onc.2016.521.
Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18(4):571–80. https://doi.org/10.1038/cdd.2010.191.
Shan Y, Li P. Long intergenic non-protein coding RNA 665 regulates viability, apoptosis, and autophagy via the MiR-186-5p/MAP4K3 axis in hepatocellular carcinoma. Yonsei Med J. 2019;60(9):842–53. https://doi.org/10.3349/ymj.2019.60.9.842.
Qiu DM, Wang GL, Chen L, Xu YY, He S, Cao XL, et al. The expression of beclin-1, an autophagic gene, in hepatocellular carcinoma associated with clinical pathological and prognostic significance. BMC Cancer. 2014;14:327. https://doi.org/10.1186/1471-2407-14-327.
Klionsky DJ, Baehrecke EH, Brumell JH, Chu CT, Codogno P, Cuervo AM et al. A comprehensive glossary of autophagy-related molecules and processes (2nd edition). Autophagy. 2011;7(11):1273–94. doi:https://doi.org/10.4161/auto.7.11.17661.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–8. https://doi.org/10.1038/nature11993.
Du H, Xu Q, Xiao S, Wu Z, Gong J, Liu C, et al. MicroRNA-424-5p acts as a potential biomarker and inhibits proliferation and invasion in hepatocellular carcinoma by targeting TRIM29. Life Sci. 2019;224:1–11. https://doi.org/10.1016/j.lfs.2019.03.028.
Zhang Y, Li T, Guo P, Kang J, Wei Q, Jia X, et al. MiR-424-5p reversed epithelial-mesenchymal transition of anchorage-independent HCC cells by directly targeting ICAT and suppressed HCC progression. Sci Rep. 2014;4:6248. https://doi.org/10.1038/srep06248.
Li B, Wu X, Chen H, Zhuang C, Zhang Z, Yao S, et al. miR199a-5p inhibits hepatic insulin sensitivity via suppression of ATG14-mediated autophagy. Cell Death Dis. 2018;9(3):405. https://doi.org/10.1038/s41419-018-0439-7.
Yu F, Dong B, Dong P, He Y, Zheng J, Xu P. Hypoxia induces the activation of hepatic stellate cells through the PVT1-miR-152-ATG14 signaling pathway. Mol Cell Biochem. 2020;465(1–2):115–23. https://doi.org/10.1007/s11010-019-03672-y.
Xiong X, Tao R, DePinho RA, Dong XC. The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J Biol Chem. 2012;287(46):39107–14. https://doi.org/10.1074/jbc.M112.412569.
Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190(4):511–21. https://doi.org/10.1083/jcb.200911141.
Fogel AI, Dlouhy BJ, Wang C, Ryu SW, Neutzner A, Hasson SA, et al. Role of membrane association and Atg14-dependent phosphorylation in beclin-1-mediated autophagy. Mol Cell Biol. 2013;33(18):3675–88. https://doi.org/10.1128/MCB.00079-13.
Wu FQ, Fang T, Yu LX, Lv GS, Lv HW, Liang D, et al. ADRB2 signaling promotes HCC progression and sorafenib resistance by inhibiting autophagic degradation of HIF1alpha. J Hepatol. 2016;65(2):314–24. https://doi.org/10.1016/j.jhep.2016.04.019.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhao, Z., He, J. & Feng, C. CircCBFB is a mediator of hepatocellular carcinoma cell autophagy and proliferation through miR-424-5p/ATG14 axis. Immunol Res 70, 341–353 (2022). https://doi.org/10.1007/s12026-021-09255-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12026-021-09255-8