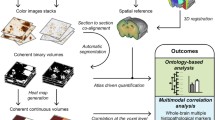
Abstract
Magnetic resonance imaging (MRI) and light-sheet fluorescence microscopy (LSFM) are technologies that enable non-disruptive 3-dimensional imaging of whole mouse brains. A combination of complementary information from both modalities is desirable for studying neuroscience in general, disease progression and drug efficacy. Although both technologies rely on atlas mapping for quantitative analyses, the translation of LSFM recorded data to MRI templates has been complicated by the morphological changes inflicted by tissue clearing and the enormous size of the raw data sets. Consequently, there is an unmet need for tools that will facilitate fast and accurate translation of LSFM recorded brains to in vivo, non-distorted templates. In this study, we have developed a bidirectional multimodal atlas framework that includes brain templates based on both imaging modalities, region delineations from the Allen’s Common Coordinate Framework, and a skull-derived stereotaxic coordinate system. The framework also provides algorithms for bidirectional transformation of results obtained using either MR or LSFM (iDISCO cleared) mouse brain imaging while the coordinate system enables users to easily assign in vivo coordinates across the different brain templates.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Uncovering complex functions of the brain for understanding disease mechanisms and developing effective therapies requires a combination of multiple neuroimaging techniques. Integration of in vivo and ex vivo tissue probing modalities, such as magnetic resonance imaging (MRI) and histology, has been described previously as it would enable researchers to study both temporal changes in a living brain as well as molecular markers in the same tissue post-mortem (Breckwoldt et al., 2016, 2019; Doerr et al., 2017; Goubran et al., 2019; Johnson et al., 2021; Leuze et al., 2017; MacKenzie-Graham et al., 2004; Morawski et al., 2018; Nie et al., 2019; Purger et al., 2009; Qu et al., 2022; Stolp et al., 2018).
MRI is a powerful imaging technique due to its non-invasive nature allowing repetitive scanning of human and animal tissue in vivo and ex vivo, with a wide range of contrast mechanisms and applications to e.g., visualize the cellular structures, neuronal activity, neuronal wiring, blood vessels, blood flow, metabolism, infarction, and malignancy (Dyrby et al., 2011, 2018; Matthews & Jezzard, 2004; Symms et al., 2004; Yousaf et al., 2018). The development of ultra-high field MRI has led to improved resolution enabling high-quality preclinical experiments (Dumoulin et al., 2018; Vaughan et al., 2001). However, MRI is still limited to its relatively low image resolution in the range of several tens of micrometers (Wang et al., 2020a, b; Wei et al., 2016). In contrast, light-sheet fluorescence microscopy (LSFM) enables direct visualization of cellular structures on a scale of a few micrometers. Recent progress in ex vivo 3D histology involving tissue clearing and immunolabeling of molecular markers, such as iDISCO (Renier et al., 2014), CUBIC (Susaki et al., 2014), or CLARITY (Chung et al., 2013) enables imaging of intact tissue and whole-organ specimens without disrupting their cytoarchitecture. Although LSFM is an ex vivo technique that lacks functionality to resolve longitudinal processes, it has become a widely applied imaging technique in preclinical studies for e.g., investigating gene and protein expression (Hansen et al., 2021; Kjaergaard et al., 2019; Skovbjerg et al., 2021), cellular architecture (di Giovanna et al., 2018; Friedmann et al., 2020), neural populations (Roostalu et al., 2019; Silvestri et al., 2015), and distribution of fluorescently labelled molecules (Gabery et al., 2020; Secher et al., 2014).
Standard processing of neuroimaging datasets involves co-registration of individual brain volumes with a reference atlas to perform group analysis (Badea et al., 2019; Friedmann et al., 2020; Goubran et al., 2019; Kirst et al., 2020; Massalimova et al., 2021; Perens et al., 2021; Renier et al., 2016; Salinas et al., 2018; Todorov et al., 2020; Tyson & Margrie, 2021; You et al., 2021). Recently, efforts have been made to combine LSFM-imaging with other neuroimaging modalities either for obtaining high-quality region delineations from existing brain atlases (Goubran et al., 2019; Murakami et al., 2018; Perens et al., 2021; Stolp et al., 2018) but also to study correlations between in vivo and ex vivo MRI-biomarkers (Breckwoldt et al., 2016, 2019; Doerr et al., 2017; Goubran et al., 2019; Stolp et al., 2018). However, currently there are no tools available that will allow fast and accurate mapping of iDISCO cleared LSFM recorded data to existing digital atlases.
Atlas coordinates are key essential tools in stereotaxic surgery, and they are widely used for procedures such as electrode implantation, injections of substances, or regional ablations. Today, several brain atlases for adult mice exist, including histology-based mouse brain atlases (Chen et al., 2019; Chon et al., 2019; Dong, 2008; Franklin & Paxinos, 1997; Hof et al., 2000; Jacobowitz & Abbott, 1997; Kaufman, 1992; Rosen et al., 2000; Sidman et al., 1971; Valverde, 2004; Wang et al., 2020a, b), MRI-based atlases (Aggarwal et al., 2009; Badea et al., 2007; Chan et al., 2007; Chuang et al., 2011; Chung et al., 2013; Dorr et al., 2008; Kovačević et al., 2005; Ma et al., 2005, 2008) and combined 2D histology and MRI-based atlases (Johnson et al., 2010; MacKenzie-Graham et al., 2004; Patel, 2018), but only three of these contain skull-derived stereotaxic coordinates (Aggarwal et al., 2009; Dong, 2008; Franklin & Paxinos, 1997). While histology-based atlases exhibit high-resolution structural information and detailed region delineations (Dong, 2008; Franklin & Paxinos, 1997), their structures cannot be translated directly into 3D in vivo space. Collection and processing of brain samples for histology may introduce anatomical deformations and lead to inaccuracies compared to in situ stereotaxic coordinates (Chan et al., 2007; Li et al., 2013). While current stereotaxic atlases are based on visually detected skull landmarks, computational detection of such landmarks has been demonstrated but not yet implemented to enhance the accuracy of stereotaxic coordinate systems (Blasiak et al., 2010).
This work aimed to develop a bidirectional multimodal atlas framework to bridge the gap between microscopic (LSFM) and macroscopic (MRI) imaging techniques. The purpose of this framework is to increase the registration throughput and versatility of LSFM imaged mouse brains. Firstly, by allowing direct translation of ex vivo LSFM results to in vivo MRI coordinates for comparison of digital brain maps derived from MRI and LSFM experiments (Perens & Hecksher-Sørensen, 2022). Secondly, the in-skull maps will allow automated stereotaxic surgery of brain regions identified using LSFM, for example c-Fos hotspots. Due to the immense size of LSFM recorded datasets and clearing-induced morphological changes it is both time consuming and imprecise to map LSFM directly to standard digital brain atlases (Perens et al., 2021). To accommodate overall throughput and accuracy of mapping we chose to keep each brain templates of different modalities in its original morphological space and then apply deformation fields to convert datasets between the templates. The multimodal atlas framework includes three mouse brain templates: an MRI template, a serial two-photon microscopy (STPT) template (Wang et al., 2020a, b) from Mouse Brain Common Coordinate Framework version 3 (CCFv3) by Allen Institute of Brain Science (AIBS), and an LSFM template (Perens et al., 2021). All maps have a isotropic voxel size of (25 μm)3. This voxel size was chosen because of the time it takes to register raw LSFM data to the reference brain. In our hands, mapping to a (25 μm)3 reference atlas takes approximately 10 min per brain while registration to a (10 μm)3 reference atlas takes roughly 3 h per brain. Finally, a stereotaxic coordinate system was developed based on semi-automatically detected skull landmarks obtained with micro-computed X-ray tomography (micro-CT) allowing precise identification of surgical locations. The AIBS CCFv3 was chosen to bridge the LSFM and MRI templates to provide access to its comprehensive resources such as region delineations, gene expression database, and tract-tracing experiments still ongoing. To enable browsing of brain anatomy in different template spaces together with region delineations and stereotaxic coordinates, an interactive web-based interface was developed and incorporated into NeuroPedia (https://www.neuropedia.dk).
Results
Concept of the Multimodal Atlas Framework
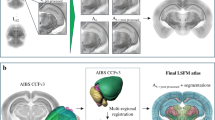
Typically, multimodal atlases combine brain templates of different imaging modalities in one common space. Here we used an alternative approach (Fig. 1), as LSFM- and STPT-imaged brains exhibit considerable morphological differences compared to in situ MR-imaged brains. The morphology of the imaged brains is dictated by sample preparation and handling during image acquisition – LSFM and STPT require that brain samples are extracted from skulls and processed either chemically or mechanically (i.e., sectioned) in contrast to MRI which can be performed on brains still located in the skull. Therefore, the current atlas framework comprises MRI-, STPT- and LSFM-based brain templates in their own respective morphological spaces describing the modality-specific average anatomy of a mouse brain. The MRI brain template was constructed from high-resolution T2-weighted images of brains imaged in the skull to mimic the in vivo setting as closely as possible. The STPT- and LSFM-based brain templates were adopted from the AIBS CCFv3 (Allen Institute for Brain Science, 2017; Wang et al., 2020a, b) and previously described iDISCO/LSFM-based atlas (Perens et al., 2021). All brain templates were resampled to an isotropic voxel size of (25 μm)3. To enable the transfer of datasets from one template space to another, applicable deformation fields were constructed from voxel displacements introduced when aligning the MRI and AIBS STPT-based templates as well as the AIBS STPT- and LSFM-based templates. The AIBS STPT-based template served as an intermediate between the in situ MRI and ex vivo LSFM spaces.
Framework for a multimodal mouse brain atlas with a stereotaxic coordinate system. Micro-CT- and MR-imaging of 12 mouse head volumes enabled the generation of a high-resolution T2-weighted in situ brain template with a coordinate system based on semi-automatically detected skull landmarks. To equip the different brain templates with the same atlas functionalities, the skull-derived coordinate system was transferred from the MRI template to the AIBS CCFv3 and LSFM templates, and region annotations from the AIBS CCFv3 to the MRI and LSFM templates (latter described in (Perens et al., 2021)). Conversion of datasets and atlas resources between the three template spaces can be performed by applying a mapping using either pre-computed deformation fields or transformation matrices. All brain templates have an isotropic voxel size of (25 μm)3 and are shown together in the figure with a micro-CT-imaged skull at the same scale.
In addition to the MRI-, STPT- and LSFM-based brain templates, the multimodal atlas framework includes detailed region delineations and stereotaxic coordinates in all template spaces. Additionally, several average diffusion tensor imaging (DTI) parameter maps obtained from diffusion-weighted MRI are likewise available in the MRI template space. Brain region delineations were adopted from the AIBS CCFv3 and transferred to the other template spaces via deformation fields (the LSFM template already included the AIBS CCFv3 region delineations as they were imported as described in (Perens et al., 2021)). Stereotaxic coordinates were generated by identifying standard reference landmarks in micro-CT-imaged skulls and creating a coordinate system related to average bregma and lambda positions in the MRI space. Finally, transformation matrices were applied to facilitate the transfer of the coordinate system from the MRI space to the AIBS STPT and LSFM spaces.
The Processing Pipeline for the Generation of the Multimodal Atlas Framework
The pipeline for setting up the multimodal atlas framework shown in Fig. 2 is divided into three main steps: data acquisition, sample level image processing, and template level image processing. For data acquisition, the main goal was to image the same brains with different image modalities. We chose to base the atlas on 10 weeks old male C57BL/6J mice. Following perfusion fixation, the whole head of each mouse was imaged using micro-CT, T2-weighted structural MRI, and diffusion-weighted MRI. The brains were then carefully dissected and processed according to the iDISCO + protocol and scanned using LSFM. Initial processing at the sample level included segmenting of total brain tissue and cranial bone from the T2-weighted MRI scans. Brain tissue segmentations were used as masks to remove the skull and superficial non-brain tissue from the T2-weighted MRI. Micro-CT-imaged skull volumes were transferred to the T2-weighted MR images obtained from the same mice by aligning them to segmentations of the cranial bone. Subsequently, exact locations of the reference landmarks, bregma and lambda, were identified from MRI-aligned micro-CT skull surfaces. In parallel, diffusion tensors were reconstructed from diffusion-weighted MRI and DTI parameter maps computed such as fractional anisotropy (FA), mean diffusivity (MD), radial diffusivity (RD), and axial diffusivity (RD). Individual LSFM-imaged brain volumes underwent pre-processing steps as described previously (Perens et al., 2021).
Computational pipeline for generating a multimodal mouse brain atlas. The computational pipeline describes the architecture and order of the image processing steps for integrating information from micro-CT, MRI, STPT-based AIBS CCFv3, and LSFM. Color-coding of the steps relates to the modality from which information has been extracted: purple denotes micro-CT, green T2-weighted MRI, light green diffusion-weighted MRI, orange LSFM, and blue STPT. Edges connecting the pipeline nodes describe the nature of processing steps: an arrow with a continuous line indicates the computation and application of a transformation matrix while an arrow with a dashed line stands for the application of an already computed transformation matrix, an arrow with several arrow-heads indicates region-wise mapping while arrow with single arrow-head stands for whole-brain mapping, and an edge with a circular tip connects to the next intermediate result in the computation pipeline achieved by other means than registration. The labels MM.IP.2 - MM.IP.8 refer to paragraphs in the Materials and Methods where the processing steps are described in detail. The figure numbers in brackets refer to the intermediate results of the processing pipeline shown in Figs. 3 and 4
Processing at the template level involved generation of a T2-weighted MRI brain template by iterative multi-resolution alignment and averaging algorithm (Kovačević et al., 2005; Kuan et al., 2015; Umadevi Venkataraju et al., 2019). A chain of transformation matrices computed as part of the iterative template creation process for aligning individual T2-weighted MR-images was applied to bregma and lambda of the same animals for transferring skull landmarks to an average T2-weighted MRI template. Subsequently, an average location was determined for the template-aligned bregma and lambda landmarks followed by the generation of the 3D coordinate system in the MRI space. DTI parameter maps of individual animals were transferred to an oriented T2-weighted MRI template and averaged. Then, the T2-weighted MRI-, AIBS STPT- and LSFM-based brain templates were linked to each other by 4D deformation fields resulting from the bi-directional alignment of the T2-weighted MRI- and AIBS STPT-based templates as well as the AIBS STPT- and LSFM-based templates. Finally, stereotaxic coordinates from the T2-weighted MRI template and region delineations from the AIBS CCFv3 template were transferred to all the templates by applying the mappings generated in the previous step.
Multimodal Imaging of a Mouse Brain Sample
The raw micro-CT, MRI, and LSFM image volumes of the same specimen are shown in Fig. 3a. The micro-CT-imaged skull volume was acquired with an isotropic voxel size of (22.6 μm)3, the T2-weighted MRI with (78 μm)3, the diffusion-weighted MRI with (125 μm)3, and the LSFM tissue autofluorescence with (4.8 μm)2 in-plane voxel size and 10 μm slice thickness. For the micro-CT images the coronal, sagittal, and lambdoidal sutures were visible on the skull surface. The T2-weighted MRI brain image showed the expected signal contrast between white matter, grey matter, and cerebrospinal fluid, and a high signal for the superficial tissue. No signal was visible in the skull area due to its short T2-relaxation time. Also, diffusion-weighted MRI exhibited the expected orientation-dependent signal decays in areas with restricted anisotropic microstructure e.g., in axon bundles of white matter. The diffusion signal was completely decayed for superficial tissue and cerebrospinal fluid because of high diffusivity. LSFM volumes of the isolated brains were scanned using an autofluorescence channel which displayed high contrast between different tissue types, despite keeping the perfusion-fixed brain sample in phosphate-buffered saline for 2–4 weeks while performing micro-CT and MRI scanning. Combining a micro-CT skull dataset with the T2-weighted MRI brain from the same mouse demonstrates how the brain fills tightly the inner volume of the skull (Fig. 3b). This suggests that the fixation process has introduced minimal shrinkage and we can expect the anatomy to be positioned as in vivo.
Semi-automatic extraction of bregma and lambda from micro-CT skull volumes. (a) Raw images of the skull and brain from the same mouse acquired sequentially via micro-CT with an isotropic voxel size of (22.6 μm)3, T2-weighted MRI with an isotropic voxel size of (78 μm)3, diffusion-weighted MRI with an isotropic voxel size of (125 μm)3 (shown for one gradient direction from 60) and LSFM with (4.8 μm)2 in-plane voxel size and 10 μm slice thickness. Both T2- and diffusion-weighted MRI were acquired from a brain in the skull. Brains were dissected from skulls for iDISCO + treatment and clearing before performing LSFM. (b) A rigidly aligned micro-CT-imaged skull (purple) to the T2-weighted MRI brain image (grayscale) of the same mouse. (c) Extraction and fitting of coronal (red), sagittal (blue), and lambdoidal (green) sutures for determining x-y-coordinates for bregma (∆) and lambda (ο). Sutures were extracted from maximum projection images of skull surfaces by manually drawn suture masks while bregma and lambda were found by identifying intersection points of the individual suture fits. (d) Determination of the z-coordinate for bregma (∆) and lambda (ο) on top of the skull
Establishing Precise Origin and Orientation for a Stereotaxic Atlas
Stereotaxic coordinate systems rely on anatomical landmarks on the skull surface which have a fixed geometric relation to underlying brain structures and have uniform locations across individuals. Typically, stereotaxic coordinate systems use cranial landmarks bregma and lambda to establish a standard orientation of the brain. Bregma is located at the intersection of the coronal suture with the cranial midline and lambda at the intersection of the lambdoidal suture with the cranial midline (Blasiak et al., 2010; Franklin & Paxinos, 1997). We applied a semi-automatic procedure to extract bregma and lambda landmarks from 12 micro-CT- imaged mouse skulls and generate the stereotaxic orientation (Fig. 3c-d).
Skull sutures made of cartilage appear as grooves between the cranial plates. Due to their different depth compared to the skull surface, the sutures can be identified from a depth image generated on the dorsal-ventral axis. Then, the skull is extracted from the depth image and the top part of the skull surface is projected onto the 2D plane by saving the voxels with the highest intensity on the dorsal-ventral axis. The skull is labeled in grey in the projected image while the sutures appear as white structures. The individual sutures were manually segmented (Fig. 3c, left) and their curves fitted (Fig. 3c, middle). Subsequently, coronal and lambdoidal suture junctions at the sagittal suture were computed to obtain xy-coordinates of the bregma and lambda (Fig. 3c, right). z-coordinates of the landmarks were determined by locating the outer edge of the skull surface at the xy-coordinate in the dorsal direction (Fig. 3d).
To generate a stereotaxic coordinate system, skull landmarks were aligned with an anatomical brain template. For this purpose, the T2-weighted MRI brain template created from 12 mouse brain images was used (Fig. 4a). Before transferring individual bregma and lambda coordinates into the template space, the visibility of anatomical structures in the T2-weighted MRI template was enhanced by gamma correction. Additionally, the hemisphere with the highest quality was mirrored to the opposite side for creating a symmetric template when viewed from coronal and horizontal orientations. Individual bregma and lambda coordinates were then averaged in the template space. To align the average bregma and lambda landmarks to the same horizontal level, the T2-weighted MRI template was rotated accordingly (Fig. 4b). The final template-space coordinates for the average bregma were x = 227.00 ± 4.73 voxels, y = 270.00 ± 5.80 voxels, z = 16.00 ± 1.85 voxels and for the average lambda were x = 227.00 ± 3.32 voxels, y = 462.00 ± 1.87 voxels, z = 16.00 ± 1.00 voxels (Fig. 4b-c). The average distance measured between bregma and lambda landmarks of individual animals was found to be Δ(bregma, lambda) = 192 ± 5.94 voxels corresponding to Δ(bregma, lambda) = 4.80 ± 0.15 mm.
Skull-derived coordinate system in MRI, AIBS CCFv3, and LSFM spaces. (a) The MRI template was created from 12 in-skull-imaged T2-weighted MR images using an iterative registration and averaging algorithm. (b) Position of the average bregma (∆) and lambda (ο) points in the MRI template space visualized in sagittal and 3D top view. The MRI template was oriented such that the average bregma and lambda points are on the same z-level (shown by the cyan dashed line). (c) Variation in bregma and lambda positions of individual skulls shown as a standard deviation from the average bregma and lambda in x-, y-, and z-dimensions. (d) Coronal slices of averaged diffusion tensor-derived parameters created from 7 in-skull-imaged diffusion-weighted MR images: fractional anisotropy (FA), b0, mean diffusivity (MD), radial diffusivity (RD), and axial diffusivity (AD). The FA values are in range [0,1] while the values of MD, AD, and RD are in range [0,0.0006] mm2/s. (e) A coordinate system was created in the bregma-lambda oriented MRI template space with an isotropic coordinate spacing of (25 μm)3 and transferred to the AIBS CCFv3 and bregma-lambda oriented LSFM template spaces by applying transformation matrices from the whole-brain mapping between the MRI template and AIBS CCFv3 and region-wise mapping between the AIBS CCFv3 and LSFM template. The coordinate system is visualized in horizontal view for x-coordinates, coronal view for z-coordinates, and sagittal view for y-coordinates. The color scale indicates coordinate values for every voxel and equidistant (step size 250 μm from origin) contour lines (black) indicate levels at which coordinate values are constant
Skull-derived Stereotaxic Coordinate System for MRI, AIBS CCFv3, and LSFM Atlases
A stereotaxic coordinate system was generated in the oriented T2-weighted MRI template space originating from the average bregma position. The coordinate system was implemented with an isotropic step size of 0.025 mm (Fig. 4e, left column) according to the coordinate convention used by Paxinos and Franklin in The Mouse Brain in Stereotaxic Coordinates (Franklin & Paxinos, 1997). The coordinate convention defines that the x-axis corresponds to the medial-lateral axis, the y-axis to the anterior-posterior axis, and the z-axis to the dorsal-ventral axis. Additionally, the convention implicates the x-coordinates are positive for both hemispheres, y-coordinates are positive anterior to the origin and negative posterior to the origin, and z-coordinates are positive ventral to the origin and negative dorsal to the origin. The resulting coordinate system was transferred to the AIBS STPT- and LSFM-based templates via pre-computed transformation matrices (Fig. 4e, middle and right columns). Both the AIBS STPT- and LSFM-based templates were reoriented to align the y-coordinate values for structures at the same coronal planes. Also, a volume with the original orientation of the AIBS CCFv3 with skull-derived coordinates was kept in the atlas framework. Conversion of the coordinate system to the AIBS STPT- and LSFM-based templates caused the coordinate system to deform when following the same anatomical structures as in the T2-weighted MRI template. Since this deformation of the coordinate system reflects the changes incurred during tissue processing it is barely visible in the AIBS space while the coordinates in the LSFM space exhibited non-equidistant spacing and extensive deformation (Fig. 4e).
Integrating Information Between the Multimodal Atlas Spaces
Conversion of the skull-derived stereotaxic coordinate system to the AIBS CCFv3 and LSFM space was performed using transformation matrices resulting from mapping the MRI template to the AIBS CCFv3 template and the AIBS CCFv3 template to the LSFM template. Identical anatomical structures were found in the proximity of the landmarks in the MRI, AIBS STPT- and LSFM-based templates indicating correspondence of the skull-derived stereotaxic coordinate system in all three template spaces (Fig. 5a).
Correspondence of the coordinate system and accuracy of deformation field mapping between the MRI, AIBS CCFv3, and LSFM spaces. (a) Two anatomical landmarks, one in the dentate gyrus (two upper panels) and one in the parabrachial nucleus (two lower panels), are shown in the MRI, AIBS CCFv3, and LSFM template spaces in coronal and sagittal view. Purple crosshair indicates the spatial location of the same x-, y-, and z-coordinate in all three template spaces. (b) Checkerboard representation of a randomly picked MRI- (upper panel) and LSFM-imaged (lower panel) brain sample registered to the MRI, AIBS CCFv3, and LSFM template spaces via constructed deformation fields. The mapped MRI- and LSFM-imaged samples are visualized in grayscale while the MRI, AIBS CCFv3, and LSFM templates are depicted in green, blue, and orange color scales, respectively
Conversion of volumetric data between the MRI, AIBS CCFv3, and LSFM spaces is enabled via deformation fields provided together with the multimodal atlas. Deformation fields are 4D matrices describing the 3-dimensional movement of every voxel in a transferable data volume which can be applied to image volumes using software for biomedical image registration (e.g., Elastix). For demonstrating mapping accuracy between the MRI-, AIBS STPT-, and LSFM-based templates, a random LSFM and a random T2-weighted MRI brain from the set of 12 brains imaged with CT, MRI, and LSFM, were aligned to all three brain templates via deformation fields (Fig. 5b). Visual inspection shows matching structures of every brain template with the overlayed sample volume which suggests high accuracy when transferring volumetric data between the brain templates of the multimodal atlas.
Interactive Platform for Accessing the Multimodal Atlas Framework
Resources of the multimodal atlas framework can be explored and downloaded via NeuroPedia (https://www.neuropedia.dk). The web interface allows users to browse region delineations and stereotaxic coordinates simultaneously in MRI, AIBS CCFv3, and LSFM template spaces. Furthermore, the interactive interface provides users with the opportunity to look up the region and coordinate of a voxel by clicking on a template as well as search for a certain region or a coordinate of interest in all three template spaces.
Discussion
Here we present a multimodal atlas framework that bridges several whole-brain modalities and incorporates a 3D stereotaxic coordinate system derived from CT-imaged mouse skulls. In contrast to previously reported multimodal atlases, which combine brain templates from different types of imaging modalities and region delineation overlay in one reference space (MacKenzie-Graham et al., 2004; Nie et al., 2019; Purger et al., 2009), the multimodal atlas framework preserves the templates in their modality-specific spaces together with region delineations and stereotaxic coordinates. Deformation fields make it possible to transfer datasets recorded with different imaging modalities to various template spaces for integration. The approach adopted in previously published multimodal atlases works well for combining modalities relying on similar sample processing protocols and result in brain volumes with comparable brain morphology. These brain volumes do not require extensive deforming upon alignment and can therefore be directly mapped to a common reference space. However, some tissue processing protocols for LSFM-imaging introduce morphological changes to the tissue due to certain clearing media (Perens et al., 2021; Qi et al., 2019; Renier et al., 2016). Given the different brain morphologies, the combination of MRI and LSFM brain templates in the same morphological space would result in error-prone or laborious region-wise registration for aligning new whole-brain datasets with respective templates. These problems can be circumvented by using modality-specific templates in the multimodal atlas, as implemented in the current work, since they allow fast and accurate alignment of new datasets with their respective templates (Perens et al., 2021).
Skull-derived Stereotaxic Coordinate System for Accurate Signal Localization and Stereotaxic Interventions
We used standard skull landmarks bregma and lambda to determine the angle and origin of the stereotaxic coordinate system for the multimodal atlas framework. In the past, few alternative landmarks have been proposed for navigating murine brains. Chan and colleagues (Chan et al., 2007) suggested a new landmark pair consisting of the lambda suture junction and the rostral confluence of the venous sinus (RCS) anterior to bregma, as they showed not only less variability between specimens but also between mouse strains compared to using paired bregma and lambda. The intersection point of the posterior edges of the cerebral hemispheres as an origin of a stereotaxic coordinate system for murine brains has also been proposed (Xiao, 2007). As both landmark systems are not visually detectable from the dorsal surface of the skull, they can only be visualized in an intersectional view of the imaged skull or brain and require exposure of the brain or image guidance during surgery. Stereotaxic surgery in rodent brains is rarely performed in combination with CT or MRI, but emerging camera-guided robotic systems for intra-cranial surgeries (Ly et al., 2021) and neural networks (Zhou et al., 2020) can help to identify less variable cranial landmarks and potentially provide additional anatomical reference points.
We detected the bregma and lambda landmarks from CT-imaged skull surfaces by identifying intersection points of fitted cranial sutures. Similar to Chan and colleagues (Chan et al., 2007), we observed higher variance when identifying bregma coordinates compared to lambda coordinates in all three dimensions, which is in agreement with more variable shape and sharper angle between coronal sutures compared to lambdoidal sutures. The highest standard deviation from the mean coordinates was found in y-dimension for both landmarks and reached maximally 145 μm. This could be attributed to the initial manual orientation of individual skulls into a flat-surface position for landmark detection, resulting in a slightly different perspective of suture lines in the maximum-projection image of the skull surface. Identification of bregma and lambda coordinates from individual animals allows the mean distance between reference landmarks to be computed. Consistently approx. 0.6 mm higher values for bregma-lambda distance were observed compared to the reported values (Aggarwal et al., 2009; Franklin & Paxinos, 1997; Zhou et al., 2020). This is probably related to the parabolic fitting of coronal sutures in this work, causing bregma to move slightly anterior from the true intersection point of the coronal and sagittal sutures.
Differences Between Brain Templates of Available Stereotaxic Atlases
The skull-derived coordinate system generated for the multimodal atlas framework allows comparing the size of the in situ-imaged T2-weighted MRI template to the size of other existing stereotaxic atlas templates (Table 1). A comparison of brain templates from different standard atlases reveals that the width and depth of the Franklin and Paxinos atlas template is at least 1 mm smaller than that of the multimodal atlas MRI template. Furthermore, the length of the AIBS CCFv1 and Franklin and Paxinos atlas template is 0.7 mm larger while the depth of Aggarwal’s template was found to be 1 mm smaller compared to the respective dimensions of the multimodal atlas MRI template. Size discrepancies between the atlas templates can be related to age and biological variance between individual brains since the T2-weighted template is based on population-averaged brain volume, whereas templates of the other stereotaxic atlases rely on a single specimen. The possible reason for the considerably smaller width and depth of the Franklin and Paxinos atlas template is due to fixation-related shrinkage of skull-extracted tissue (Lee et al., 2021). The increased length of the Franklin and Paxinos atlas and the AIBS CCFv1 template could be caused by variations in microtome section thickness of a few-µm range accumulating for hundreds of collected sections.
Limitations of the Multimodal Atlas Framework
The multimodal atlas described in this study was generated with a 25 μm isotropic voxel size for transferring the coordinate system from the MRI to the LSFM template via the AIBS CCFv3 space. Re-sampling of the atlas to other voxel sizes can be accomplished by creating all templates with the new voxel size, calculating mapping fields between the templates, and aligning a newly generated coordinate system with updated grid-spacing with other templates. Assigning coordinate values at ventricle borders posed a challenge in bridging the different imaging modalities due to the observed enlargement of ventricles in cleared brains. To overcome this, we used interpolation of coordinates from neighbouring regions. This can potentially result in minor inaccuracies, as one ventricular voxel in the MRI space can be mapped to several voxels in the LSFM space.
Conclusion
Here we report a multimodal adult mouse brain atlas framework that allows convenient translation between the LSFM (iDISCO + processed and cleared), AIBS CCFv3, and in situ MRI brain templates using deformation fields. The purpose of these deformation fields is to facilitate the integration of in vivo and ex vivo whole-brain datasets via co-registration which is essential for cross-modality image analysis. Importantly, the multimodal atlas provides a stereotaxic coordinate system in all template spaces reflecting in vivo in-skull coordinates.
The coordinates allow identification of the exact spatial location of neuronal populations in vivo for performing accurate stereotaxic targeting for e.g., brain site-specific drug delivery, viral tract-tracing in connectome studies, or electrode implantation. Compatibility of the LSFM space with the MRI and AIBS CCFv3 space permits large-scale integration of LSFM-imaged neuronal populations with AIBS gene expression and connectivity atlases. The applicability extends also to other open-source MRI, STPT, and LSFM datasets available at Neuroimaging Tools & Resources Collaboratory (NITRC) repository, OpenNeuro platform by Stanford Center for Reproducible Neuroscience, and EBRAINS data repository. The multimodal atlas framework can be extended with additional brain templates (e.g., based on various MRI contrasts, clearing methods, imaging modalities), region delineation volumes (e.g., (Chon et al., 2019)), coordinate systems based on a different origin (e.g., (Chan et al., 2007; Xiao, 2007)), anatomical maps (e.g., vascular trees (di Giovanna et al., 2018; Todorov et al., 2020)), and structural connectivity (Friedmann et al., 2020). An interactive browser of the multimodal templates is available at NeuroPedia (https://www.neuropedia.dk), where all atlas files and deformation fields can also be downloaded.
Materials and Methods
Animals and Sample Preparation
Male C57Bl/6J mice (n = 12) were obtained from Janvier Labs (Le Genest-Saint-Isle, France) and housed in a controlled environment (12 h light/dark cycle, 21 ± 2˚C humidity 50 ± 10%) with ad libitum access to tap water and chow (Altromin 1324, Hørsholm, Denmark). 10-week-old mice were sacrificed via transcardial perfusion of heparinized phosphate-buffered saline (PBS) and 40 ml of 10% neutral buffered formalin (CellPath, Newtown, UK) while under 2–4% isoflurane/O2 (Attane Vet., ScanVet Animal Health, Fredensborg, Denmark) anesthesia. Mouse skulls were removed and cleaned from superficial tissue, then post-fixed in 10% neutral buffered formalin for four days at 4˚C and washed to remove excess fixative in PBS/NaN3 for 2–4 weeks until micro-CT and MRI (Dyrby et al., 2011, 2018). After micro-CT and MR-imaging, brains were carefully dissected from skulls and processed according to the iDISCO+ (immunolabeling-enabled three-dimensional imaging of solvent-cleared organs) protocol (Renier et al., 2014) as described in (Perens et al., 2021) using reagents from the same vendors. In contrast to the original iDISCO + protocol, no antibodies were included in the staining buffers.
Data Acquisition
Micro-computed Tomography (micro-CT)
For obtaining bregma and lambda locations from the skull surface, 3D mouse skull volumes were imaged using high-resolution micro-CT. Bregma and lambda are visually detectable landmarks on the skull, which are conventionally used to navigate in the brain of a living mouse, for example during stereotaxic surgeries. Image volumes were acquired with the ZEISS XRadia Versa XRM-410 scanner at the 3D Imaging Centre, at the Technical University of Denmark, by collecting 1601 projections of the skull with an exposure time of 2 s per projection and tube voltage set to 50 kVp. The resulting skull volumes exhibited an isotropic voxel size of (22.6 μm)3.
Magnetic Resonance Imaging (MRI)
In situ MRI mouse brain scanning was done at the Danish Research Centre for Magnetic Resonance using a 7.0 T Bruker Biospec preclinical MRI system equipped with a maximum strength of 660 mT/m (Dyrby et al., 2011, 2018). Transmit/receive used a dual cryogenic radiofrequency surface coil optimized for mouse brain MRI (CryoProbe, Bruker Biospin MRI GmbH, Ettlingen, Germany). The imaging protocol included a 3 h-long high-resolution structural T2-weighted MRI and a subsequent 13 h-long diffusion-weighted scan. For acquiring the T2-weighted MRI, a True 3D FISP sequence (i.e., gradient balanced steady-state coherent sequence along three axes) was used with the following settings: flip angle = 30˚, TE = 2.5 ms, TR = 5.1 ms, number of repetitions = 1, number of averages = 60, bandwidth = 125 kHz, image size = 256 × 256 × 128 pixels, field of view = 20 mm × 20 mm × 10 mm, and isotropic voxel size of (78 μm)3. For acquiring the diffusion-weighted MRI, a spin echo sequence with single-line readout was used with the following settings: flip angle = 90˚, TE = 26 ms, TR = 5700 ms, number of repetitions = 1, number of averages = 1, bandwidth = 20 kHz, matrix size = 128 × 128, field of view = 16 mm × 16 mm, number of slices = 55, slice thickness = 0.125 μm, isotropic voxel size of (125 μm)3, gradient strength = 456 mT/m, gradient duration = 5 ms, gradient separation = 13 ms, encoding duration = 0.8 ms, and number of directions = 60. A b-value of 4000 s/mm2 adjusted to ex vivo tissue with decreased diffusivity was used as compared with in vivo.
Light-sheet Fluorescence Microscopy (LSFM)
Skull-dissected and cleared brain samples were imaged in dibenzyl ether in axial orientation on a LaVision ultramicroscope II (Miltenyi Biotec, Bergisch Gladbach, Germany) equipped with a Zyla 4.2P-CL10 sCMOS camera (Andor Technology, Belfast, UK), SuperK EXTREME supercontinuum white-light laser EXR-15 (NKT Photonics, Birkerød, Denmark), and MV PLAPO 2XC (Olympus, Tokyo, Japan) objective lens. Version 7 of the Imspector microscope controller software was used. Images from tissue the structure were acquired at an excitation wavelength of 560 nm ± 20 nm and emission wavelength of 620 nm ± 30 nm with 80% laser power, 1.2X total magnification, 257 ms exposure time, 9 horizontal focusing steps, and blend-blend mode in a z-stack at 10 μm intervals. Resulting brain volumes (16 bit-tiff) had a (4.8 μm)2 in-plane and 3.8 μm axial voxel size (NA = 0.156).
Brain Atlases Bridged in the Current Work
Mouse Common Coordinate Framework by Allen Institute of Brain Science (AIBS CCF)
The latest version of the AIBS CCF, CCF version 3 (CCFv3) released in 2017 includes a 3D template brain based on tissue autofluorescence volumes and an annotation volume with 662 region delineations (Allen Institute for Brain Science, 2017; Wang et al., 2020a, b). Raw data of the template stems from 1675 specimens collected with a serial two-photon tomography (STPT) in the red channel (excitation at 925 nm) in coronal 2D sections with an in-plane voxel size of (0.35 μm)2 at every 100 μm through the anterior-posterior axis. The CCFv3 is accessible with isotropic voxel sizes of (10 μm)3 and (25 μm)3 which could be realized in the anterior-posterior dimension due to slight offsets in positions of vibratome-cut sections for each brain. Region annotations provided by the CCFv3 are manually drawn delineations in 3D space based on features from structural, transgenic, tracing, cytoarchitectonic, chemoarchitectonic, and in situ hybridization datasets.
Light-sheet Fluorescence Microscopy (LSFM) Based Mouse Brain Atlas
The LSFM-based atlas was made publicly available in 2020 and includes a 3D template brain based on tissue autofluorescence volumes of iDISCO + processed brains and an annotation volume transferred region-wise from the AIBS CCFv3 (Perens et al., 2021). An anatomical template of the LSFM atlas was created from 139 brain volumes acquired in the red channel (excitation at 560 nm ± 20 nm, emission at 650 nm ± 25 nm) by optically sectioning samples in the axial orientation with (4.8 μm)2 in-plane voxel size, 3.8 μm slice thickness, and a 10 μm distance between the slices. The final voxel size of the LSFM atlas is (20 μm)3 in all three dimensions. The atlas is fully dedicated for mapping cleared and LSFM-imaged brain samples as the chemicals used in the iDISCO + protocol cause brain samples to deform resulting in a different morphology than the AIBS CCFv3 template.
Image Processing
Computational Tools (MM.IP.1)
Most of the data processing was performed in Python 3.7 except for the extraction of brain tissue from T2-weighted MRI scans which was performed in MATLAB R2020a. All the scripts used for data processing were custom-made and based on publicly available packages such as Numpy (Harris et al., 2020), Scikit-image (van der Walt et al., 2014), SciPy (Virtanen et al., 2020), and SimpleITK (Beare et al., 2018; Lowekamp et al., 2013; Yaniv et al., 2018) for Python, and NIfTI (Shen, 2021) for MATLAB. The Elastix toolbox 4.9 (Klein et al., 2010; Shamonin et al., 2014) was deployed to implement registrations and transformations.
Generation of Brain and Skull Masks (MM.IP.2)
As the MR images were acquired from mouse brains in the skull, skull stripping was performed before co-registration and averaging of the brain samples for generating a T2-weighted template brain. For extracting the brain from the surrounding tissue, a T2-weighted structural image was binarized such that all voxels belonging to the brain tissue were given the value 1 and all voxels belonging to the background the value 0. As several voxels in the tissue around the skull showed intensity values in the same range as the voxels in the brain tissue, the binarized image underwent morphological opening and erosion with a disk-formed structuring element (radius = 2). Subsequently, the biggest connected component was found from the image and dilated with the same structuring element as used in previous morphological operations. Finally, left-over holes in the brain mask were filled and manual corrections were made in the hindbrain area where the signal intensity of the original image was the lowest. ITK-SNAP 3.8 (Yushkevich et al., 2006) was used for performing manual corrections to individual tissue masks.
For mitigating co-registration of micro-CT-imaged skull volumes to MR images of the same individuals, a coarse skull mask was generated for individual T2-weighted images. First, the T2-weighted image was binarized at the threshold found by the Otsu’s method. Then, the brain mask of the same T2-weighted image was dilated using a cubic structuring element until the mask reached the outer edge of the skull. Finally, voxels of the binarized T2-weighted image which have positive intensity values outside of the skull surface are set to zero using the dilated brain mask.
Registration at the Sample Level (MM.IP.3)
The registration procedures at the sample level were initialized by the sampling of micro-CT skull volumes, MR images, and tissue masks to (25 μm)3 voxel size followed by multi-resolution rigid alignment of MR images to the AIBS CCFv3 template for orienting every sample volume to the standard orientation. Subsequently, micro-CT skull volumes were registered to the corresponding MR images by multi-resolution rigid registration via skull masks extracted from T2-weighted images. A multi-resolution registration strategy for mapping micro-CT skulls was realized by blurring fixed and moving image volumes with smoothing kernels of decreasing size before performing registration at every resolution level. Both rigid registration procedures were performed by maximizing normalized correlation for fixed and moving image pairs and using the stochastic gradient descent as an optimization method.
The T2-weighted MRI mouse brain template was generated by applying the same registration procedure used for creating the LSFM mouse brain template (Perens et al., 2021) inspired by (Chan et al., 2007; Kuan et al., 2015; Umadevi Venkataraju et al., 2019). The algorithm involved one multi-resolution affine and five uni-resolution B-spline transformation steps at increasing resolutions. An increase in resolution was realized by decreasing the size of the smoothing kernel, down-sampling, and spacing of control points of the deformation grid. After every registration step, resulting datasets were intensity averaged to generate an intermediate average brain which served as a reference brain volume in the following registration step. All registration steps used to create the T2-weighted MRI mouse brain template deployed mattes mutual information as a similarity metric and gradient descent as an optimization method. For B-Spline registrations, the following optimization parameters were specified: gain factor a = 10,000, α = 0.6, A = 100. For realizing symmetry between the hemispheres of the resulting average T2-weighted MRI brain, a final very coarse multi-resolution B-spline registration to the AIBS CCFv3 was performed using the same similarity metric, optimization method, and parameters as for the previous registrations except that registration was only performed at 2 lower resolutions with a = 5000.
Registration at the Template Level (MM.IP.4)
Registration procedures at the template level involved computing transformation matrices to enable mapping between the T2-weighted MRI and AIBS CCFv3 templates and between the AIBS CCFv3 and LSFM templates (all templates with an isotropic voxel size of (25 μm)3). While mapping between the T2-weighted MRI and AIBS CCFv3 templates was performed in a whole-brain manner, alignment of the AIBS CCFv3 and LSFM templates required the region-wise approach as shown in (Perens et al., 2021). The first step of the region-wise approach was to register the AIBS CCFv3 template and segmentation volume to the LSFM template in the whole-brain manner. Then the AIBS CCFv3 and the LSFM templates were divided into six parental brain regions (cerebral cortex, cerebral nuclei, interbrain, midbrain, hindbrain, and cerebellum) using the segmentation volume and region hierarchy of the AIBS CCFv3. Mapping of the AIBS CCFv3 template to the LSFM template was finally conducted by registering the six parental regions of the AIBS CCFv3 separately to the corresponding regions of the LSFM template. While the previous work focused only on the registration of the AIBS CCFv3 template to the LSFM template, in the current work registration was also conducted in the opposing direction. All between-template registrations included multi-resolution affine and B-spline transformations with the mattes mutual information similarity metric and the gradient descent optimization method. For B-spline registrations, the following optimization parameters were specified: a = 10,000 (in case of hindbrain a = 40,000 and septum a = 50,000), α = 0.6, A = 100. The multi-resolution strategy was realized by decreasing the size of the smoothing kernel, down-sampling, and spacing of the control points of the deformation grid.
Detection of Skull Landmarks (MM.IP.5)
Skull landmarks bregma and lambda were determined from every micro-CT skull volume by a semi-automatic computational algorithm. First, a subvolume constituting only the dorsal half of the skull with sutures was sampled from a micro-CT skull volume aligned to the corresponding T2-weighted MR image (described in Registration at the Sample Level). Then an intensity threshold (Iglobal = 1) was found for distinguishing the skull from the background, a depth image was computed by summing up all voxels belonging to the background along the z-axis (dorsal-ventral axis), and the depth image was smoothed with a Gaussian kernel (σdepth = 3). Subsequently, the maximum projection of the extracted skull surface along the z-axis was calculated, slightly smoothed with a Gaussian kernel (σprojection = 0.5), and thresholded (Isuture = 2.5) for visualizing the sutures clearly. Suture curves were extracted from the final maximum projection image by applying the coronal, sagittal, and lambdoidal suture masks which were manually drawn in ITK-SNAP 3.8 (Yushkevich et al., 2006). The mask for the lambdoidal suture only included straight horizontally oriented grooves and excluded the triangular part of the suture. While the coronal suture was fitted by quadratic least squares regression, both sagittal and lambdoidal sutures were fitted by linear least squares regression. In-plane (x and y) coordinates for the bregma were identified by calculating the intersection of coronal and sagittal suture fits and for the lambda by calculating the intersection of sagittal and lambdoidal suture fits. For determining the z-coordinate of the bregma and lambda, which is by definition located on top of the skull, a small subvolume (5–10 voxels x 10 voxels x number of voxels on the z-axis) was extracted from the whole micro-CT skull volume. Subsequently, intensity values on the x- and y-axis were averaged resulting in an intensity profile of the skull on the z-axis in close vicinity to the xy-locations of bregma and lambda. z-coordinate was finally found by identifying the outer boundary between the skull surface and background from the intensity profile.
Generating and Mapping a Stereotaxic Coordinate System (MM.IP.6)
The chain of transformation matrices computed for the individual T2-weighted MR images as described in Registration at the Sample Level were applied to the bregma and lambda landmarks extracted from the MRI-aligned micro-CT skulls of the corresponding animals for transforming the landmarks to the T2-weighted MRI template space. Positions of the landmarks for the individual animals were averaged and the final T2-weighted MRI template together with the average landmarks was rotated such that the bregma and lambda were aligned horizontally (i.e., positioned at the same z-level). A coordinate system could then be generated by using the average bregma position as an origin for all three dimensions and a step size equal to voxel size (step size = 0.025 mm). A coordinate volume was generated with the same matrix shape as the horizontally aligned T2-weighted MRI template and every element in the coordinate volume was assigned a vector containing x-, y- and z-coordinate. The coordinate values assigned to the elements in the coordinate volume describe the distance of the voxel edge nearest to the origin from the origin in millimeters (e.g. when the x-coordinate of the voxel overlapping with bregma is 0, then the x-coordinate of the neighboring voxel in the lateral direction is 0.0125 (mm) and the next neighboring voxel is 0.0125 (mm) + 0.0250 (mm) = 0.0375 (mm)).
The transformation matrix resulting from the mapping of the T2-weighted MRI template to the AIBS CCFv3 (described in Registration at the Template Level) was applied to transfer the average bregma- and lambda-derived coordinate system from the MRI template space to the AIBS CCFv3 space. Followingly, six transformation matrices resulting from mapping the AIBS CCFv3 region-wise to the LSFM template (described in Registration at the Template Level) were applied to the coordinate system parcellated into cortical, cerebral nuclei, interbrain + midbrain, hindbrain, cerebellar, and septal subvolumes to transfer the coordinate system from the AIBS CCFv3 space to the LSFM template space. For reconstructing the whole coordinate system in the LSFM template space, subvolumes were merged into one volume. While original coordinates were kept in the non-overlapping areas, coordinates in the overlapping and gap areas needed to be interpolated. Interpolation was performed in 2D on planes showing gradient in coordinate values (x- and z-coordinates in coronal planes, y-coordinates in the axial planes). Post-processing of the coordinate system involved assigning reasonable coordinate values to left-over voxels based on the coordinate values of neighboring voxels.
Generation of Deformation Fields (MM.IP.7)
Deformation fields were generated for providing a possibility to transform datasets fast and accurately between the T2-weighted MRI, AIBS CCFv3, and LSFM template spaces in all directions. The Transformix program (Klein et al., 2010; Shamonin et al., 2014) which comes as part of the Elastix toolbox 4.9 was used to generate the deformation fields and can also be utilized for applying them. As the mapping between the T2-weighted MRI template and the AIBS CCFv3 was performed in the whole-brain manner (described in Registration at the Template Level), deformation fields provided by Transformix did not require further processing. However, as the mapping between the AIBS CCFv3 and LSFM templates was performed in the region-wise manner (described in Registration at the Template Level), Transformix created six deformation field volumes, one for mapping every region. These six region-specific deformation fields needed to be combined into one volume for facilitating the transfer of whole-brain datasets between the AIBS CCFv3 and LSFM templates. For reconstructing the deformation field, vector fields of every region were extracted from the corresponding deformation field volumes via masking and then merged into one volume. While the original vector elements were kept in the non-overlapping areas, vector elements in the overlapping and gap areas were interpolated in 2D planes (x and z vector elements in the coronal planes, y vector elements in axial planes). Post-processing of the deformation field involved setting y and z vector elements outside the brain tissue to a very high value for avoiding ghost images in the mapped volumes. One of the generated deformation fields was applied for transferring region delineations from the AIBS CCFv3 to the MRI template.
Processing of Diffusion-weighted MR Images (MM.IP.8)
Diffusion-weighted MRI datasets were successfully acquired for 7 animals. Pre-processing of diffusion-weighted MRI involved coarse rotation of the volumes and gradient directions to the orientation of the T2-weighted MRI template, denoising (Veraart et al., 2016), and removal of Gibbs ringing (Kellner et al., 2016). DTI parameter maps were calculated with the MRtrix toolbox 3.0 (Tournier et al., 2019). Diffusion tensors were estimated from pre-processed diffusion-weighted MR images for every voxel in the brain according to the standard MRtrix methodology and settings. Diffusion metrics, such as fractional anisotropy (FA), mean diffusivity (MD), axial diffusivity (AD), and radial diffusivity (RD), were derived from diffusion tensors (Basser et al., 1994). Additionally, b0-volumes (n = 5) collected without diffusion-sensitizing gradients were averaged. Diffusion tensor datasets sampled to a voxel size of (25 μm)3 were registered to the T2-weighted MRI template by mapping individual b0-volumes to the template using multi-resolution affine and high-order B-spline registration and applying computed transformation matrices to the other diffusion parameter maps. Finally, the maps were averaged in the T2-weighted MRI template.
References
Aggarwal, M., Zhang, J., Miller, M. I., Sidman, R. L., & Mori, S. (2009). Magnetic resonance imaging and micro-computed tomography combined atlas of developing and adult mouse brains for stereotaxic surgery. Neuroscience, 162(4), 1339–1350. https://doi.org/10.1016/j.neuroscience.2009.05.070.
Allen Institute for Brain Science. (2017). Allen Mouse Common coordinate Framework and Reference Atlas. Technical White Paper.
Badea, A., Ali-Sharief, A. A., & Johnson, G. A. (2007). Morphometric analysis of the C57BL/6J mouse brain. Neuroimage, 37(3), 683–693. https://doi.org/10.1016/j.neuroimage.2007.05.046.
Badea, A., Ng, K. L., Anderson, R. J., Zhang, J., Miller, M. I., & O’Brien, R. J. (2019). Magnetic resonance imaging of mouse brain networks plasticity following motor learning. Plos One, 14(5), e0216596. https://doi.org/10.1371/JOURNAL.PONE.0216596.
Basser, P. J., Mattiello, J., & Lebihan, D. (1994). Estimation of the effective self-diffusion Tensor from the NMR spin Echo. Journal of Magnetic Resonance Series B, 103(3), 247–254. https://doi.org/10.1006/JMRB.1994.1037.
Beare, R., Lowekamp, B., & Yaniv, Z. (2018). Image Segmentation, Registration and characterization in R with SimpleITK. Journal of Statistical Software, 86(8), https://doi.org/10.18637/JSS.V086.I08.
Blasiak, T., Czubak, W., Ignaciak, A., & Lewandowski, M. H. (2010). A new approach to detection of the bregma point on the rat skull. Journal of Neuroscience Methods, 185(2), 199–203. https://doi.org/10.1016/j.jneumeth.2009.09.022.
Breckwoldt, M. O., Bode, J., Kurz, F. T., Hoffmann, A., Ochs, K., Ott, M., et al. (2016). Correlated magnetic resonance imaging and ultramicroscopy (MR-UM) is a tool kit to assess the dynamics of glioma angiogenesis. eLife, 5, 1–17. https://doi.org/10.7554/eLife.11712.
Breckwoldt, M. O., Bode, J., Sahm, F., Krüwel, T., Solecki, G., Hahn, A., et al. (2019). Correlated MRI and ultramicroscopy (MR-UM) of brain tumors reveals vast heterogeneity of tumor infiltration and neoangiogenesis in preclinical models and human disease. Frontiers in Neuroscience, 13, 1–10. https://doi.org/10.3389/fnins.2018.01004.
Chan, E., Kovacevíc, N., Ho, S. K. Y., Henkelman, R. M., & Henderson, J. T. (2007). Development of a high resolution three-dimensional surgical atlas of the murine head for strains 129S1/SvImJ and C57Bl/6J using magnetic resonance imaging and micro-computed tomography. Neuroscience, 144(2), 604–615. https://doi.org/10.1016/j.neuroscience.2006.08.080.
Chen, Y., McElvain, L. E., Tolpygo, A. S., Ferrante, D., Friedman, B., Mitra, P. P., et al. (2019). An active texture-based digital atlas enables automated mapping of structures and markers across brains. Nature Methods, 16(4), 341–350. https://doi.org/10.1038/s41592-019-0328-8.
Chon, U., Vanselow, D. J., Cheng, K. C., & Kim, Y. (2019). Enhanced and unified anatomical labeling for a common mouse brain atlas. Nature communications, 10(1), 5067. https://doi.org/10.1038/s41467-019-13057-w.
Chuang, N., Mori, S., Yamamoto, A., Jiang, H., Ye, X., Xu, X., et al. (2011). An MRI-based atlas and database of the developing mouse brain. Neuroimage, 54(1), 80–89. https://doi.org/10.1016/j.neuroimage.2010.07.043.
Chung, K., Wallace, J., Kim, S. Y., Kalyanasundaram, S., Andalman, A. S., Davidson, T. J., et al. (2013). Structural and molecular interrogation of intact biological systems. Nature, 497(7449), 332–337. https://doi.org/10.1038/nature12107.
di Giovanna, A. P., Tibo, A., Silvestri, L., Müllenbroich, M. C., Costantini, I., Mascaro, A., A. L., et al. (2018). Whole-brain vasculature Reconstruction at the single Capillary Level. Scientific Reports, 8(1), 12573. https://doi.org/10.1038/s41598-018-30533-3.
Doerr, J., Schwarz, M. K., Wiedermann, D., Leinhaas, A., Jakobs, A., Schloen, F., et al. (2017). Whole-brain 3D mapping of human neural transplant innervation. Nature Communications, 8, 1–7. https://doi.org/10.1038/ncomms14162.
Dong, H. W. (2008). The Allen Reference Atlas: a Digital Color Brain Atlas of the C57BL/6J male mouse. Wiley.
Dorr, A. E., Lerch, J. P., Spring, S., Kabani, N., & Henkelman, R. M. (2008). High resolution three-dimensional brain atlas using an average magnetic resonance image of 40 adult C57Bl/6J mice. Neuroimage, 42(1), 60–69. https://doi.org/10.1016/J.NEUROIMAGE.2008.03.037.
Dumoulin, S. O., Fracasso, A., van der Zwaag, W., Siero, J. C. W., & Petridou, N. (2018). Ultra-high field MRI: advancing systems neuroscience towards mesoscopic human brain function. Neuroimage, 168, 345–357. https://doi.org/10.1016/J.NEUROIMAGE.2017.01.028.
Dyrby, T. B., Baaré, W. F. C., Alexander, D. C., Jelsing, J., Garde, E., & Søgaard, L. v (2011). An ex vivo imaging pipeline for producing high-quality and high-resolution diffusion-weighted imaging datasets. Human Brain Mapping, 32(4), 544–563. https://doi.org/10.1002/HBM.21043.
Dyrby, T. B., Innocenti, G. M., Bech, M., & Lundell, H. (2018). Validation strategies for the interpretation of microstructure imaging using diffusion MRI. Neuroimage, 182, 62–79. https://doi.org/10.1016/J.NEUROIMAGE.2018.06.049.
Franklin, K. B. J., & Paxinos, G. (1997). The mouse brain in stereotaxic coordinates (1st ed.). San Diego: Academic Press.
Friedmann, D., Pun, A., Adams, E. L., Lui, J. H., Kebschull, J. M., Grutzner, S. M. (2020). Mapping mesoscale axonal projections in the mouse brain using a 3D convolutional network. Proceedings of the National Academy of Sciences, 117(20), 11068–11075. https://doi.org/10.1073/PNAS.1918465117
Gabery, S., Salinas, C. G., Paulsen, S. J., Ahnfelt-Rønne, J., Alanentalo, T., Baquero, A. F., et al. (2020). Semaglutide lowers body weight in rodents via distributed neural pathways. JCI Insight, 5(6), https://doi.org/10.1172/jci.insight.133429.
Goubran, M., Leuze, C., Hsueh, B., Aswendt, M., Ye, L., Tian, Q., et al. (2019). Multimodal image registration and connectivity analysis for integration of connectomic data from microscopy to MRI. Nature Communications, 10(1), 1–17. https://doi.org/10.1038/s41467-019-13374-0.
Hansen, H. H., Perens, J., Roostalu, U., Skytte, J. L., Salinas, C. G., Barkholt, P., et al. (2021). Whole-brain activation signatures of weight-lowering drugs. Molecular Metabolism, 47(January), 101171. https://doi.org/10.1016/j.molmet.2021.101171.
Harris, C. R., Millman, K. J., van der Walt, S. J., Gommers, R., Virtanen, P., Cournapeau, D., et al. (2020). Array programming with NumPy. Nature 2020, 585:7825(7825), 357–362. https://doi.org/10.1038/s41586-020-2649-2. 585.
Hof, P. R., Young, W. G., Bloom, F. E., Belichenko, P., & Celio, M. R. (2000). Comparative cytoarchitectonic atlas of the C57BL/6 and 129/Sv mouse brains (1st ed.). Amsterdam: Elsevier.
Jacobowitz, D. M., & Abbott, L. C. (1997). Chemoarchitectonic atlas of the developing mouse brain (1st ed.). Boca Raton: CRC Press.
Johnson, G. A., Badea, A., Brandenburg, J., Cofer, G., Fubara, B., Liu, S., & Nissanov, J. (2010). Waxholm Space: an image-based reference for coordinating mouse brain research. Neuroimage, 53(2), 365–372. https://doi.org/10.1016/J.NEUROIMAGE.2010.06.067.
Johnson, G. A., Laoprasert, R., Anderson, R. J., Cofer, G., Cook, J., Pratson, F., & White, L. E. (2021). A multicontrast MR atlas of the Wistar rat brain. NeuroImage, 242(December 2020), 118470. https://doi.org/10.1016/j.neuroimage.2021.118470
Kaufman, M. (1992). Atlas of Mouse Development (1st ed.). London: Academic Press.
Kellner, E., Dhital, B., Kiselev, V. G., & Reisert, M. (2016). Gibbs-ringing artifact removal based on local subvoxel-shifts. Magnetic Resonance in Medicine, 76(5), 1574–1581. https://doi.org/10.1002/MRM.26054.
Kirst, C., Skriabine, S., Vieites-Prado, A., Topilko, T., Bertin, P., Gerschenfeld, G., et al. (2020). Mapping the Fine-Scale Organization and plasticity of the Brain vasculature. Cell, 180(4), 780–795e25. https://doi.org/10.1016/j.cell.2020.01.028.
Kjaergaard, M., Salinas, C. B. G., Rehfeld, J. F., Secher, A., Raun, K., & Wulff, B. S. (2019). PYY(3–36) and exendin-4 reduce food intake and activate neuronal circuits in a synergistic manner in mice. Neuropeptides, 73, 89–95. https://doi.org/10.1016/j.npep.2018.11.004.
Klein, S., Staring, M., Murphy, K., Viergever, M. A., & Pluim, J. P. W. (2010). Elastix: a toolbox for intensity-based Medical Image Registration. IEEE Transactions on Medical Imaging, 29(1), 196–205. https://doi.org/10.1109/TMI.2009.2035616.
Kovačević, N., Henderson, J. T., Chan, E., Lifshitz, N., Bishop, J., Evans, A. C., et al. (2005). A three-dimensional MRI atlas of the mouse brain with estimates of the average and variability. Cerebral Cortex, 15(5), 639–645. https://doi.org/10.1093/cercor/bhh165.
Kuan, L., Li, Y., Lau, C., Feng, D., Bernard, A., Sunkin, S. M., et al. (2015). Neuroinformatics of the Allen Mouse Brain Connectivity Atlas. Methods, 73, 4–17. https://doi.org/10.1016/j.ymeth.2014.12.013.
Lee, B. C., Lin, M. K., Fu, Y., Hata, J., Miller, M. I., & Mitra, P. P. (2021). Multimodal cross-registration and quantification of metric distortions in marmoset whole brain histology using diffeomorphic mappings. Journal of Comparative Neurology, 529(2), 281–295. https://doi.org/10.1002/CNE.24946.
Leuze, C., Aswendt, M., Ferenczi, E., Liu, C. W., Hsueh, B., Goubran, M. (2017). The separate effects of lipids and proteins on brain MRI contrast revealed through tissue clearing. NeuroImage, 156(December 2016), 412–422. https://doi.org/10.1016/j.neuroimage.2017.04.021
Li, X., Aggarwal, M., Hsu, J., Jiang, H., & Mori, S. (2013). AtlasGuide: Software for stereotaxic guidance using 3D CT/MRI hybrid atlases of developing mouse brains. Journal of Neuroscience Methods, 220(1), 75–84. https://doi.org/10.1016/j.jneumeth.2013.08.017.
Lowekamp, B. C., Chen, D. T., Ibanez, L., & Blezek, D. (2013). The design of SimpleITK. Frontiers in Neuroinformatics, 7, 45. https://doi.org/10.3389/FNINF.2013.00045.
Ly, P. T., Lucas, A., Pun, S. H., Dondzillo, A., Liu, C., Klug, A., & Lei, T. C. (2021). Robotic stereotaxic system based on 3D skull reconstruction to improve surgical accuracy and speed. Journal of Neuroscience Methods, 347, 108955. https://doi.org/10.1016/J.JNEUMETH.2020.108955.
Ma, Y., Hof, P. R., Grant, S. C., Blackband, S. J., Bennett, R., Slatest, L., et al. (2005). A three-dimensional digital atlas database of the adult C57BL/6J mouse brain by magnetic resonance microscopy. Neuroscience, 135(4), 1203–1215. https://doi.org/10.1016/j.neuroscience.2005.07.014.
Ma, Y., Smith, D., Hof, P. R., Foerster, B., Hamilton, S., Blackband, S. J., et al. (2008). In vivo 3D digital atlas database of the adult C57BL/6J mouse brain by magnetic resonance microscopy. Frontiers in Neuroanatomy, 2(APR), 1–10. https://doi.org/10.3389/neuro.05.001.2008.
MacKenzie-Graham, A., Lee, E. F., Dinov, I. D., Bota, M., Shattuck, D. W., Ruffins, S., et al. (2004). A multimodal, multidimensional atlas of the C57BL/6J mouse brain. Journal of Anatomy, 204(2), 93–102. https://doi.org/10.1111/J.1469-7580.2004.00264.X.
Massalimova, A., Ni, R., Nitsch, R. M., Reisert, M., von Elverfeldt, D., & Klohs, J. (2021). Diffusion Tensor Imaging reveals whole-brain microstructural changes in the P301L mouse model of Tauopathy. Neurodegenrative diseases, 20, 173–184. https://doi.org/10.1159/000515754.
Matthews, P. M., & Jezzard, P. (2004). Functional magnetic resonance imaging. Journal of Neurology Neurosurgery & Psychiatry, 75, 6–12.
Morawski, M., Kirilina, E., Scherf, N., Jäger, C., Reimann, K., Trampel, R. (2018). Developing 3D microscopy with CLARITY on human brain tissue: Towards a tool for informing and validating MRI-based histology. NeuroImage, 182(November 2017), 417–428. https://doi.org/10.1016/j.neuroimage.2017.11.060
Murakami, T. C., Mano, T., Saikawa, S., Horiguchi, S. A., Shigeta, D., Baba, K., et al. (2018). A three-dimensional single-cell-resolution whole-brain atlas using CUBIC-X expansion microscopy and tissue clearing. Nature Neuroscience, 21(4), 625–637. https://doi.org/10.1038/s41593-018-0109-1.
Nie, B., Wu, D., Liang, S., Liu, H., Sun, X., Li, P. (2019). A stereotaxic MRI template set of mouse brain with fine sub-anatomical delineations: Application to MEMRI studies of 5XFAD mice. Magnetic Resonance Imaging, 57(June 2018), 83–94. https://doi.org/10.1016/j.mri.2018.10.014
Patel, J. (2018). The mouse brain: a 3D atlas registering MRI, CT, and histological sections in three cardinal planes. John Hopkins University.
Perens, J., & Hecksher-Sørensen, J. (2022). Digital Brain Maps and Virtual Neuroscience: An Emerging Role for Light-Sheet Fluorescence Microscopy in Drug Development. Frontiers in Neuroscience, 16. https://doi.org/10.3389/fnins.2022.866884
Perens, J., Salinas, C. G., Skytte, J. L., Roostalu, U., Dahl, A. B., Dyrby, T. B., et al. (2021). An optimized mouse brain Atlas for Automated Mapping and quantification of neuronal activity using iDISCO + and light sheet fluorescence Microscopy. Neuroinformatics, 19, 433–446. https://doi.org/10.1007/S12021-020-09490-8/FIGURES/5.
Purger, D., McNutt, T., Achanta, P., Quĩones-Hinojosa, A., Wong, J., & Ford, E. (2009). A histology-based atlas of the C57BL/6J mouse brain deformably registered to in vivo MRI for localized radiation and surgical targeting. Physics in Medicine and Biology, 54(24), 7315–7327. https://doi.org/10.1088/0031-9155/54/24/005.
Qi, Y., Yu, T., Xu, J., Wan, P., Ma, Y., Zhu, J., et al. (2019). FDISCO: advanced solvent-based clearing method for imaging whole organs. Science Advances, 5(1), eaau8355. https://doi.org/10.1126/sciadv.aau8355.
Qu, L., Li, Y., Xie, P., Liu, L., Wang, Y., Wu, J., et al. (2022). Cross-modal coherent registration of whole mouse brains. Nature Methods, 19(1), 111–118. https://doi.org/10.1038/s41592-021-01334-w.
Renier, N., Adams, E. L., Kirst, C., Dulac, C., Osten, P., & Tessier-Lavigne, M. (2016). Mapping of brain activity by automated volume analysis of Immediate Early genes. Cell, 165(7), 1789–1802. https://doi.org/10.1016/j.cell.2016.05.007.
Renier, N., Wu, Z., Simon, D. J., Yang, J., Ariel, P., & Tessier-Lavigne, M. (2014). iDISCO: a simple, Rapid Method to Immunolabel large tissue samples for volume imaging. Cell, 159(4), 896–910. https://doi.org/10.1016/j.cell.2014.10.010.
Roostalu, U., Salinas, C. B. G., Thorbek, D. D., Skytte, J. L., Fabricius, K., Barkholt, P., et al. (2019). Quantitative whole-brain 3D imaging of tyrosine hydroxylase-labeled neuron architecture in the mouse MPTP model of Parkinson’s disease. Disease Models and Mechanisms, 12(11), https://doi.org/10.1242/dmm.042200.
Rosen, G. D., Williams, A. G., Capra, J. A., Connolly, M. T., Cruz, B., Lu, L. (2000). The Mouse Brain Library. Int Mouse Genome Conference 14, (166). www.mbl.org
Salinas, C. B. G., Lu, T. T. H., Gabery, S., Marstal, K., Alanentalo, T., Mercer, A. J., et al. (2018). Integrated Brain Atlas for unbiased mapping of Nervous System Effects following Liraglutide Treatment. Scientific Reports, 8(1), 10310. https://doi.org/10.1038/s41598-018-28496-6.
Secher, A., Jelsing, J., Baquero, A. F., Cowley, J. H. S. M. A., Dalbøge, L. S., Hansen, G., et al. (2014). The arcuate nucleus mediates GLP-1 receptor agonist liraglutide-dependent weight loss. The Journal of Clinical Investigation, 124(10), 4473–4488. https://doi.org/10.1172/JCI75276.
Shamonin, D. P., Bron, E. E., Lelieveldt, B. P., Smits, M., Klein, S., & Staring, M. (2014). Fast parallel image registration on CPU and GPU for diagnostic classification of Alzheimer’s disease. Frontiers in Neuroinformatics, 7. https://doi.org/10.3389/FNINF.2013.00050
Shen, J. (2021). Tools for NIfTI and ANALYZE. MATLAB Central File Exchange. https://www.mathworks.com/matlabcentral/fileexchange/8797-tools-for-nifti-and-analyze-image.
Sidman, R. L., Angevine, J. B., & Taber-Pierce, E. (1971). Atlas of the mouse brain and spinal cord (Commonwealth Fund Publications). Cambridge: Harvard University Press.
Silvestri, L., Paciscopi, M., Soda, P., Biamonte, F., Iannello, G., Frasconi, P., & Pavone, F. S. (2015). Quantitative neuroanatomy of all Purkinje cells with light sheet microscopy and high-throughput image analysis. Frontiers in Neuroanatomy, 9(MAY), 1–11. https://doi.org/10.3389/fnana.2015.00068.
Skovbjerg, G., Roostalu, U., Hansen, H. H., Lutz, T. A., le Foll, C., Salinas, C. G., et al. (2021). Whole-brain mapping of amylin-induced neuronal activity in receptor activity–modifying protein 1/3 knockout mice. European Journal of Neuroscience, 54(1), 4154–4166. https://doi.org/10.1111/EJN.15254.
Stolp, H. B., Ball, G., So, P. W., Tournier, J. D., Jones, M., Thornton, C., & Edwards, A. D. (2018). Voxel-wise comparisons of cellular microstructure and diffusion-MRI in mouse hippocampus using 3D bridging of optically-clear histology with Neuroimaging Data (3D-BOND). Scientific Reports, 8(1), 1–12. https://doi.org/10.1038/s41598-018-22295-9.
Susaki, E. A., Tainaka, K., Perrin, D., Kishino, F., Tawara, T., Watanabe, T. M., et al. (2014). Whole-brain imaging with single-cell resolution using Chemical Cocktails and computational analysis. Cell, 157(3), 726–739. https://doi.org/10.1016/j.cell.2014.03.042.
Symms, M., Jäger, H. R., Schmierer, K., & Yousry, T. A. (2004). A review of structural magnetic resonance neuroimaging. Journal of Neurology Neurosurgery and Psychiatry, 75(9), 1235–1244. https://doi.org/10.1136/jnnp.2003.032714.
Todorov, M. I., Paetzold, J. C., Schoppe, O., Tetteh, G., Shit, S., Efremov, V., et al. (2020). Machine learning analysis of whole mouse brain vasculature. Nature Methods, 17(4), 442–449. https://doi.org/10.1038/s41592-020-0792-1.
Tournier, J. D., Smith, R., Raffelt, D., Tabbara, R., Dhollander, T., Pietsch, M., et al. (2019). MRtrix3: a fast, flexible and open software framework for medical image processing and visualisation. Neuroimage, 202, 116137. https://doi.org/10.1016/J.NEUROIMAGE.2019.116137.
Tyson, A. L., & Margrie, T. W. (2021). Mesoscale microscopy and image analysis tools for understanding the brain. Progress in Biophysics and Molecular Biology, (xxxx). https://doi.org/10.1016/j.pbiomolbio.2021.06.013
Umadevi Venkataraju, K. U., Gornet, J., Murugaiyan, G., Wu, Z., & Osten, P. (2019). Development of brain templates for whole brain atlases. Progress in Biomedical Optics and Imaging - Proceedings of SPIE, 10865, 1086511. https://doi.org/10.1117/12.2505295
Valverde, F. (2004). Golgi atlas of the postnatal mouse brain. New York: Springer-Verlag.
van der Walt, S., Schönberger, J. L., Nunez-Iglesias, J., Boulogne, F., Warner, J. D., Yager, N., et al. (2014). scikit-image: image processing in Python. PeerJ, 2(1), https://doi.org/10.7717/PEERJ.453.
Vaughan, J. T., Garwood, M., Collins, C. M., Liu, W., DelaBarre, L., Adriany, G., et al. (2001). 7T vs. 4T: RF power, homogeneity, and signal-to-noise comparison in head images. Magnetic resonance in medicine, 46(1), 24–30. https://doi.org/10.1002/MRM.1156.
Veraart, J., Novikov, D. S., Christiaens, D., Ades-aron, B., Sijbers, J., & Fieremans, E. (2016). Denoising of diffusion MRI using random matrix theory. Neuroimage, 142, 394–406. https://doi.org/10.1016/J.NEUROIMAGE.2016.08.016.
Virtanen, P., Gommers, R., Oliphant, T. E., Haberland, M., Reddy, T., Cournapeau, D., et al. (2020). SciPy 1.0: fundamental algorithms for scientific computing in Python. Nature Methods 2020, 17:3(3), 261–272. https://doi.org/10.1038/s41592-019-0686-2. 17.
Wang, N., White, L. E., Qi, Y., Cofer, G., & Johnson, G. A. (2020a). Cytoarchitecture of the mouse brain by high resolution diffusion magnetic resonance imaging. NeuroImage, 216 (November 2019). https://doi.org/10.1016/j.neuroimage.2020a.116876
Wang, Q., Ding, S. L., Li, Y., Royall, J., Feng, D., Lesnar, P., et al. (2020b). The Allen Mouse Brain Common coordinate Framework: a 3D reference Atlas. Cell, 181(4), 936–953e20. https://doi.org/10.1016/J.CELL.2020b.04.007.
Wei, H., Xie, L., Dibb, R., Li, W., Decker, K., Zhang, Y., et al. (2016). Imaging whole-brain cytoarchitecture of mouse with MRI-based quantitative susceptibility mapping. Neuroimage, 137, 107–115. https://doi.org/10.1016/j.neuroimage.2016.05.033.
Xiao, J. (2007). A new coordinate system for rodent brain and variability in the brain weights and dimensions of different ages in the naked mole-rat. Journal of Neuroscience Methods, 162(1–2), 162–170. https://doi.org/10.1016/j.jneumeth.2007.01.007.
Yaniv, Z., Lowekamp, B. C., Johnson, H. J., & Beare, R. (2018). SimpleITK Image-Analysis Notebooks: a collaborative environment for Education and Reproducible Research. Journal of Digital Imaging, 31(3), 290–303. https://doi.org/10.1007/S10278-017-0037-8.
You, T., Im, G. H., & Kim, S. G. (2021). Characterization of brain-wide somatosensory BOLD fMRI in mice under dexmedetomidine/isoflurane and ketamine/xylazine. Scientific Reports, 11(1), 13110. https://doi.org/10.1038/s41598-021-92582-5.
Yousaf, T., Dervenoulas, G., & Politis, M. (2018). Advances in MRI methodology. International Review of Neurobiology (1st ed., 141 vol., pp. 31–76). Elsevier Inc. https://doi.org/10.1016/bs.irn.2018.08.008.
Yushkevich, P. A., Piven, J., Hazlett, H. C., Smith, R. G., Ho, S., Gee, J. C., & Gerig, G. (2006). User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage, 31(3), 1116–1128. https://doi.org/10.1016/J.NEUROIMAGE.2006.01.015.
Zhou, P., Liu, Z., Wu, H., Wang, Y., Lei, Y., & Abbaszadeh, S. (2020). Automatically detecting bregma and lambda points in rodent skull anatomy images. Plos One, 15(12 December), 1–11. https://doi.org/10.1371/journal.pone.0244378.
Acknowledgements
We would like to thank Hanne Duus Lautsen and Mette Musfelth Nebbelunde for their skillful assistance in the lab.
Funding
The work was funded by Gubra ApS and Innovation Fund Denmark, grant number 8053-00121B.
Author information
Authors and Affiliations
Contributions
Johanna Perens, Casper Gravesen Salinas, Urmas Roostalu, Jacob Lercke Skytte, Tim B. Dyrby and Carsten Gundlach designed and performed the experiments. Johanna Perens, Tim B. Dyrby, Anders Bjorholm Dahl and Jacob Hecksher-Sørensen wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Johanna Perens, Casper Gravesen Salinas, Urmas Roostalu, Jacob Lercke Skytte, and Jacob Hecksher-Sørensen are currently employed at Gubra ApS and own shares of Gubra ApS.
Competing interests
Johanna Perens, Casper Gravesen Salinas, Urmas Roostalu, Jacob Lercke Skytte and Jacob Hecksher-Sørensen are employees at Gubra and own shares in Gubra.
Ethics Approval
Animal experiments were conducted in compliance with internationally accepted principles for the use of laboratory animals and approved by the Danish Animal Experiments Inspectorate (license #2013-15-2934-00784).
Information sharing statement
Resources of the multimodal mouse brain atlas can be found at https://www.neuropedia.dk. The source code used for generating the atlas and the parameter files used for registrations are accessible at https://github.com/Gubra-ApS.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Perens, J., Salinas, C.G., Roostalu, U. et al. Multimodal 3D Mouse Brain Atlas Framework with the Skull-Derived Coordinate System. Neuroinform 21, 269–286 (2023). https://doi.org/10.1007/s12021-023-09623-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12021-023-09623-9