Abstract
Glioma is a kind of brain tumor with low overall survival and treatment success rates in the advanced stage. Evidence has shown microRNA-575 (miR-575) plays an important role in the generation and development of various cancers. This study aimed to explore the function of miR-575 in the prognosis and cell biological behavior of glioma. qRT-PCR was used to evaluate the expression of miR-575 in glioma tissues and cells, Kaplan–Meier survival analysis and Cox regression analysis were used to evaluate the prognostic value. The proliferation ability of glioma cells was determined by MTT assay; the invasion and migration abilities were determined by transwell assays. Compared with normal brain tissues, the expression of miR-575 in glioma tissue cells was significantly up-regulated (P < 0.001). The survival rate of patients in the miR-575 high expression group was significantly lower than that in the low expression group (P = 0.020). In addition, the overexpression of miR-575 promoted the proliferation, migration, and invasion of glioma cells. The results of this study suggested that miR-575 may be a new biomarker for the prognosis of glioma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioma is one of the most frequent primary malignant tumors that originated in the brain (Wirsching et al., 2016). Nearly 30% of primary brain tumors and 80% of malignant tumors are gliomas, with six cases per 100,000 cancer patients each year in the United States in 2009–2013 (Winkler et al., 2018; Wirsching et al., 2016). Glioma is the most invasive and always erodes the surrounding normal glial cells, so it has a high mortality rate, which is also the cause of poor prognosis (Jiang & Uhrbom, 2012). According to the 2007 World Health Organization (WHO) classification, gliomas are divided into astroglioma, oligodendroglioma, mixed oligodendroglioma, or ependymoma, with grades of I–IV, and grade IV glioma is the most aggressive (Chen et al., 2017). With the advancement of medical conditions, various methods such as surgery, radiotherapy, and chemotherapy have shown effectiveness in the treatment of gliomas, but the survival rate of patients is still low (Lapointe et al., 2018; Shi et al., 2017). Previous studies have found there are few specific markers for the diagnosis, treatment, and prognosis of gliomas so far (Camelo-Piragua & Kesari, 2016). Therefore, developing more novel and effective biomarkers for detecting patients’ gliomas and monitoring prognosis remains crucial.
MicroRNAs (miRNAs) are short non-coding RNAs that regulate gene expression mainly by inhibiting the degradation of messenger RNA (mRNA) (Ban et al., 2017; Kappel & Keller, 2017). Some studies have found that miRNAs have involved in different physiological and pathological processes, including metabolism, cell proliferation, differentiation and apoptosis, embryo formation and development, and virus infection (Correia de Sousa et al., 2019; Shao et al., 2018). Dysregulation of miRNA expression caused by gene mutations or enzymes regulates miRNA is associated with many diseases, including neurological diseases, cardiovascular diseases, and various cancers (Lodewijk et al., 2012). In the evolution of glioma research, many miRNAs are abnormally expressed in tissues or cell lines, such as miR-211, miR-222, miR-124, and miR-19 (Lv & Zhao, 2017; Wang et al., 2018; Xue et al., 2017). Previous studies have found that miR-575 is abnormally expressed in non-small lung cancer, gastric cancer, and gallbladder cancer tissues, and the introduction of miR-575 significantly affects the proliferation, migration, and invasion of these cell lines (Qin et al., 2020; Wang et al., 2015, 2019). Piwecka et al. have found that 35 kinds of miRNAs, including miR-575, express abnormality in GBM patients (Piwecka et al., 2015). However, the expression mechanism and clinical significance of miR-575 in the pathogenesis of glioma are still unclear.
This study explored the abnormal expression of miR-575 in glioma by comparing it between glioma and normal brain tissues. Then, the relationship between miR-575 expression and patient pathological characteristics and survival time was explored. In addition, we also studied the effect of miR-575 overexpression on the proliferation, migration, and invasion of glioma cells, and explored whether miR-575 can be used as a new biomarker for glioma.
Materials and Methods
Patients and Specimens
This study was approved and supported by the Research Ethics Committee of Weifang People’s Hospital Brain Hospital. We selected 107 glioma patients who underwent surgery in Weifang People’s Hospital Brain Hospital from January 2012 to December 2014. All the patients were enrolled according to the inclusion criteria: (i) patients were pathologically diagnosed as glioma; (ii) None of them had received other treatments such as radiotherapy or chemotherapy before surgery; (iii) patients were followed up for 5-year survival outcomes; (iv) patients with complete clinical characteristic information. Besides, 33 normal cortex tissues were obtained from the patients who suffered traffic injury. Each patient provided informed consent and agreed to use their tissues for scientific research. The newly obtained glioma tissue and normal brain tissue samples were immediately snap-frozen in liquid nitrogen and stored at − 80 °C until the experiment was performed. A 5-year telephone follow-up of all glioma patients was used to record the survival status of the patients.
Cell Lines and Transfection
Human glioma cell lines (LN229, U251, A172, and T98G) and a normal human astrocytes cell line NHAs, were obtained from the Shanghai Cell Bank of the Chinese Academy of Sciences. These cell lines were inoculated into a DMEM medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS and were cultured at 37 °C in a humidified incubator containing 5% CO2. Cells (2 × 106 cells/well) were seeded into six-well plates for subsequent experiments. According to the manufacturer’s protocol, the cells were transfected with 50 nM final concentration of miR-575 mimic (5ʹ-GAGCCAGUUGGACAGGAGC-3ʹ), mimic negative control (NC; 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ), miR-575 inhibitor (5ʹ-GCUCCUGUCCAACUGGCUC-3ʹ), or inhibitor NC (5ʹ-CAGUACUUUUGUGUAGUACAA-3ʹ) (GenePharma, Shanghai, China) using Lipofectamine 2000 Reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The cells without any treatment were used as blank control. Then we used qRT-PCR to monitor the transfection efficiency.
RNA Extraction and qRT-PCR Analysis
Grind the broken cells in liquid nitrogen, and use TRIzol reagent (TaKaRa, Otsu, Shiga, Japan) to extract total RNA from tissues or cell lines according to the manufacturer’s instructions. Using NanoDrop to measure the absorbance of the prepared samples to calculate the concentration of extracted RNA. Using the cDNA reverse transcription kit (Applied Biosystems, Foster City, CA) to obtain Complementary DNA reversed transcription from the extracted RNA. We used the SYBR Green I real-time PCR kit (GenePharma, Shanghai, China) to quantify the level of miR-575 in cell lines and tissues, and then used the Applied Biosystems 7900 real-time PCR system (Applied Biosystems, Foster City, CA) to perform QRT-PCR. U6 is used as a standardization control for miRNA quantification. The 2−∆∆Ct method was used for calculating the relative expression levels.
Cell Proliferation Assay
According to the manufacturer’s recommendations, MTT (MTT; Aladdin, Shanghai, China) was used to determine cell proliferation. Briefly, glioma cells (2 × 103/well) were seeded in 96-well plates and cultured in a 37 °C 5% CO2 saturated humidity incubator for 24 h. MTT was added to each well and continue to incubate for 4 h after the cells adhere to the wall. Adding DMSO into each well and measuring the absorbance value of each well at the 490 nm wavelength of the microplate reader, then measuring it every 24 h (24, 48, 72 h).
Cell Migration and Invasion Assay
The migration and invasion ability of cells was checked by transwell kit (BD Biosciences, San Jose, CA, USA). In the invasion assay, the bottom layer of the upper chamber of the transwell chamber was pre-coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA), and the upper chamber of the migration assay did not need to add Matrigel. The cells were seeded in a serum-free medium after transfection. Cell suspension (5 × 104 cells for U251 and 3 × 104 cells for A172) was added to the upper chamber of the Transwell chamber, and 600–800 μL basal medium containing 10% FBS was added to the lower chamber. Incubating at 37 °C and 5% CO2 for 24 h. Then we took out the transwell chamber, removed Matrigel and non-penetrating cells in the upper chamber. The bottom membrane of the upper chamber was fixed with methanol for 20 min and stained with 0.1% crystal violet for 15 min. the cells were observed and counted under the microscope after air-drying.
Statistical Analysis
The statistical software SPSS version 19.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 7.0 (GraphPad Software, San Diego, CA, USA) were used to analyze the experimental data. All the experiments were repeated three or more times with triple replicates. χ2 test, Student’s t test, or one-way ANOVA were used for comparison between groups; Kaplan–Meier analysis was used for survival analysis, and Cox regression analysis was used to evaluate prognostic significance. The measurement data is expressed as the mean ± SD, and P < 0.05 is considered statistically significant.
Results
miR-575 Expression in Glioma Tissues and Cell Lines
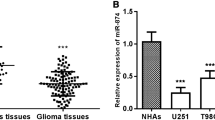
We first used qRT-PCR to detect the expression level of miR-575 in glioma tissues and cells. Real-time PCR analysis showed that compared with normal brain tissue, the expression level of miR-575 in glioma tissues was significantly up-regulated (P < 0.001, Fig. 1A). Among the 107 glioma patients, there are 12 WHO I grade patients, 68 WHO II grade patients, 25 WHO III grade patients, and 6 WHO IV grade patients. Furthermore, the expression of miR-575 was compared in tumor tissues in different WHO grades. The results in Fig. 1B showed that the expression of miR-575 was higher in WHO III and WHO IV grade tissues compared with that in WHO I grade tissues (P < 0.05). Although the expression of miR-575 was higher in WHO II grade tissues, there is no significant difference. Further analysis was carried out by clustering WHO grade I and II and WHO grades III and IV to analyze the relevance of miR-575 and WHO grade.
The expression of miR-575 in glioma tissues and cell lines, miR-575 was upregulated in tissues and cells. A The expression of miR-575 in glioma cells and adjacent normal liver tissues was evaluated by qRT-PCR. The assay was repeated three times and analyzed using Student’s t test (***P < 0.001 vs. normal tissues). B The expression of miR-575 in different WHO grades. The data was analyzed using one-way ANOVA followed by Tukey’s post-hoc test. (*P < 0.05, **P < 0.01 vs. WHO I grade) C The expression of miR-575 in four glioma cells (LN229, U251, A172, and T98G) and normal brain cells was evaluated by qRT-PCR. The assays were repeated three times and analyzed using one-way ANOVA followed by Tukey’s post-hoc test. (**P < 0.01, ***P < 0.001 vs. NHAs)
As shown in Fig. 1C, the expression levels of miR-575 in the four glioma cell lines (LN229, U251, A172, and T98G) were significantly up-regulated compared with the expression level in the normal cell line NHAs (P < 0.01). At the same time, we can perceive that among the four cell lines, A172 has the largest upregulation, followed by U251. Therefore, we chose these two groups of cell lines for later experiments.
The Correlation Between the miR-575 Expression and the Clinicopathological Features of Glioma Patients
In order to find out the changes of miR-575 in the progression of glioma, we analyzed the relationship between miR-575 expression and clinicopathological characteristics of patients with glioma (Table 1). According to the mean expression level of miR-575 (1.5276) in tumor tissues as cut-off value, 107 glioma samples were divided into two categories: (1) miR-575 low expression group (n = 52); (2) miR-575 high expression group (n = 55). As shown in Table 1, we found a significant positive correlation between miR-575 level and tumor size (P = 0.032) as well as WHO grade (P = 0.008), but no significant correlation with other characteristics (such as age, gender, preoperative KPS score, and IDH mutation) (P > 0.05).
miR-575 was Correlated with Poor Prognosis in Glioma Patients
To verify whether the upregulation of miR-575 expression has the prognostic value of glioma, the Kaplan–Meier method was used to calculate the survival curve based on the relative expression of miR-575 in glioma and survival information of the patients postoperation. As can be seen from Fig. 2, the 5-year survival rate of patients in the higher miR-575 expression group was significantly lower than that of the lower miR-575 expression group (log-rank test P = 0.020). Then the multivariate Cox’s hazard regression model was used to analyze the correlation between the expression level of miR-575 and the overall survival of patients with glioma. It can be obtained from Table 2 that the expression level of miR-575 (HR = 2.589, 95% CI 1.258–5.326, P = 0.010) and WHO grade (HR = 2.248, 95% CI 1.158–4.632, P = 0.017) were independent prognostic factors of glioma patients.
miR-575 Promoted Glioma Cell Proliferation, Migration, and Invasion
Then, we used the MTT assay to detect the changes in the proliferation of glioma cells with miR-575 overexpression, and the transwell assays to evaluate the effects of miR-575 on the migration and invasion of glioma cells. In the proliferation experiment, miR-575 mimic was transfected into U251 and A172 cells, which showed a significant increase; U251 and A172 cells transfected with miR-575 inhibitor showed a significant decrease (P < 0.001) (Fig. 3A). It can be seen from Fig. 3B that miR-575 mimic up-regulated the expression of miR-575 in cells and promoted cell proliferation (P < 0.01), while miR-575 inhibitor inhibited U251 (P < 0.05) and A172 (P < 0.01) cells proliferation. Through the transwell migration experiment, it can be concluded that miR-575 mimic significantly increased the migration of U251 cell (P < 0.001) and A172 cell (P < 0.01), while miR-575 inhibitor decreased the migration of U251 cell (P < 0.01) and A172 cell (P < 0.001) (Fig. 4A). It can be seen from the transwell invasion experiment that the overexpression of miR-575 significantly increased the invasion of U251 cell (P < 0.001) and A172 cell (P < 0.01), while the inhibition of miR-575 significantly reduced cell invasion (P < 0.001) (Fig. 4B). In summary, these results indicate that miR-575 promotes the proliferation, migration, and invasion of glioma cells.
The effects of miR-575 expression level on the proliferation of glioma cells, its overexpression promoted cell proliferation. A miR-575 mimic promoted the expression of miR-575, and miR-575 inhibitor suppressed the expression of miR-575 in U251 and A172 cells (***P < 0.001). The transfection efficiency experiments were repeated five times and analyzed using one-way ANOVA followed by Tukey’s post-hoc test. B The effects of miR-575 mimic and inhibitor on the proliferation of U251 and A172 cells were evaluated by MTT assay within 72 h. The experiments were repeated three times and analyzed using one-way ANOVA followed by Tukey’s post-hoc test. (**P < 0.05, ***P < 0.01)
Transwell analysis of U251 and A172 cells with miR-575 mimic, inhibitor, or no transfection, the overexpression of miR-575 promoted the migration and invasion. The experiments were repeated three times and results were analyzed using one-way ANOVA followed by Tukey’s post-hoc test. A The effect of miR-575 on glioma cell migration (** P < 0.01, *** P < 0.001). B The effect of miR-575 on the invasion of glioma cells (** P < 0.01, *** P < 0.001)
Discussion
Glioma is the most common primary tumor of the central nervous system, while its efficacy is not optimistic, especially the 5-year survival rate of high-grade glioma is extremely low (Ostrom et al., 2014, 2015; Perry & Wesseling, 2016). It is necessary to find new oncogenes for glioma detection, treatment, and prognosis (Dong et al., 2019; Jiang et al., 2018). Recent studies have revealed that miRNAs regulation involves the gene expression of many cancers, and has effects on cell differentiation, cell proliferation, migration, and invasion (Aghdam et al., 2019; Hayes & Chayama, 2016; Saliminejad et al., 2019). For example, studies have found that the expression of miR-339-5p was significantly reduced in HCC patients, and was related to the invasion of HCC cells, indicating that miR-339-5p is a biomarker for the prognosis of HCC (Wang et al., 2016). Yan et al. have revealed that the expression of miR-1290 was significantly up-regulated in glioma, indicating it was involved in the occurrence and development of glioma (Yan et al., 2018). Many studies have proved the abnormal expression of miR-575 in diseases and cancers, as well as its influence on the biological behavior of cells (Qin et al., 2020; Wang et al., 2015, 2019).
Many studies have shown that miR-575 can be used as an oncogene in a variety of cancers. Wang et al. have found that the expression of miR-575 in non-small cell lung cancer tissue was higher than that in normal controls (Camelo-Piragua & Kesari, 2016). Mikkelsen et al. have revealed that nine miRNAs, including miR-575, were significantly upregulated in primary conjunctival melanoma (Mikkelsen et al., 2019). Our research has obtained results consistent with the above research. We compared the expression of miR-575 in normal brain tissues and cells with glioma tissues and cell lines in this study and found that the expression of miR-575 in glioma tissues and cell lines was significantly up-regulated. At the same time, through considering the relationship between miR-575 overexpression and the pathological characteristics of patients, it can be concluded that miR-575 overexpression is significantly related to WHO grade. These results indicate that miR-575 may be an oncogene of glioma, which plays a role in the occurrence and development of glioma.
Previous research results show that miR-575 can be used as a new molecular target for cancer diagnosis and prognosis. Wang et al. have confirmed that among GC patients, the survival rate of the miR-575 high expression level group was significantly lower than that of the low expression level group (Wang et al., 2019). While our study also concluded that miR-575 may be an important prognostic biomarker for glioma. Kaplan–Meier curve and Cox regression analysis can reflect the relationship between the expression of miR-575 and the survival of glioma patients. It can be concluded that the expression of miR-575 in glioma tissues of patients with lower survival rates is upregulated. It can also be concluded that the expression level of miR-575 can be an independent prognostic factor for overall survival.
Xia et al. have confirmed that after transfecting JEG‑3 cells with miR-575 mimic, the expression of miR-575 increased significantly compared with the control group (Xia et al., 2017). A previous study has proved that miR-575 mimic significantly inhibited the migration and proliferation of human umbilical vein endothelial cells, and their apoptosis was also accelerated (Zhao et al., 2019). The introduction of mir-575 significantly promoted the proliferation, migration, and invasion of NSCLC cells (Wang et al., 2015). Knockdown of miR-575 can reduce the proliferation and invasion of Gallbladder Cancer cells (Qin et al., 2020). These studies show that miR-575 participates in the generation and development of cancer by participating in cell biological behaviors such as proliferation and migration, which is an obvious signal of cancers. Our experiments have proved that miR-575 also plays the same role in glioma cells. Through MTT proliferation experiments and Transwell migration and invasion experiments, it can be concluded that miR-575 mimic accelerates the proliferation, migration, and invasion of glioma cells significantly, while inhibitors have the opposite results. Previous studies revealed that miR-575 could regulate tumor progression by targeting p27 kip1 (CDKN1B) (Qin et al., 2020), PTEN (Wang et al., 2019), and BLID (Wang et al., 2015). Interestingly, PTEN was reported to be a tumor suppressor gene in glioma (Chai et al., 2018; Ma et al., 2019). Based on the above studies and present results, we speculate that miR-575 may promote the progression of glioma through targeting PTEN. Whereas, whether miR-575 could regulate PTEN expression and its detailed mechanism of action in glioma are still unclear. This hypothesis will be investigated in our future researches.
In conclusion, this research found that the expression of miR-575 is upregulated in the tissues of patients with glioma. The overexpression of miR-575 indicated a poor prognosis, and miR-575 promoted the proliferation, migration, and invasion of glioma cells. In summary, miR-575 may be used as a prognostic biomarker for glioma.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Code Availability
Not applicable.
References
Aghdam, S. G., Ebrazeh, M., Hemmatzadeh, M., Seyfizadeh, N., Shabgah, A. G., Azizi, G., Ebrahimi, N., Babaie, F., & Mohammadi, H. (2019). The role of microRNAs in prostate cancer migration, invasion, and metastasis. Journal of Cellular Physiology, 234(7), 9927–9942. https://doi.org/10.1002/jcp.27948
Ban, E., Chae, D. K., Yoo, Y. S., & Song, E. J. (2017). An improvement of miRNA extraction efficiency in human plasma. Analytical and Bioanalytical Chemistry, 409(27), 6397–6404. https://doi.org/10.1007/s00216-017-0580-7
Camelo-Piragua, S., & Kesari, S. (2016). Further understanding of the pathology of glioma: Implications for the clinic. Expert Review of Neurotherapeutics, 16(9), 1055–1065. https://doi.org/10.1080/14737175.2016.1194755
Chai, C., Song, L. J., Han, S. Y., Li, X. Q., & Li, M. (2018). MicroRNA-21 promotes glioma cell proliferation and inhibits senescence and apoptosis by targeting SPRY1 via the PTEN/PI3K/AKT signaling pathway. CNS Neuroscience & TherapeutIcs, 24(5), 369–380. https://doi.org/10.1111/cns.12785
Chen, R., Smith-Cohn, M., Cohen, A. L., & Colman, H. (2017). Glioma subclassifications and their clinical significance. Neurotherapeutics, 14(2), 284–297. https://doi.org/10.1007/s13311-017-0519-x
de Sousa, M. C., Gjorgjieva, M., Dolicka, D., Sobolewski, C., & Foti, M. (2019). Deciphering miRNAs’ action through miRNA editing. International Journal of Molecular Sciences, 20(24), 6249. https://doi.org/10.3390/ijms20246249
Dong, Q., Li, Q., Wang, M., Hu, J., Dai, J., Niu, L., Yuan, G., & Pan, Y. (2019). Elevated CD44 expression predicts poor prognosis in patients with low-grade glioma. Oncology Letters, 18(4), 3698–3704. https://doi.org/10.3892/ol.2019.10728
Hayes, C. N., & Chayama, K. (2016). MicroRNAs as biomarkers for liver disease and hepatocellular carcinoma. International Journal of Molecular Sciences, 17(3), 280. https://doi.org/10.3390/ijms17030280
Jiang, Y., & Uhrbom, L. (2012). On the origin of glioma. Upsala Journal of Medical Sciences, 117(2), 113–121. https://doi.org/10.3109/03009734.2012.658976
Jiang, Y., Wang, X., Zhang, J., & Lai, R. (2018). MicroRNA-599 suppresses glioma progression by targeting RAB27B. Oncology Letters, 16(1), 1243–1252. https://doi.org/10.3892/ol.2018.8727
Kappel, A., & Keller, A. (2017). miRNA assays in the clinical laboratory: Workflow, detection technologies and automation aspects. Clinical Chemistry and Laboratory Medicine, 55(5), 636–647. https://doi.org/10.1515/cclm-2016-0467
Lapointe, S., Perry, A., & Butowski, N. A. (2018). Primary brain tumours in adults. Lancet, 392(10145), 432–446. https://doi.org/10.1016/s0140-6736(18)30990-5
Lodewijk, L., Prins, A. M., Kist, J. W., Valk, G. D., Kranenburg, O., Rinkes, I. H. M., & Vriens, M. R. (2012). The value of miRNA in diagnosing thyroid cancer: A systematic review. Cancer Biomarkers, 11(6), 229–238. https://doi.org/10.3233/cbm-2012-0273
Lv, Z., & Zhao, Y. (2017). MiR-124 inhibits cell proliferation, invasion, and migration in glioma by targeting Smad2. International Journal of Clinical and Experimental Pathology, 10(11), 11369–11376.
Ma, J., Benitez, J. A., Li, J., Miki, S., de Albuquerque, C. P., Galatro, T., Orellana, L., Zanca, C., Reed, R., Boyer, A., Koga, T., Varki, N. M., Fenton, T. R., NagahashiMarie, S. K., Lindahl, E., Gahman, T. C., Shiau, A. K., Zhou, H., DeGroot, J., … Furnari, F. B. (2019). Inhibition of nuclear PTEN tyrosine phosphorylation enhances glioma radiation sensitivity through attenuated DNA repair. Cancer Cells, 35(3), 504-518.e7. https://doi.org/10.1016/j.ccell.2019.01.020
Mikkelsen, L. H., Andersen, M. K., Andreasen, S., Larsen, A. C., Tan, Q., Toft, P. B., Wadt, K., & Heegaard, S. (2019). Global microRNA profiling of metastatic conjunctival melanoma. Melanoma Research, 29(5), 465–473. https://doi.org/10.1097/cmr.0000000000000606
Ostrom, Q. T., Bauchet, L., Davis, F. G., Deltour, I., Fisher, J. L., Langer, C. E., Pekmezci, M., Schwartzbaum, J. A., Turner, M. C., Walsh, K. M., Wrensch, M. R., & Barnholtz-Sloan, J. S. (2014). The epidemiology of glioma in adults: A “state of the science” review. Neuro-Oncology, 16(7), 896–913. https://doi.org/10.1093/neuonc/nou087
Ostrom, Q. T., Gittleman, H., Stetson, L., Virk, S. M., & Barnholtz-Sloan, J. S. (2015). Epidemiology of gliomas. Cancer Treatment and Research, 163, 1–14. https://doi.org/10.1007/978-3-319-12048-5_1
Perry, A., & Wesseling, P. (2016). Histologic classification of gliomas. Handbook of Clinical Neurology, 134, 71–95. https://doi.org/10.1016/b978-0-12-802997-8.00005-0
Piwecka, M., Rolle, K., Belter, A., Barciszewska, A. M., Żywicki, M., Michalak, M., Nowak, S., Naskręt-Barciszewska, M. Z., & Barciszewski, J. (2015). Comprehensive analysis of microRNA expression profile in malignant glioma tissues. Molecular Oncology, 9(7), 1324–1340. https://doi.org/10.1016/j.molonc.2015.03.007
Qin, Y., Mi, W., Huang, C., Li, J., Zhang, Y., & Fu, Y. (2020). Downregulation of miR-575 inhibits the tumorigenesis of gallbladder cancer via targeting p27 Kip1. Oncotargets and Therapy, 13, 3667–3676. https://doi.org/10.2147/ott.s229614
Saliminejad, K., Khorram Khorshid, H. R., Soleymani Fard, S., & Ghaffari, S. H. (2019). An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. Journal of Cellular Physiology, 234(5), 5451–5465. https://doi.org/10.1002/jcp.27486
Shao, Q., Xu, J., Guan, X., Zhou, B., Wei, W., Deng, R., Li, D., Xu, X., & Zhu, H. (2018). In vitro and in vivo effects of miRNA-19b/20a/92a on gastric cancer stem cells and the related mechanism. International Journal of Medical Sciences, 15(1), 86–94. https://doi.org/10.7150/ijms.21164
Shi, J., Dong, B., Cao, J., Mao, Y., Guan, W., Peng, Y., & Wang, S. (2017). Long non-coding RNA in glioma: Signaling pathways. Oncotarget, 8(16), 27582–27592. https://doi.org/10.18632/oncotarget.15175
Wang, H., Yan, C., Shi, X., Zheng, J., Deng, L., Yang, L., Yu, F., Yang, Y., & Shao, Y. (2015). MicroRNA-575 targets BLID to promote growth and invasion of non-small cell lung cancer cells. FEBS Letters, 589(7), 805–811. https://doi.org/10.1016/j.febslet.2015.02.013
Wang, W., Zhang, A., Hao, Y., Wang, G., & Jia, Z. (2018). The emerging role of miR-19 in glioma. Journal of Cellular and Molecular Medicine, 22(10), 4611–4616. https://doi.org/10.1111/jcmm.13788
Wang, Y. L., Chen, C. M., Wang, X. M., & Wang, L. (2016). Effects of miR-339-5p on invasion and prognosis of hepatocellular carcinoma. Clinics and Research in Hepatology and Gastroenterology, 40(1), 51–56. https://doi.org/10.1016/j.clinre.2015.05.022
Wang, Y. N., Xu, F., Zhang, P., Wang, P., Wei, Y. N., Wu, C., & Cheng, S. J. (2019). MicroRNA-575 regulates development of gastric cancer by targeting PTEN. Biomedicine & Pharmacotherapy, 113, 108716. https://doi.org/10.1016/j.biopha.2019.108716
Winkler, F., Osswald, M., & Wick, W. (2018). Anti-angiogenics: Their role in the treatment of glioblastoma. Oncology Research and Treatment, 41(4), 181–186. https://doi.org/10.1159/000488258
Wirsching, H. G., Galanis, E., & Weller, M. (2016). Glioblastoma. Handbook of Clinical Neurology, 134, 381–397. https://doi.org/10.1016/b978-0-12-802997-8.00023-2
Xia, S., Zhen, Y., Ma, H., & Wang, A. (2017). Abnormal expression of microRNA-575 leads to missed abortion through regulating apoptosis and angiogenesis. Experimental and Therapeutic Medicine, 14(5), 3993–4000. https://doi.org/10.3892/etm.2017.5086
Xue, L., Wang, Y., Yue, S., & Zhang, J. (2017). The expression of miRNA-221 and miRNA-222 in gliomas patients and their prognosis. Neurological Sciences, 38(1), 67–73. https://doi.org/10.1007/s10072-016-2710-y
Yan, L., Cai, K., Sun, K., Gui, J., & Liang, J. (2018). MiR-1290 promotes proliferation, migration, and invasion of glioma cells by targeting LHX6. Journal of Cellular Physiology, 233(10), 6621–6629. https://doi.org/10.1002/jcp.26381
Zhao, X., Yi, Y., Meng, C., & Fang, N. (2019). MiRNA-575 suppresses angiogenesis by targeting Rab5-MEK-ERK pathway in endothelial cells. Bioscience Reports, 39(1), BSR20181218. https://doi.org/10.1042/BSR20181218
Funding
No funds, grants, or other support was received.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Ethical Approval
This study was approved and supported by the Research Ethics Committee of Weifang People’s Hospital Brain Hospital.
Consent to Participate
Informed consent was obtained from legal guardians.
Consent for Publication
The participant has consented to the submission of the research to the journal.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wei, G., Li, S., Wang, P. et al. Altered Expression of miR-575 in Glioma is Related to Tumor Cell Proliferation, Migration, and Invasion. Neuromol Med 24, 224–231 (2022). https://doi.org/10.1007/s12017-021-08679-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12017-021-08679-7