Abstract
Background
Stem cells from human exfoliated deciduous teeth (SHED) hold promise in regenerative medicine owing to their multipotent capabilities resembling mesenchymal stem cells (MSCs). Despite their potential, SHED have not been extensively investigated because their limited lifespan and unavailability of cell-lines pose challenges for therapeutic applications. This study investigated the effect of ectopic human telomerase reverse transcriptase (hTERT) expression on SHEDs’ proliferation while preserving stemness and genomic integrity.
Methods
Deciduous teeth were collected from children aged 6–10 years. After isolation and characterization, the SHED were transduced with pBabe-puro-hTERT retrovirus to establish SHED cell-line, which was evaluated and compared with pBabe-puro (mock control) for stemness, multipotency and growth attributes through flow cytometry, trilineage differentiation, and growth kinetics. We also estimated hTERT gene expression, genomic integrity, and validated cell-line through STR analysis.
Results
Following hTERT transduction, SHED displayed elevated hTERT gene expression while retaining fibroblast-like morphology and mesenchymal stem cell markers. Moreover, after hTERT transduction cellular shape remained same along with increased replicative lifespan and proliferation potential. SHED-hTERT cells exhibited multi-potency and maintained stemness, as evidenced by surface marker expression and multilineage differentiation. Furthermore, genomic integrity was not affected by hTERT integration, as confirmed by STR analysis and CDKN2A gene assessment.
Conclusion
Ectopic hTERT expression in SHED successfully prolonged their replicative lifespan and improved their ability to proliferate and migrate, while preserving their stemness, multipotency and genomic integrity, suggesting minimal carcinogenic risk. Establishment of SHED cell-line holds potential in regenerative medicine applications, especially in cell-based drugs and tissue engineering experiments.
Graphical Abstract

Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
Stem cells derived from the pulp of human exfoliated deciduous teeth, commonly referred to as SHED, possess unique attributes such as specific cell surface markers associated with mesenchymal stem cells (MSCs), fibroblast-like cellular morphology, plastic adherence, high proliferation, and clonogenicity [1]. These cells demonstrated remarkable multipotency, being capable of differentiating into multiple lineages, including odontoblasts, adipocytes, and chondrocytes. The potential therapeutic applications of SHED, including stem cell-based tooth engineering, tissue regeneration, and neural regeneration, have garnered significant interest in recent years [2]. Moreover, SHED offer several advantages as ideal stem cell sources for therapeutics because of their accessibility, affordability, ethical considerations, noninvasive procurement, substantial progenitor populations, enhanced clonogenic potential, and robust proliferative capabilities [3].
Irrespective of their biological source, stem cells grown in a laboratory setting are naturally inclined to undergo senescence, known as the Hayflick limit and is the main impediment to stem cell culture [4]. With successive passages, stem cells experience reduced multipotency, necessitating their use at lower passages in clinical applications. The gradual structural and functional decline of stem cells over time compromises their self-renewal ability and multipotency, thereby limiting the efficacy of stem cell-based therapies [5]. Stem cells sourced from primary human tissues exhibit a finite lifespan similar to somatic cells owing to the substantial cultivation challenges encountered during in vitro cultivation. Moreover, the availability of in vitro models to study tooth development and conduct comprehensive cell-based assays is limited. Cell lines, however, play a pivotal role in scientific research, serving as revolutionary assets for elucidating molecular mechanisms such as gain or loss of function, assessing drug cytotoxicity, producing antibodies or vaccines, and facilitating various cell-based assays. Establishing appropriate cell lines is a promising solution to the limitations associated with using primary cells. A cell line is a specific population of cells capable of indefinite propagation under in vitro culture conditions in appropriate media while retaining physiological attributes such as those of primary cells [6].
Various techniques exist for immortalizing cells, including chemical, physical, and ectopic telomerase expression. The most efficient approach involves the virus-mediated ectopic expression of the catalytic subunit of human telomerase reverse transcriptase (hTERT). The hTERT enzyme plays a pivotal role in maintaining telomere length, essential for a prolonged cell lifespan, as telomere shortening has been implicated in replicative senescence [7]. The introduction of human telomere reverse transcriptase via ectopic expression has gained widespread recognition as an effective method for cell line establishment. Lin et al. successfully established a Hutchinson–Gilford progeria fibroblast line via retrovirus-mediated hTERT transformation [8]. Multiple studies have shown that ectopic TERT expression provides substantial benefits compared to viral oncogenes.
Given the established safety and efficacy of telomerase-based approaches in cell line generation, we employed retroviral-mediated hTERT integration to immortalize stem cells from human deciduous teeth. We chose retroviral systems for hTERT transduction because of their ability to stably integrate the transgene into the host genome, ensuring long-term expression. In addition, they offer high transduction efficiency and sustained transgene expression with relatively low immunogenicity, making them ideal for achieving persistent telomerase activity. To the best of our knowledge, retrovirus-mediated ectopic expression in SHED has yet to be successfully accomplished, despite the abundance of available cell lines. The establishment of this cell line is essential for assessing the impact of retroviral-based hTERT immortalization on stemness attributes and growth characteristics. Moreover, establishing an immortalized SHED cell line may alleviate the challenge of cell scarcity in cell-based assays. The development of SHED-based cell lines is pivotal not only for deciphering the intricate molecular mechanisms underlying stem cell factors influencing attributes such as stemness and multipotency but also for elucidating the potential therapeutic applications of SHED in stem cell-based interventions.
Materials and Methods
Subjects Recruitment
Children aged 6–10 years who underwent exfoliated deciduous tooth extraction were recruited for this study. The samples were collected according to the inclusion and exclusion criteria. Exfoliating deciduous teeth that were indicated for extraction from normal healthy children and teeth with caries without pulpal involvement were included in the study, whereas children with special health care needs and teeth with necrotic pulp were excluded.
In the event of a scheduled procedure, a freshly extracted tooth was examined to verify the presence of viable dental tissue. A qualified dentist under standard anesthesia performed the tooth collection procedure under sterile conditions. Samples (n = 50) were procured following the National Stem Cell Guidelines, and written assent and consent were obtained from the children or legitimate representatives. This study was approved by the Institutional Committee of Stem Cell Research. IC-SCR/2023/05, and Biomedical and Health Research Ethics Committee (PGIDS/BHRC/24/10).
Stem Cell Isolation and Characterization
The exfoliated deciduous teeth designated for removal were collected in phosphate-buffered saline (PBS) supplemented with penicillin, streptomycin, and amphotericin B solution. Following collection, deciduous teeth were processed according to the protocol outlined by Kerkis and Caplan (2012) with minor modifications [9].
Briefly, the pulp tissue extracted from the teeth was mechanically minced into 2–3 mm pieces and cultured in a 6-well plate (30,006, Genetix) to initiate outgrowth. The culture was maintained in mesenchymal stem cell expansion medium (MSCEM) (AL519, HiMedia) supplemented with 10% fetal bovine serum (FBS) (RM1112, HI Media), antibiotics, and antimycotic solution (ABM) (A002, HiMedia). The medium was replenished every alternate day to facilitate cell growth and expansion. Isolated cells were characterized to determine their stem cell potential. For MSC assessment, disaggregated cells from explants were examined for cellular morphology and the expression of cell surface markers, such as cluster of differentiation (CD)-90, CD-105, and STRO-1. Additionally, the multilineage ability of the cells was explored by inducing their differentiation into osteocytes, chondrocytes, and adipocytes, which were subsequently stained with Alizarin Red S, Alcian Blue, and Oil Red O.
Transfection of Plasmids for Retrovirus Generation
The mammalian retroviral expression vectors pBabe-puro-hTERT (1771, Addgene) and pBabe-puro (1764, Addgene) were procured from Addgene (Cambridge, MA, USA) in bacterial stab cultures (DH5α strain), and the amphotropic packaging plasmid pCL-Ampho (NBP2-29541, Imgenex) was acquired from Imgenex. The plasmids were amplified in DH5α medium and selected using an ampicillin marker. Plasmid DNA was purified using a Qiagen plasmid purification kit (12,143) and its quantity and purity were verified using a NanoDrop analyzer. The amphotropic retroviral packaging plasmid pCL-Ampho produced mock control (pBabe-puro) and test (pBabe-puro-hTERT) retroviruses in the HEK293T cells.
One day prior to transfection, 2 × 105 HEK-293T cells were seeded in a 6-well plate containing Dulbecco’s modified Eagle’s medium (DMEM) (AL007S, HiMedia) supplemented with 10% fetal bovine serum (FBS) and ABM solution. Upon reaching 70% confluency, the cells were co-transfected with Lipofectamine™ Stem Transfection Reagent (STEM00001, Thermo Fisher Invitrogen). The transfection cocktail comprised 1.5 µg of packaging plasmid, 1.5 µg of expression plasmid (either test or mock control), P3000, and Lipofectamine™ Stem Transfection Reagent, all of which were suspended in Opti-MEM. Transfection was performed according to the manufacturer’s guidelines, with dropwise addition of the transfection cocktail. To mitigate potential toxic effects, 2 ml of fresh DMEM was added 24 h post transfection. Viral supernatants were collected 48 h after replenishment of fresh medium, followed by harvesting, centrifugation, and filtration using a 0.45 μm syringe filter (MG13045-CASL, Genetix). The filtered supernatant was preserved at -80 °C for subsequent transduction experiments.
Before transduction, we determined the optimal concentration of puromycin dihydrochloride for selecting stable transformation in NIH3T3 cells using the antibiotic kill curve method. Briefly, NIH3T3 cells (1.2 × 10^5 cells/well) were plated in a 6-well plate with DMEM medium. Upon reaching 70–80% confluency, the cells were treated with varying concentrations of puromycin (0, 1, 1.5, 2, 2.5, and 3 µg/ml) for two weeks, with medium changes every two days. Cell death was monitored daily, and the lowest concentration of puromycin that resulted in massive cell death within the first week was selected.
A standard gene transducing unit assay was performed to determine the viral titer of test and mock control. Briefly, 1 × 105 NIH 3T3 cells were seeded in 24-well plates and transduced with different dilutions of viral supernatant collected at 48 h (viz. 10− 4, 10− 3, 10− 2, and 10− 1 dilutions). Transduction was performed using 10 µg/ml Hexadimethrine Bromide (Polybrene). Following a 48-hour transduction period, cells were treated with 2.5 µg/ml puromycin (kill-curve method) to select transduced cells, until all the un-transduced cells were dead. The un-transduced SHED was used as control for the viral titer determination. Puromycin-resistant colonies were counted to determine colony-forming units (cfu/ml).
Transduction of SHED
Stem cells derived from deciduous teeth were cultured using MSCEM. Cells at passages three to six (P3–P6) were used to generate stable SHED-hTERT cells. We used the early passage of SHED owing to their enhanced stability, genetic consistency, reduced risk of cellular senescence and functional characteristics compared to higher passaged cells. One day prior to transduction, 1.5 × 105 SHED were seeded into 6-well culture dishes. Upon determination of the viral titer, SHED cells, approximately 60% confluent, were transduced with 48-hour viral supernatant at a dilution of 10− 3 from either the test (pBabe-puro-hTERT) or the mock control (pBabe-puro) using 10 µg/ml polybrene. Following a 48-hour transduction period, cells were treated with 2.5 µg/ml puromycin to select the transduced SHED until the un-transduced SHED cells were completely dead. Cells transduced with pBabe-puro and pBabe-puro-hTERT were labeled SHED-puro and SHED-hTERT cells, respectively, whereas non-transduced cells were referred to as SHED.
Characterization of the Transduced SHED
Cellular Morphology Evaluation
Cultured SHED-hTERT cells from passages 4 to 40 were analyzed to discern any alterations in cellular morphology. The cells were observed and compared with SHED-puro and SHED.
Detection of SHED-hTERT, SHED-puro, and SHED Cell Surface Markers
To assess the stemness of transduced cells, flow cytometry was performed using a panel of mesenchymal cell surface markers [10], including CD-90, CD105, and STRO-1, and compared with untransduced cells. Single-cell suspensions were washed with sheath fluid and subsequently incubated with fluorophore-tagged antibodies, CD-90-FITC (561969, BD Pharmingen™), CD105-APC (562408, BD Pharmingen™), and STRO-1-PE (MA5-28636, Thermo Fisher Scientific), using a flow cytometer (BD Biosciences), following the manufacturer’s guidelines. FlowJo software was used for data analysis.
Analysis of Multipotency of Transduced Cells
Mesenchymal stem cells are characterized by their potential to differentiate into multiple lineages [11]. The cultivated SHED-hTERT cells were cultured in a specialized medium that induced differentiation into osteocytes, adipocytes, and chondrocytes for a duration of three weeks to ascertain multilineage properties. Following differentiation, cells were stained using specific methods: Alizarin Red S (CCK030, HiMedia) for osteocytes, Oil Red O (CCK013, HiMedia) for adipocytes, and Alcian Blue (CCK013, HiMedia) for chondrocytes. The stained differentiated cells were observed using an inverted microscope.
Investigation of Growth Kinetics
Cell proliferation ability was assessed using various parameters, including population doubling level (PDL), population doubling time (PDT), [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] (MTT) assay, and scratch assay.
Population Doubling Level (PDL)
The population doubling level of cells was determined by counting the cells using trypan blue once they reached 70–80% confluency at each passage throughout the culture experiment. Population Doubling level (PDL) was calculated using the following formula.
\(\text{P}\text{D}\text{L}=\frac{\text{L}\text{o}\text{g}\text{N}\text{t}-\text{L}\text{o}\text{g}\text{N}0}{\text{L}\text{o}\text{g}2}\) Here, N0 represents the initial number of cells and Nt represents the final number of cells at a specific passage or time point.
Population Doubling Time (PDT)
PDT was evaluated using the modified protocol described by Kunimatsu et al., 2018) [12]. First, 1 × 103 cells were counted and seeded into a 12-well plate. After 48 h of incubation, cell quantification was performed using the trypan blue exclusion method across three independent samples. The quantification process was systematically repeated every 48 h for 20 successive passages, with triplicate evaluations performed at each time point. The arithmetic mean of the daily cell counts was plotted to generate a growth curve, facilitating the determination of the PDT. The following formula was employed to calculate the PDT, which was computationally ascertained using Assymetrex.
\(\text{P}\text{D}\text{T}=\text{t}-\text{t}\text{o}\times \frac{\text{l}\text{o}\text{g} \left(2\right)}{\text{l}\text{o}\text{g} (\text{N}\text{t} - \text{N}0)}\) ; where t-to represents the culture time duration, Nt represents the number of cells at a particular time point, and N0 represents the initial number of cells introduced into the culture.
Cell proliferation: To assess cell proliferation, we seeded 21 wells of a 96-well microtiter plate with a cell density of 0.3 × 104/well in MSCEM, and the cells were maintained for a week. Cell proliferation was assessed by MTT assay by incubating triplicate wells at each time point (days 1, 2, 3, 4, 5, 6, and 7 of culture) with MTT solution (Himedia, Cat: CCK003) for three hours. Following incubation, a solubilizing buffer was added to dissolve the formazan crystals. The optimal density was spectrophotometrically quantified according to the stipulated incubation period. The absorbance of the cells was measured at various time points, collated, and visually depicted to establish cellular growth trajectory.
Scratch Assay: To evaluate the migratory ability of transduced and un-transduced SHED, a scratch assay was conducted [13]. An optimal seeding density of 1 × 105 cells per well was maintained in a 24-well plate to achieve approximately 70% confluence, ensuring optimal conditions for the scratch assay. Subsequently, a linear scratch was created using a 200 µl pipette tip. The dislodged cells and debris were removed by aspiration and replenished with fresh medium. Systemic cell observations were conducted at 24 and 48-hours intervals post-scratch using an inverted microscope to evaluate the migratory potential of the cells. The wound closure rate, indicative of cell migration effectiveness, was computed using the following formula: \(\frac{{(At = 0h - At = \Delta h)}}{{{\rm{At}} = \Delta {\rm{h}}}} \times 100\%\), where At = 0 h represents the initial area and At = Δh represents the area covered at a given time point. The cell migration rate was calculated using the equation: \(\frac{Wi-Wf}{t}\). where Wi and Wf denotes the initial and final scratch widths, respectively, and t represents time.
Quantitative Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR) Analysis of the hTERT Gene
Total RNA was isolated from SHED-hTERT and SHED-puro cells using a hybrid method involving TRIzol and a kit, in accordance with the manufacturer’s instructions. Reverse transcription of 100 ng RNA was performed using the iScriptTM cDNA Synthesis Kit (1,708,891; Bio-Rad) in accordance with the manufacturer’s protocol. hTERT expression was analyzed by qRT-PCR using a Bio-Rad CRX Maestro thermal cycler and the iTaq SYBR Green (1725120 Bio-Rad) detection method. The qRT-PCR protocol included an initial denaturation step at 95 °C;30 s, primer annealing at 59.2o C; 30 s, and a final extension at 72 °C ;30 s. Primers utilized were as follows: Forward Primer (F): CTACGGCGACATGGAGAAC; Reverse Primer (R): GACACTTCAGCCGCAAGAC for hTERT; Forward Primer (F): GTGGGGCGCCCCAGGCACCA; Reverse Primer (R): CTCCTTAATGCACGCACGATTTC for β-actin [14]. The cycle threshold (Ct) values were estimated, and the mRNA expression levels of the hTERT genes were normalized using the housekeeping β-actin gene. The relative gene expression was estimated using the 2–ΔΔCt method.
Short Tandem Repeat Analysis
DNA extraction from SHED, SHED-puro, and SHED-hTERT cells was performed manually. Subsequently, amplification of 16 short tandem repeat (STR) loci were done by AmpFISTR Identifier Plus PCR Kit (Applied Biosystems) following the manufacturer’s guidelines. Subsequently, samples were analyzed by Applied Biosystems 3500 Genetic Analyzer. The acquired raw data was further processed and interpreted using GeneMapper ID-X software.
Evaluation of Cyclin-dependent Kinase Inhibitor 2 A (CDKN2A) for Genome Integrity Assessment
To estimate the effect of hTERT on cell cycle regulation and potential carcinogenicity, the genomic DNA of SHED, SHED-puro, and SHED-hTERT cells was isolated using a genomic isolation kit (2115500031730, Genei) according to the manufacturer’s instructions. CDKN2A amplification was performed using the following primers: forward (F): CCTGGCTCTGACCATTCTGTTC and Reverse (R): GCTTTGGAAGCTCTCAGGGTAC [15]. Additionally, the housekeeping gene β-actin was amplified using the following primer sequences: forward (F), GTGGGGCGCCCCAGGCACCA; Reverse (R), CTCCTTAATGCACGCACGATTTC. PCR reactions consisted of 25 µl of PCR master mix (PGK017-A; Puregene), 0.5 µM of each forward and reverse primer, and 100 ng of target DNA. The reactions were incubated in a Bio-Rad thermal cycler with the following cycling parameters: initial denaturation at 95 °C for 3 min, followed by denaturation at 95 °C–30 s, annealing at 57 °C (CDKN2A) and 56 °C (β-actin) − 30 s, and extension at 70 °C–1 min, with a final extension at 72 °C–10 min. Subsequently, the amplified CDKN2A (386 bp) and β-actin (500 bp) genes were separated by electrophoresis on 2% agarose gel (Lonza, Basel, Switzerland) and visualized using a Bio-Rad XR+ gel documentation system.
Statistical Analysis
A significance threshold of p < 0.05 was set, and the data are shown as mean ± standard deviation. Kruskal-Wallis and nonparametric one-way ANOVA analyses were performed using PRISM software (GraphPad Software, San Diego, CA, USA) to estimate the statistical significance of the differences between groups. Flow cytometry data were analyzed using FlowJo version 10 (BD Biosciences).
Results
SHED Have Fibroblast-Like Morphology and are Identical to Mesenchymal Stem Cells
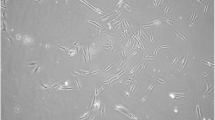
In the present study, we employed an explant-based method to procure SHED, achieving successful isolation of SHED regardless of the degree of tooth root reabsorption. The cellular disintegration process of the explants occurred over a period of 15 ± 8 days, initiating at the periphery once they were attached to the culture vessel (Fig. 1A). This timeline of cell outgrowth from explants aligns with the existing data [16, 17].
SHED isolation and Mesenchymal stem cells characterization. A: Illustrates the outgrowth of deciduous tooth pulp cells from the explant on day 8. B: Represented the fibroblast-like morphology of sub-cultured cells stained with giemsa (P-2). C: The image demonstrates the osteocytes (Alizarin Red-S), chondrocytes (Alcan-blue) and adipocytes (Oil-Red-O), depicting the multi-potency of SHED, after 21 days of in-vitro differentiation medium (left to right). D: The image illustrates the gating of SHED; upper panel represent un-stained cells while lower panel represent stained cells with CD-90-FITC, CD-105-APC, and STRO-1-PE. E: The graph represents the percentage of positive population of SHED (CD-90, CD-105, and STRO-1) at P-4. The data is presented as mean ± standard deviation (SD) based on three replicates (n = 3). Cellular observations were performed under a phase contrast inverted microscope, and images were captured at a magnification of 10X and 100X, with a scale bar of 100 μm
SHED, recognized as mesenchymal stem cells, displayed fibroblast-like morphology (Fig. 1B), consistent with previous research [1]. Notably, after cultivating SHED for three weeks in specialized differentiation media, our analysis confirmed their ability to differentiate into osteogenic, chondrogenic, and adipogenic lineages, as depicted in Fig. 1C. Staining of the cultured cells with Alizarin Red S, Alcian Blue, and Oil Red O corroborated the presence of red-stained mineralized nodules in osteocytes, blue-stained proteoglycans in chondrocytes, and red-stained fat globules in round adipocytes. Flow cytometry analysis revealed that 87.5%, ± 7.14, 91.4%, ± 3.90 and 9.68 ± 5.63% of the population were positive for the human mesenchymal stem cell markers CD-90, CD-105, and STRO-1, respectively. (Fig. 1D).
Ectopic Expression of hTERT in SHED Does Not Alter Cellular Morphology
SHED were transduced with either the pBabe-puro-hTERT retrovirus or a mock control (pBabe-puro) after assessing viral titration in NIH3T3 cells (Fig. 2A and Supplementary Table 1). Following transduction, SHED-hTERT and SHED-puro underwent a nine-day selection process with 2.5 µg/ml puromycin, as the untransduced cells underwent apoptosis (Fig. 2B). Consistent with previous reports, SHED, a mesenchymal stem cell, exhibited fibroblast-like morphology [1]. Transduced cells displayed morphology similar to that of fibroblasts, with no discernible changes in physical characteristics observed in either SHED-hTERT or SHED-puro cells (Fig. 2C). Notably, SHED-puro cells displayed altered cellular morphology at passage 20, whereas SHED-hTERT-transformed cells maintained stable cellular morphology until passage 40 (Fig. 2C).
Viral titration, SHED transduction and phenotypic alteration:
A: Titration of retroviral (supernatant of 48 h.) at different dilutions: (a) un-transduced, (b)10− 4, (c) 10− 3, (d) 10− 2, and (e)10− 1 on NIH3T3 cell line. The cells were treated with 2.5 µg/ml puromycin until the cells in the un-transduced well were dead, and images were captured at day 9 after 0.1% crystal violet staining of colonies
B: Puromycin selection at day 9 (a). Debris in un-transduced SHED; (b). Puromycin-resistant transduced SHED-puro cells(c). Puromycin-resistant transduced SHED-hTERT cells
C: (a) SHED-puro and (c) SHED-hTERT cells exhibit fibroblast-like morphology (post-transduction P-3). (b) Morphological alteration at P-20 (day 70) in SHED-puro (d); no morphological alteration at P-20 in SHED-hTERT (e); minimal morphological alteration at P-40 (day 170) in SHED-hTERT. Cellular observations were performed under a phase-contrast inverted microscope, and images were captured at a magnification of 10X, with a scale bar of 100 μm
Transduced SHED- hTERT Cells Have an Extended Lifespan and Proliferative Ability
To evaluate the effect of hTERT, we examined the proliferative potential of SHED-hTERT and SHED-puro cells using the MTT assay, cell proliferation test, and cell counting (trypan blue method) at each passage. The cell growth curve of the population doubling level showed an increase in the proliferative ability of the hTERT-transformed cells (Fig. 3A). SHED-puro cells underwent 33 population-doubling cycles over 70 days. Thereafter, the cells developed cellular senescence, with modification in cellular morphology and cessation of cell division. In contrast to SHED-puro cells, SHED-hTERT-transduced cells underwent 100 population doubling cycles over a 170-day culture period prior to cryopreservation. Population doubling times differed significantly between the SHED-Puro and SHED-hTERT cohorts, with PDT efficacy of 25.57± 0.8 and 24.89 ± 0.60 respectively (Fig. 3B). Concurrently, the results of the MTT assay performed on P-3 SHED-hTERT cells for seven consecutive days showed a significant increase in cell proliferation compared to SHED-puro (Fig. 3C).
Assessment of the growth kinetics of transduced SHED
A: Population doubling levels of SHED (70 days/P-19), SHED-puro (70 days/P-20) and SHED-hTERT (170 days/P-40)
B: Mean value of the population doubling time of SHED (P1-P19), SHED-puro (P-1 to P-20) and SHED-hTERT (P-1 to P-20)
C: Cell growth curve of SHED-hTERT with respect to SHED-puro cells and SHED at P-3
D: The migration potential of SHED-hTERT cells in the scratch assay after 24 and 48 h. of cell culture
E: The plot depicts the areas covered by SHED, SHED-hTERT, and SHED-puro after 24 and 48 h of the scratch assay. The experiments were conducted in triplicate (n = 3), and the data are represented as the mean ± standard deviation (SD) with a p-value (P < 0.05) determined by performing the Kruskal–Wallis test. Cellular observations were performed under a phase-contrast inverted microscope, and images were captured at a magnification of 10X, with a scale bar of 100 μm
The migration potential of SHED-puro and SHED-hTERT cells was assessed using the scratch assay. The cell migration rate in SHED-puro was 3.9 mm/hr, with the ability to cover the half-scratched region after 25.47 h., and 94.2% of the area was covered after 48 h. In contrast to SHED-puro cells, SHED-hTERT cells exhibited a higher migratory potential (4.1 μm/hr. The half-scratched area was covered after 24.4 h., and the extra area was covered after 48 h (98.2%) (Fig. 3D and E).
Stemness and Multipotency Maintenance in SHED-hTERT Cell Line
The assessment of cell surface markers and trilineage differentiation highlighted the ability of SHED-hTERT cells to preserve stem cell characteristics and differentiation in various cell types. Analysis of the MSC surface markers CD-90, CD-105, and STRO-1 in SHED-hTERT and SHED-puro cells showed no significant alterations compared to those in SHED. As depicted in Fig. 4A, the SHED-hTERT population exhibited positivity rate of 87.1 ± 0.80% for CD-90, 90.46± 3.03% for CD-105, and 11.3 ±0.51% for STRO-1, while SHED-puro cells showed positivity rates of 87.23 ± 3.57, 89.36 ± 7.62% and 9.96 ± 1.97% for the CD-90, CD-105, and STRO-1 populations, respectively. Figure 4B depicts cell gating and the positive population of stained cells.
Mesenchymal stem cells surface markers evaluation and trilineage potential of the generated SHED-hTERT:
A: Histogram of stained cells, viz., CD-90-FITC, CD-105-APC, and STRO-1-PE: Left panel, SHED-puro and Right panel, SHED-hTERT.
B: Percentage of MSC cell surface markers viz. CD-90, CD-105, and STRO-1, on SHED, SHED-puro (P-6), and SHED-hTERT (P-6)
C: SHED-hTERT trilineage differentiation in (a) chondrocytes (Alcian blue), (b) osteocytes (Alizarin Red S), and (c) adipocytes (Oil Red O)
Data are presented as the mean ± standard deviation (SD) based on three replicates (n = 3). One-way analysis of variance (ANOVA) was performed to analyze significant differences between groups, with p-values indicated. Cellular observations were performed under a phase-contrast inverted microscope, and images were captured at a magnification of 10X, with a scale bar of 100 μm
The multipotent potential of SHED in osteogenic, odontogenic, adipogenic, neurogenic, and chondrogenic differentiation has been extensively documented [18]. Our study further confirmed the osteogenic, chondrogenic, and adipogenic differentiation capabilities of SHED-hTERT cells after three weeks of cultivation in specific differentiation media. The chondrogenic capability of SHED was confirmed by blue staining, which indicated the presence of proteoglycans (Fig. 4C (a)). Furthermore, Alizarin Red S staining revealed calcium carbonate and phosphate deposits, as evidenced by the red staining of cells in Fig. 4C (b). The presence of round adipocytes filled with fat globules was validated by Oil Red O staining, as illustrated in Fig. 4C (c). These findings indicate that SHED-hTERT cells maintain stemness and multipotency similar to primary SHED cells.
Increased hTERT Expression without Affecting CDKN2A Gene
We assessed the integrity of genomic cyclin-dependent kinase inhibitor 2 A (CDKN2A), which encodes the INK4 family (p16INK4a) and p14arf protein. To analyze the effect of hTERT integration, genomic DNA was extracted from SHED, SHED-puro, and SHED-hTERT cells and subjected to PCR amplification targeting the CDKN2A locus. Gel electrophoresis following PCR amplification showed bands of the anticipated size, with regular band intensity (Fig. 5A). To assess alterations in hTERT expression in genetically modified cells, hTERT mRNA levels were quantified at different stages of cell culture using quantitative polymerase chain reaction (qPCR) with specific hTERT primers. The results demonstrated a statistically significant upregulation of hTERT expression, showing a seven-fold change (p < 0.005) in transduced SHED-hTERT cells compared to that in SHED-puro cells (Fig. 5B).
Evaluation of CDKN2A and hTERT gene expression:
A: PCR amplification of CDKN2A (cyclin-dependent kinase inhibitor 2 A) implies that there is no evidence of disruption or alteration in CDKN2A as a consequence of hTERT integration
B: The graph illustrates the fold change of the hTERT gene in SHED-puro and SHED-hTERT cells, analyzed by qRT-PCR.
The data are represented as the mean ± standard deviation (SD), with a p-value (P < 0.05) determined by performing the t-test
STR Analysis Authenticates SHED-hTERT Cell Line
Short tandem repeat (STR) analysis is a cost-effective and reliable method for authenticating cell lines and identifying potential cross-contaminations. Using a panel of 16 STR loci, the retroviral-mediated SHED-hTERT was validated and compared with that of SHED-puro and un-transduced SHED. The results revealed that all three cell types, SHED, SHED-puro, and SHED-hTERT, exhibited homozygosity for 15 STR loci and were heterozygous for the CSF1PO locus. Notably, a consistent allele pattern was observed across 15 STR loci, and both SHED-puro and SHED-hTERT displayed distinct deletions of allele 13 at the CSF1PO locus, in contrast to the Tri allelic pattern observed in the untransduced SHED. Although the absence of CSF1PO in both SHED-puro and SHED-hTERT cells suggests a deletion, this alteration was unrelated to hTERT integration (Supplementary Fig. 1 and Supplementary Table 2).
Discussion
This study aimed to investigate the effect of ectopic hTERT expression on SHED using a retrovirus-mediated approach. SHED, which are found in the residual pulp of exfoliated teeth, have been well-documented for their multipotent stem cell properties and regenerative potential. The extensive expansion of SHED is required to assess their efficiency and efficacy in clinical trials and cell-based assays under in vitro culture conditions. However, challenges, such as clonal variability, multipotency, and cellular senescence under culture conditions, hinder their expansion [4, 5, 19]. Transformation techniques, including cell-line creation, have been widely used to overcome these limitations. Numerous techniques have been used for cell immortalization including chemical transduction, simian virus 40-mediated gene expression, and lentiviral- or retrovirus-mediated ectopic hTERT production. Unlike other methods that may lead to chromosomal changes, ectopic hTERT expression preserves normal chromosomal configuration [20]. Human telomerase reverse transcriptase is a specialized ribonuclear protein comprising two crucial components, an integral RNA template and a reverse transcriptase enzyme responsible for telomere synthesis [7]. Telomerase activity is gradually repressed during cell development, contributing to cellular senescence [4]. Elevation of telomerase activity induces upregulation of hTERT mRNA, leading to enhanced proliferative capacity and delayed cell senescence, offering promising implications for SHED expansion and clinical applications [21, 22].
In our study, we used retrovirus-mediated ectopic expression of human telomerase in SHED to achieve sustained telomerase integration. Our findings revealed a substantial effect of hTERT expression on the longevity of primary-cultivated SHED, which was accompanied by enhanced cell proliferation and prevention of senescence. Based on previous studies by Gao et al. and Yin et al., which immortalized human periodontal ligament stem cells and SHED, respectively, via ectopic hTERT expression [15, 21], we successfully cultured SHED-hTERT cells for 40 passages and cryopreserved them for future use. Transduced SHED demonstrated robust proliferative capacity owing to ectopic expression of the hTERT component, which remained stable even in subsequent passages. Furthermore, our investigation highlighted the migratory potential of these cells, with the SHED-hTERT cells exhibiting a statistically significant (p > 0.005) increase in migration compared with the SHED and SHED-puro cells.
Ectopic expression of the hTERT catalytic subunit, known for its role in cell immortalization, has been shown to maintain multipotency, cellular morphology, and the expression of cell surface markers characteristic of stem cells [23, 24]. Previous studies have shown that ectopic expression of hTERT in dental pulp stem cells, mesenchymal stem cells, and periodontal ligament stem cells preserves the morphology and multipotency of the immortalized cells [21, 23,24,25]. Consistent with these findings, our research demonstrated that SHED transduced with hTERT exhibited fibroblast-like morphology similar to un-transduced SHED and retained the ability to differentiate into chondrocytes, osteocytes, and adipocytes when cultured in a specific differentiation medium. We observed that differentiated chondrocytes contained proteoglycans and adipocytes with lipid droplets, and osteocytes contained calcium and phosphate deposits. Furthermore, immortalized SHED-hTERT cells displayed expression of mesenchymal cell surface markers, including STRO-1, CD-105, and CD-90, similar to primary SHED cells, with no discernible differences in morphology, surface marker expression, or multipotency among SHED-hTERT, SHED-puro, and SHED. These findings highlight the potential of hTERT-mediated immortalization to maintain the key qualities of SHED for a range of applications in regenerative medicine.
The Cellosaurus database reported 1137 instances of misidentified cell lines [26], emphasizing the importance of accurate cell line characterization. Characterization of the DNA hypervariable regions confirmed the single donor origin of a cell line, and a variable number of tandem repeats in the hypervariable regions’ functions in DNA fingerprinting [27]. Short tandem repeat (STR) analysis is a universally accepted and robust forensic technique that is used to authenticate human cell lines. During the culture process, intra- or inter-species cross-contamination is a potential risk for generating immortalized cell lines, particularly when co-cultured cells have similar cellular morphologies [28]. Therefore, STR profiling is recommended for the validation of cell lines. In our study, STR profiling of transduced SEHD confirmed their genetic integrity and revealed allele profiles closely resembling those of primary SHED, with minimal discrepancies. SHED-hTERT cells had a single deletion of allele 13 in CSF1PO. Nevertheless, identical deletions were detected in SHED-puro, thereby excluding the involvement of hTERT-mediated deletions at the STR loci. Cyclin-dependent kinase inhibitor 2 A (CDKN2A) encodes the INK4 family, comprising the p16INK4a and p14arf proteins, and its deletion indicates the carcinogenic potential of transduction. Although previous studies, including Serakinchi et al. (2004), reported that hMSC-TERT4 acquired a KRAS mutation and deletion at the Ink4a/ARF locus in both hMSC-TERT4- and hMSC-TERT20-generated cell lines [29], we did not identify any deletions in CDKN2A, suggesting that hTERT transduction did not cause carcinogenesis.
While previous studies have illustrated the effectiveness of viral-mediated hTERT expression in achieving cell immortality [15, 21, 22, 25], our research offers a distinctive approach by utilizing retroviruses to express hTERT, which provides advantages such as stable integration, a broader host range, and ease of production [30,31,32]. Although retroviral vectors are commonly used for gene expression, there is a lack of comprehensive literature on retrovirus-mediated hTERT expression, which underscores the novelty and significance of our study.
Conclusion
In summary, we successfully established a stem cell line derived from human exfoliated deciduous teeth (SHED) via retrovirus-mediated ectopic expression of the catalytic subunit of human telomerase reverse transcriptase (hTERT). Immortalized SHED-hTERT cells exhibited fibroblast-like morphology and enhanced hTERT gene expression, accompanied by accelerated population doubling time, increased cell proliferation rate, and improved migratory capacity compared with SHED-puro cells.
Importantly, our extensive characterization revealed that SHED-hTERT maintained stemness and multipotency in primary cells, as evidenced by the similar trilineage differentiation potential and expression of mesenchymal stem cell surface markers. Short Tandem Repeat (STR) profiling demonstrated genetic stability in transduced cell lines. Furthermore, no notable alteration in CDKN2A expression was observed, suggesting a minimal carcinogenic risk associated with retroviral transduction. These findings emphasize the safety characteristics of immortalized SHED-hTERT cells and provide support for their prospective use in regenerative medicine.
Overall, our study underscored the stability and safety of retrovirally transduced and immortalized SHED-hTERT cells by demonstrating that they enhance hTERT expression while retaining stemness, multipotency, and growth characteristics similar to those of primary cells, highlighting their suitability for use in various research and therapeutic applications and emphasizing their stability and safety in the context of continuous cell line growth.
Data Availability
All relevant data generated or analyzed during this study are included in the article.
Abbreviations
- SHED:
-
Stem cells from human-exfoliated deciduous teeth
- MSCs:
-
Mesenchymal stem cells
- hTERT:
-
Human telomerase reverse transcriptase
- PBS:
-
Phosphate buffer saline
- MSCEM:
-
Mesenchymal stem cell expansion medium
- FBS:
-
Fetal bovine serum
- ABM:
-
Antibiotic and antimycotic solution
- CD:
-
Cluster of differentiation
- PDL:
-
Population doubling level
- PDT:
-
Population doubling time
- MTT:
-
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
- qPCR:
-
Quantitative polymerase chain reaction
- STR:
-
Short tandem repeat
- CDKN2A:
-
Cyclin-dependent kinase inhibitor 2 A
References
Miura, M., Gronthos, S., Zhao, M., Lu, B., Fisher, L. W., Robey, P. G., & Shi, S. (2003). SHED: Stem cells from human exfoliated deciduous teeth. Proceedings of the National Academy of Sciences of the United States of America, 100(10), 5807–5812. https://doi.org/10.1073/PNAS.0937635100/ASSET/C629822E-B3FE-4064-8133-F9F737084D0F/ASSETS/GRAPHIC/PQ0937635006.JPEG
Yadav, P., Vats, R., Bano, A., Namdev, R., & Bhardwaj, R. (2023). Ameliorative potential of stem cells from human exfoliated deciduous teeth (SHED) in preclinical studies: A meta-analysis. Regenerative Therapy, 24, 117–134. https://doi.org/10.1016/j.reth.2023.06.004
Alge, D. L., Zhou, D., Adams, L. L., Wyss, B. K., Shadday, M. D., Woods, E. J., & Goebel, W. S. (2010). Donor-matched comparison of dental pulp stem cells and bone marrow-derived mesenchymal stem cells in a rat model. Journal of Tissue Engineering and Regenerative Medicine, 4(1), 73–81. https://doi.org/10.1002/term.220
Liu, J., Ding, Y., Liu, Z., & Liang, X. (2020). Senescence in mesenchymal stem cells: Functional alterations, Molecular mechanisms, and rejuvenation strategies. Frontiers in Cell and Developmental Biology. https://doi.org/10.3389/FCELL.2020.00258/FULL. 8.
Zhou, X., Hong, Y., Zhang, H., & Li, X. (2020). Mesenchymal stem cell senescence and rejuvenation: Current Status and challenges. Frontiers in Cell and Developmental Biology. https://doi.org/10.3389/FCELL.2020.00364/FULL. 8.
Ulrich, A. B., & Pour, P. M. (2001). Cell lines. Brenner’s Encyclopedia of Genetics: Second Edition, 481–482. https://doi.org/10.1016/B978-0-12-374984-0.00212-6
Cong, Y. S., Wright, W. E., & Shay, J. W. (2002). Human telomerase and its regulation. Microbiology and Molecular Biology Reviews, 66(3), 407. https://doi.org/10.1128/MMBR.66.3.407-425.2002
Lin, H., Mensch, J., Haschke, M., Jäger, K., Köttgen, B., Dernedde, J., & Walter, M. (2022). Establishment and characterization of hTERT Immortalized Hutchinson-Gilford Progeria Fibroblast Cell lines. Cells, 11(18). https://doi.org/10.3390/CELLS11182784
Kerkis, I., Caplan, A. I., & Reviews (2012). 18(2), 129. https://doi.org/10.1089/TEN.TEB.2011.0327
Dominici, M., Le Blanc, K., Mueller, I., Slaper-Cortenbach, I., Marini, F. C., Krause, D. S., & Horwitz, E. M. (2006). Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy, 8(4), 315–317. https://doi.org/10.1080/14653240600855905
Zhang, N., Chen, B., Wang, W., Chen, C., Kang, J., Deng, S. Q., & Han, F. (2016). Isolation, characterization and multi-lineage differentiation of stem cells from human exfoliated deciduous teeth. Molecular Medicine Reports, 14(1), 95–102. https://doi.org/10.3892/mmr.2016.5214
Kunimatsu, R., Nakajima, K., Awada, T., Tsuka, Y., Abe, T., Ando, K., & Tanimoto, K. (2018). Comparative characterization of stem cells from human exfoliated deciduous teeth, dental pulp, and bone marrow-derived mesenchymal stem cells. Biochemical and Biophysical Research Communications, 501(1), 193–198. https://doi.org/10.1016/J.BBRC.2018.04.213
Martinotti, S., & Ranzato, E. (2020). Scratch wound healing assay. Methods in Molecular Biology, 2109, 225–229. https://doi.org/10.1007/7651_2019_259/COVER
Yadav, P., Vats, R., Bano, A., Vashishtha, A., & Bhardwaj, R. (2022). A Phytochemicals Approach towards the treatment of Cervical Cancer using polyphenols and flavonoids. Asian Pacific Journal of Cancer Prevention, 23(1), 261–270. https://doi.org/10.31557/APJCP.2022.23.1.261
Yin, Z., Wang, Q., Li, Y., Wei, H., Shi, J., & Li, A. (2016). A novel method for banking stem cells from human exfoliated deciduous teeth: Lentiviral TERT immortalization and phenotypical analysis. Stem cell Research & Therapy, 7(1). https://doi.org/10.1186/S13287-016-0309-0
Bhandary, M., Rao, S., Shetty, A. V., Kumar, B. M., Hegde, A. M., & Chhabra, R. (2021). Comparison of stem cells from human exfoliated deciduous posterior teeth with varying levels of root resorption. Stem Cell Investigation, 8. https://doi.org/10.21037/SCI-2020-039
Suchánek, J., Visek, B., Soukup, T., El-Din Mohamed, S. K., Ivancaková, R., Mokrỳ, J., & Omran, A. (2010). Stem cells from human exfoliated deciduous teeth–isolation, long term cultivation and phenotypical analysis. Acta Medica (Hradec Kralove), 53(2), 93–99. https://doi.org/10.14712/18059694.2016.66
Guo, R., & Yu, J. (2022). Multipotency and Immunomodulatory benefits of stem cells from human exfoliated deciduous teeth. Frontiers in Dental Medicine, 3, 805875. https://doi.org/10.3389/FDMED.2022.805875/BIBTEX
Smith, J. R., & Whitney, R. G. (1980). Intraclonal variation in proliferative potential of human diploid fibroblasts: Stochastic mechanism for cellular aging (Vol. ,207 N.Y.), 207, pp. 82–84). Science (New York. 4426https://doi.org/10.1126/SCIENCE.7350644
Xie, F., Gong, K., Li, K., Zhang, M., Chang, J. C., Jiang, S., & Kan, Y. W. (2016). Reversible immortalization enables Seamless Transdifferentiation of primary fibroblasts into other Lineage cells. Stem Cells and Development, 25(16), 1243–1248. https://doi.org/10.1089/SCD.2016.0035
Gao, L., Zhao, F., Jiang, W., Niu, C., Yuan, K., Lin, W., & Huang, Z. (2019). Immortalization of human periodontal ligament stem cells by transferring human telomerase reverse transcriptase gene. Frontiers of Oral and Maxillofacial Medicine, 1(0), 6. https://doi.org/10.21037/FOMM.2019.09.02
Smith, M. C., Goddard, E. T., Lanfranca, P., M., & Davido, D. J. (2013). hTERT extends the life of human fibroblasts without compromising type I Interferon Signaling. Plos One, 8(3). https://doi.org/10.1371/JOURNAL.PONE.0058233
Moffatt-Jauregui, C. E., Robinson, B., de Moya, A. V., Brockman, R. D., Roman, A. V., Cash, M. N., & Lamont, R. J. (2013). Establishment and characterization of a telomerase immortalized human gingival epithelial cell line. Journal of Periodontal Research, 48(6), 713–721. https://doi.org/10.1111/JRE.12059
Huang, G., Zheng, Q., Sun, J., Guo, C., Yang, J., Chen, R., & Wang, J. (2008). Stabilization of cellular properties and differentiation mutilpotential of human mesenchymal stem cells transduced with hTERT gene in a long-term culture. Journal of Cellular Biochemistry, 103(4), 1256–1269. https://doi.org/10.1002/JCB.21502
Skårn, M., Noordhuis, P., Wang, M. Y., Veuger, M., Kresse, S. H., Egeland, E. V., & Myklebost, O. (2014). Generation and characterization of an immortalized human mesenchymal stromal cell line. Stem Cells and Development, 23(19), 2377. https://doi.org/10.1089/SCD.2013.0599
Bairoch, A. (2018). The Cellosaurus, a cell-line knowledge resource. Journal of Biomolecular Techniques: JBT, 29(2), 25–38. https://doi.org/10.7171/JBT.18-2902-002
Nakamura, Y., Leppert, M., O’Connell, P., Wolff, R., Holm, T., Culver, M., & White, R. (1987). Variable number of tandem repeat (VNTR) markers for human gene mapping (Vol. ,235 N.Y.), 235, pp. 1616–1622). Science (New York. 4796https://doi.org/10.1126/SCIENCE.3029872
Lilly, E., Almeida, J. L., & Korch, C. T. (2004). Authentication of Human and Mouse Cell Lines by Short Tandem Repeat (STR) DNA Genotype Analysis. Retrieved from https://www.ncbi.nlm.nih.gov/books/
Serakinci, N., Guldberg, P., Burns, J. S., Abdallah, B., Schrødder, H., Jensen, T., & Kassem, M. (2004). Adult human mesenchymal stem cell as a target for neoplastic transformation. Oncogene, 23(29), 5095–5098. https://doi.org/10.1038/SJ.ONC.1207651
Daniel, R., & Smith, J. A. (2008). Integration site selection by retroviral vectors: Molecular mechanism and clinical consequences. Human gene Therapy, 19(6), 557–568. https://doi.org/10.1089/HUM.2007.148
Sandrin, V., Russell, S. J., & Cosset, F. L. (2003). Targeting retroviral and lentiviral vectors. Current Topics in Microbiology and Immunology, 281, 137–178. https://doi.org/10.1007/978-3-642-19012-4_4
Kurian, K. M., Watson, C. J., & Wyllie, A. H. (2000). Retroviral vectors. Molecular Pathology, 53(4), 173. https://doi.org/10.1136/MP.53.4.173
Funding
We thank the Centre for Medical Biotechnology, Maharshi Dayanand University, Rohtak, and Council of Scientific & Industrial Research (CSIR) Pusa, New Delhi, for providing financial support for this work.
Author information
Authors and Affiliations
Contributions
PY set up the experiments, analyzed and interpreted the data, collected the literature, and drafted, designed, wrote, and edited the manuscript. RV, AB, SW, RN, and MG edited the manuscript. RB conceptualized the outline and article topic, designed the study, and edited the manuscript for submission. All authors have read and approved the final manuscript. All authors have read and approved the manuscript for submission.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
This study was approved by the Institutional Committee for Stem Cell Research (vide letter no. IC-SCR/2023/05), and the Biomedical and Health Research Ethics Committee (vide letter no. PGIDS/BHRC/24/10).
Consent to Participate
The consent of the participants was obtained according to the ethical guidelines.
Consent for Publication
Consent for publication was obtained from the participants through an ethical consent and assent form.
Competing Interests
The authors declare no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1:
The figure represented the short tandem repeat analysis of SHED, SHED-puro, and SHED-hTERT to elucidate the contamination of other cells with established cell-line.
Supplementary Table 1: Viral Titer of mock and test was determined by counting puromycin-resistant colonies.
Supplementary Table 2: STR profiles of 16 short tandem repeat loci analyzed in SHED, SHED-puro, and SHED-hTERT. The result depicted out of the 16 loci, 15 loci shared similarities, while CSF1PO depicted one missing allele in transduced cells with respect to un-transduced cells.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yadav, P., Vats, R., Wadhwa, S. et al. Enhancing Proliferation of Stem Cells from Human Exfoliated Deciduous Teeth (SHED) through hTERT Expression while Preserving Stemness and Multipotency. Stem Cell Rev and Rep (2024). https://doi.org/10.1007/s12015-024-10746-y
Accepted:
Published:
DOI: https://doi.org/10.1007/s12015-024-10746-y