Abstract
ADP ribosylation factor guanylate kinase 1 (ASAP1), a key protein regulating cell migration and invasion, has attracted extensive attention in oncological research in recent years. This study aims to explore the effects of ASAP1 inhibition on lung cancer metastasis and its potential mechanisms, particularly how it modulates the tumor immune microenvironment through the p-STAT3 signaling pathway. In this study, shRNA technology was employed to specifically inhibit ASAP1 expression in lung cancer cell lines A549, NCI-H1299, and PC-9. The effects of ASAP1 inhibition on lung cancer cell viability, apoptosis, migration, and invasion were evaluated using CCK-8, TUNEL apoptosis detection, and cell migration and invasion assays. Furthermore, animal experiments were conducted to assess the in vivo effects of ASAP1 inhibition on lung cancer metastasis, and immunohistochemical analysis was performed to investigate changes in immune cells in lung metastasis models, further exploring its impact on the tumor immune microenvironment. The experimental results demonstrated that ASAP1 inhibition significantly reduced lung cancer cell viability, induced apoptosis in A549, NCI-H1299, and PC-9 cells, and suppressed the migration and invasion abilities of these cells. In vivo experiments revealed that ASAP1 inhibition effectively suppressed lung cancer metastasis and altered the tumor immune microenvironment by regulating immune cells. Moreover, we found that ASAP1 inhibition could decrease tumor cell proliferation and induce tumor apoptosis in lung metastasis models by inhibiting the p-STAT3 signaling pathway. This study confirms that ASAP1 inhibition can suppress lung cancer metastasis by modulating the tumor immune microenvironment through the inhibition of the p-STAT3 signaling pathway. These findings provide new targets for lung cancer treatment and a theoretical basis for developing novel strategies against lung cancer metastasis. Future research will further explore the mechanisms of ASAP1 in lung cancer metastasis and how to optimize treatment strategies for lung cancer patients by targeting ASAP1.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the current field of medical research, the treatment and prevention of lung cancer remain pivotal areas of study [1, 2]. Lung cancer is one of the most common cancers worldwide, with persistently high mortality rates primarily due to its propensity for metastasis [3, 4]. The metastasis of lung cancer is a complex, multi-step process involving the invasion, migration, and colonization of tumor cells at distant organs [5]. Recent advances in molecular biology and cell biology have increasingly recognized the significant role of the tumor microenvironment in the metastasis of lung cancer [6, 7]. The tumor microenvironment, comprising tumor cells, immune cells, extracellular matrix, blood vessels, and lymphatic vessels, plays a crucial role in tumor growth, invasion, and metastasis due to the interactions among these components [8,9,10].
One key factor in lung cancer metastasis is the alteration of the tumor microenvironment [11]. ASAP1 is a protein closely associated with cytoskeletal reorganization, cell adhesion, and migration [12, 13]. Both ASAP1 and the p-STAT3 signaling pathway are important in regulating the tumor microenvironment [14, 15]. Existing studies have shown that overexpression of ASAP1 is associated with increased tumor aggressiveness and poor prognosis, while promotes tumor cell proliferation and survival [16,17,18]. However, the specific mechanisms by which ASAP1 regulates the tumor microenvironment through the p-STAT3 signaling pathway, thereby affecting lung cancer metastasis, remain unclear. This represents a critical gap in the field of lung cancer research.
In recent years, studies on the role of ASAP1 in tumor development have increased. For instance, ASAP1 promotes tumor cell migration and invasion, potentially through mechanisms involving cytoskeletal reorganization [19, 20]. Additionally, the p-STAT3 signaling pathway, a key pathway for tumor cell survival and proliferation, has been closely linked to malignant progression in various cancers [21, 22]. p-STAT3 in lung cancer cells promotes evasion of immune surveillance, thereby facilitating tumor growth and metastasis [23, 24]. Although these studies provide a preliminary understanding of the roles of ASAP1 and p-STAT3 in tumor development, their interaction and specific roles within the tumor microenvironment remain underexplored.
Given these research gaps, this study aims to explore the mechanisms by which ASAP1, through the p-STAT3 signaling pathway, alters the tumor microenvironment and thereby inhibits lung cancer metastasis. We will employ molecular biology and cell biology techniques to thoroughly investigate the regulatory effects of ASAP1 on the p-STAT3 signaling pathway and how this process impacts the tumor microenvironment, particularly changes in the immune microenvironment, and subsequently affects lung cancer metastasis. Moreover, we will explore potential therapeutic strategies targeting the ASAP1 and p-STAT3 signaling pathways, offering new insights for lung cancer treatment. The significance of this study lies in its potential to provide a new theoretical basis for the mechanisms of lung cancer metastasis and to identify new targets and strategies for treatment, potentially improving survival rates and quality of life for lung cancer patients.
Methods
Bioinformatics Analysis
To investigate the expression and prognostic significance of ASAP1 in lung cancer, we performed a comprehensive bioinformatics analysis using publicly available datasets. We utilized The Human Protein Atlas database to analyze the expression levels of ASAP1 in lung cancer tumor tissues and adjacent non-tumor tissues. Immunohistochemical data were sourced from the Human Protein Atlas website (https://www.proteinatlas.org/). To assess the prognostic value of ASAP1 expression in lung cancer patients, we analyzed data from The Cancer Genome Atlas (TCGA) database. We used the GEPIA (Gene Expression Profiling Interactive Analysis) platform (http://gepia.cancer-pku.cn/index.html) to correlate ASAP1 expression levels with patient survival outcomes.
Cell Culture
The human lung cancer cell lines used in this study (A549, NCI-H1299, PC-9) and Mouse Lewis lung cancer cells were all purchased from Wuhan PunoCai Life Science Co., Ltd. All cells were authenticated by STR genotyping and tested negative for mycoplasma. Cells were cultured in medium containing 10% fetal bovine serum at 37 °C in a 5% CO2 incubator. DMEM, fetal bovine serum (Gibco), trypsin, and double antibiotics were purchased from Beijing Solarbio Science & Technology Co., Ltd.
Cell Transfection
The experiments were divided into control plasmid (shNC) and ASAP1-silenced (sh-ASAP1) groups. Both the control and ASAP1 silencing plasmids were synthesized by Wuhan Jinkairui Bioengineering Co., Ltd. Transfections were performed in 6-well plates when cells reached 70% confluence. Each well received 5 µg of plasmid diluted in 250 µL of serum-free medium, which was mixed by pipetting. LipofectamineTM 2000 (10 µL) was diluted in 250 µL of serum-free medium and allowed to sit at room temperature for 5 min. After mixing the two solutions, they were incubated at room temperature for 20 min before being added to the cells. The medium was changed after 6 h, and cells were cultured for 48 h before subsequent experiments.
MTT Assay for Cell Proliferation
Transfected cells were seeded into a 96-well plate at 4 × 104 cells per well. Each condition included six replicates, and a blank group containing only dimethyl sulfoxide solution was also set up. After 24 h of incubation in a CO2 incubator, 20 µL of MTT solution was added to each well and incubated at 37 °C in 5% CO2 for 4 h. The supernatant was discarded, and 150 µL of dimethyl sulfoxide was added. The plate was shaken for 10 min, and the absorbance at 490 nm (OD) was measured using a microplate reader. Cell viability was calculated as follows: Viability = 100% × (OD of the transfected group - OD of the blank group) - (OD of the control group - OD of the blank group).
EdU Assay
The EdU assay was performed using an EdU kit according to the manufacturer’s instructions. EdU solution (1 µL per mL of complete medium) was added and incubated at 37 °C for 2 h. Cells were then washed three times with PBS and fixed with 150 µL of 4% paraformaldehyde for 30 min. After removing the fixative, 150 µL of 2 mg/mL glycine solution was added and incubated at room temperature for 5 min. Cells were washed three times with PBS, then permeabilized with 250 µL of 0.5% Triton X-100 for 10 min. After one PBS wash, 250 µL of Apollo staining solution was added per well and incubated in the dark at room temperature for 30 min. After three washes with permeabilization solution, cells were stained with DAPI for 30 min and washed twice with PBS. Cells were finally examined and photographed using an Olympus BX51 fluorescence microscope.
Colony Formation Assay
Cells from each group were seeded into 6-well plates at a density of 1.0 × 104 cells per well. The medium was changed every 2 days. After 14 days of culture, the medium was discarded. Cells were fixed with paraformaldehyde and stained with crystal violet. Colonies consisting of more than 50 cells were counted under a microscope.
TUNEL Assay
A549 cell samples were pre-plated on coverslips and the medium was discarded. Cells were washed three times with PBS. Following the TUNEL assay kit instructions, cells were fixed with 4% paraformaldehyde at room temperature for 30 min and washed three times with PBS. Cells were then incubated with 0.2% Triton X-100 in PBS at room temperature for 30 min, followed by three PBS washes. Each sample received 100 µL of TdT reaction buffer and was incubated at room temperature for 10 min. After preparing the TdT mixture according to the instructions, the reaction buffer was absorbed with filter paper, and 50 µL of the TdT mixture was added and incubated in the dark at 37 °C for 60 min. Cells were washed twice with PBS containing 3% BSA. Each well received 100 µL of Alexa Fluor™ 594 fluorescent stain, incubated in the dark at room temperature for 30 min, and washed twice with 3% BSA in PBS. Cells were then stained with 100 µL of DAPI fluorescent stain per well, incubated in the dark at room temperature for 5 min, followed by two PBS washes. Anti-fade mounting medium was added, and samples were sealed. The experiment was repeated three times, and cells were observed and photographed using a fluorescence microscope.
PCR
RNA was extracted according to the RNA extraction kit instructions, and cDNA of ASAP1 was synthesized using a reverse transcription kit. The expression level of ASAP1 in cells was detected using an RT-PCR kit. The RT-PCR program was as follows: 95 °C for 5 min for initial denaturation, followed by 30 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 1 min. The upstream primer for GAPDH was 5′-GAGTCAACGGATTTGGTCGT-3′, and the downstream primer was 5′-TTGATTTTGGAGGGATCTCG-3′. GAPDH was used as the internal reference gene, and the expression level of ASAP1 was calculated using the 2-ΔΔCt method.
Western Blot
After collecting cells, 400 µL of lysis buffer was added, and cells were lysed on ice for 30 min. The lysate was transferred to a centrifuge tube and centrifuged at 12,000 rpm at 4 °C for 20 min. The supernatant was transferred to an Eppendorf tube. The protein concentration was determined using a BCA protein assay kit. Protein samples were mixed with 5× loading buffer at a 4:1 ratio and boiled at 100 °C for 5 min for denaturation. Denatured protein samples were loaded into wells, 40 µL per well. Electrophoresis was performed at 80 V for 30 min until the bromophenol blue reached the separating gel, then the voltage was increased to 120 V until the end of electrophoresis. Proteins were transferred to a nitrocellulose membrane at 4 °C, blocked with 5% non-fat milk at room temperature for 60 min, and sequentially incubated with primary antibody, including anti-Bax antibody (ab32503), anti-Bcl-2 antibody (ab182858), anti-STAT3 antibody (ab68153), anti-STAT3 (phospho Y705) antibody (ab76315) (Abcam, diluted 1:1000, overnight at 4 °C) and secondary antibody (diluted 1:1000, for 60 min at room temperature). The membrane was then moved to an exposure device, and detection reagent was added. After exposure, the relative expression levels of the target proteins were analyzed with GAPDH as the internal reference.
Mitochondrial Membrane Potential Assay Using JC-1 Kit
After culturing cells in a 5% CO2, 37 °C incubator with saturated humidity for 24 h, the culture medium was aspirated, and cells were washed once with PBS. Subsequently, 0.5 mL of culture medium and 0.5 mL of JC-1 staining solution were added and thoroughly mixed. Cells were incubated at 37 °C for 20 min. Following incubation, the supernatant was removed, and cells were washed twice with JC-1 staining buffer (1×). After adding 1 mL of culture medium, cells were examined under a fluorescence microscope using the FL1 (green) and FL2 (red) channels for imaging.
ROS Detection with DCFH-DA Fluorescent Probe
After 48 h of treatment, when cell confluence reached 70 to 80%, 1 mL of PBS containing DCFH-DA probe (10 µM) was added. Cells were incubated at room temperature for 30 min and imaged under a laser scanning confocal microscope (Ex: 488 nm, Em: 525 nm).
Transwell Invasion Assay
Cell suspensions were prepared at a concentration of 6 × 105 cells/mL. Each well received 100 µL of cell suspension. The lower chamber was filled with 700 µL of medium containing 10% FBS. Cells were incubated at 37 °C for 24 h. Transwell inserts were washed three times with PBS, and non-invading cells and Matrigel matrix on the upper surface were removed with a cotton swab. Cells were fixed with 4% paraformaldehyde for 30 min and air-dried. Staining was performed with 0.1% crystal violet for 40 min at room temperature, followed by two PBS washes. Cells were observed under a microscope, and cell counting was performed in five random fields (10× magnification).
Tail Vein Injection for Lung Metastasis Animal Model
Mice were housed in SPF conditions within the experimental animal facility, with the room temperature maintained at 20–25 °C and humidity between 40 and 70%. Cage cleaning and sterilization were performed weekly. All experimental procedures strictly adhered to the 3Rs principle and animal welfare guidelines, approved by the Institutional Animal Care and Use Committee. Cells in logarithmic growth phase were digested with 0.25% trypsin to prepare a cell suspension. When the cell concentration reached 2 × 107 cells/mL, 0.2 mL of the suspension was injected into the tail vein of nude mice. From the fourth week post-injection, one mouse from each group was euthanized every three days, and their lungs were examined macroscopically for metastatic lesions. Lung tissues were fixed in 10% formaldehyde for 24 h, dehydrated sequentially in 80, 90, and 100% ethanol and butanol, embedded in paraffin at 60 °C, sectioned, and stored for further analysis. The experiment met the ethical requirements and was approved by the Ethics Committee of the Fourth Hospital of Hebei Medical University (No. 2022KY252).
H&E Staining
Formalin-fixed, paraffin-embedded tissue sections were cut at a thickness of 4 µm. Sections were baked at 60 °C for 2 h followed by routine deparaffinization: xylene I for 20 min, xylene II for 10 min, absolute ethanol for 30 s, 95% ethanol for 30 s, 75% ethanol for 30 s, and distilled water for 5 min. Hematoxylin staining was performed for 4 min, followed by differentiation in 0.5% hydrochloric acid alcohol for 9 s. Sections were then washed under running water for 15 min, followed by 75% ethanol for 10 s and 85% ethanol for 10 s, and eosin staining for 10 s. Dehydration was subsequently carried out with 95% ethanol for 10 s, absolute ethanol for 10 s, xylene I for 10 s, and xylene II for 10 s. Slides were then coverslipped and observed.
Flow Cytometry
A small piece of lung tumor tissue from a lung metastasis animal model was aseptically excised and placed in a filter. After an ice bath for 10 min and two washes with 1 × PBS, the tumor was minced and digested with collagenase II (1 mg/mL), collagenase IV (1 mg/mL), hyaluronidase (100 U/mL), and DNase I (100 U/mL) for 30 min. The digested tissue was filtered through a 70 µm mesh, washed once with 1× PBS to terminate digestion, and washed again with 1 × PBS. A 10 µL aliquot of the cell suspension was diluted tenfold with 1 × PBS for cell counting. The single-cell suspension was adjusted to a cell density of 1 × 107 cells/mL, and 100 µL per tube was prepared for flow cytometric staining with CD11b and Gr1 antibodies; CD8 and CD69 antibodies. Antibodies were obtained from BD Biosciences (San Diego, CA, USA). Each sample tube was adjusted to a volume of 100 µL and a cell density of 1 × 106 cells/100 µL. Each sample tube received 0.5 µL of flow cytometry antibody and was incubated in the dark at 4 °C for 30 min. After incubation, 1 mL of precooled phosphate-buffered saline was added to each tube for washing, centrifuged at 570 × g for 5 min, and the supernatant was discarded. Cells were resuspended in 500 µL of phosphate-buffered saline, filtered through a 200-mesh sieve, and analyzed on the instrument. Flow cytometric data were analyzed using FlowJo-V10 software.
Immunohistochemistry
Immunohistochemical methods were used to detect tumor-related markers in lung tissue. After routine deparaffinization and rehydration of paraffin sections, the sections were microwaved in PBS for 10 min and treated with 3% H2O2 in the dark at room temperature for 10 min. Sections were washed three times with PBS, incubated with normal sheep serum at room temperature for 30 min, and then incubated with polyclonal antibodies (dilutions of 1:40 and 1:100) overnight at 4 °C. Subsequent steps were strictly followed according to the ABC kit instructions. Each experiment included both positive and negative controls.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 7 software. Comparisons between two groups were conducted using the Student’s t-test. Comparisons among multiple groups were performed using one-way ANOVA, with multiple comparisons using the LSD-t method. A p-value < 0.05 was considered statistically significant.
Results
Inhibition of ASAP1 reduces the activity of lung cancer cells
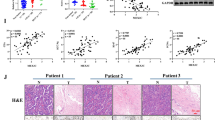
To confirm the role of ASAP1 in lung cancer patients, we analyzed the expression of ASAP1 and its impact on the survival of lung cancer patients. Immunohistochemical analysis of ASAP1 in tumor tissues and normal lung tissues of lung cancer patients showed that ASAP1 is highly expressed in lung cancer tumor tissues (Fig. S1A). Prognostic analysis of ASAP1 expression in lung cancer patients indicated that patients with high ASAP1 expression have shorter survival periods (Fig. S1B). The above results suggest that ASAP1 may play an important role in lung cancer. Using PCR technology, we confirmed that in lung cancer cell lines A549, NCI-H1299, and PC-9, the expression of ASAP1 was significantly reduced in the ASAP1 knockdown group (sh-ASAP1) compared to the control group (shNC) (Fig. 1A). This result indicates that we successfully inhibited the expression of ASAP1 through specific interference RNA technology. In MTT and EdU assays, 48 h after ASAP1 knockdown treatment, the proliferation capacity of A549, NCI-H1299, and PC-9 cells was significantly reduced compared to the control group (Fig. 1B, C). This result suggests that the inhibition of ASAP1 reduces the activity of lung cancer cells, potentially inhibiting tumor growth. The colony formation assay is a method to assess cell growth ability and survival capacity. In this experiment, the number of colonies formed by lung cancer cells in the ASAP1 knockdown group was significantly reduced compared to the control group, indicating that the inhibition of ASAP1 significantly reduces the growth and survival ability of lung cancer cells (Fig. 1D).
Suppression of ASAP1 reduces the viability of lung cancer cells. A PCR validation of sh-ASAP1 silencing efficiency in lung cancer cell lines A549, NCI-H1299, and PC-9. Cell viability of lung cancer cell lines A549, NCI-H1299, and PC-9 treated with sh-ASAP1 for 48 h, measured by MTT (B) and EdU (C) assay. D Effect of ASAP1 inhibition on colony-forming ability of the three lung cancer cell lines over 14 days. Bars show mean ± S.D. n = 3, *P < 0.05, **P < 0.01, ***P < 0.001
Inhibition of ASAP1 Induces Apoptosis in A549, NCI-H1299, and PC-9 Cells
In this experiment, using TUNEL staining technology, we analyzed A549, NCI-H1299, and PC-9 cells treated with sh-ASAP1 for 24 h. The results showed that compared to the control group (shNC), the apoptosis rate in the ASAP1 knockdown group (sh-ASAP1) was significantly increased. This indicates that inhibition of ASAP1 can induce apoptosis in lung cancer cells (Fig. 2A, B). Bax and Bcl-2 are two key apoptotic regulatory proteins, where Bax promotes apoptosis and Bcl-2 inhibits apoptosis. By Western blot analysis, we examined the expression levels of the apoptotic-related proteins Bcl-2 and Bax in A549 cells treated with sh-ASAP1. The results showed that compared to the control group (shNC), the expression of Bax increased, while the expression of Bcl-2 decreased in the ASAP1 knockdown group (sh-ASAP1) (Fig. 2C & Fig. S2). An increased Bax/Bcl-2 ratio is often considered an indicator of apoptotic tendency. In this experiment, compared to the control group, the Bax/Bcl-2 ratio in the ASAP1 knockdown group was significantly increased, further confirming that inhibition of ASAP1 can promote apoptosis in lung cancer cells by regulating the expression of apoptotic-related proteins (Fig. 2D).
Inhibition of ASAP1 induces apoptosis in A549, NCI-H1299, and PC-9 cells. A Analysis of A549, NCI-H1299, and PC-9 cells treated with sh-ASAP1 for 24 h using TUNEL staining. B Statistical results of the apoptosis assays. C Western blot analysis of A549 cells treated with sh-ASAP1 for 24 h. D The ratio of Bax to Bcl-2 expression presented as a percentage in bar graphs. Bars show mean ± S.D. n = 3, **P < 0.01, ***P < 0.001
Effects of Inhibition of ASAP1 on the Intrinsic Apoptosis Pathway
Changes in mitochondrial membrane potential are an important early indicator of apoptosis. Using the JC-1 probe, changes in mitochondrial membrane potential can be measured; a decrease in the ratio of JC-1 monomers to aggregates indicates a drop in mitochondrial membrane potential, an early sign of apoptosis. JC-1 probe activity demonstrated that, compared to the control group, the ratio of JC-1 monomers to aggregates increased in A549 cells with inhibited ASAP1, indicating a reduction in mitochondrial membrane potential, suggestive of increased apoptosis (Fig. 3A). An increase in reactive oxygen species (ROS) levels is another important marker of cellular stress and apoptosis. Excessive production of ROS can damage cellular structures, leading to apoptosis. Compared to the control group, ROS levels significantly increased in A549 cells with inhibited ASAP1, suggesting elevated intracellular stress levels, which may promote apoptosis (Fig. 3B). NAC (N-acetylcysteine) is an antioxidant that can reduce ROS production, thereby protecting cells from oxidative stress damage. After pretreatment with NAC, regardless of ASAP1 inhibition, there was no significant difference in ROS levels in A549 cells, indicating that NAC can effectively inhibit the increase in ROS levels caused by ASAP1 knockdown, thereby potentially slowing the apoptosis process (Fig. 3C).
Impact of ASAP1 inhibition on the intrinsic apoptotic pathway. A Induction of mitochondrial membrane potential changes in A549 cells upon ASAP1 inhibition, measured by the ratio of JC-1 monomers to aggregates. B Increase in ROS levels following treatment with sh-ASAP1 in A549 cells. Cells were incubated with 10 µM DCFH-DA at 37 °C for 30 min post-treatment and analyzed using fluorescence microscopy. C A549 cells pre-treated with 2 mM NAC for 1 h prior to sh-ASAP1 treatment. Cells were then incubated with 10 µM DCFH-DA at 37 °C for 30 min and analyzed using fluorescence microscopy. Bars show mean ± S.D. n = 3, ns: no significant, ***P < 0.001
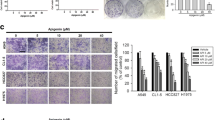
Inhibition of ASAP1 Suppresses Migration and Invasion of A549, NCI-H1299, and PC-9 Cells
Transwell assays were used to assess the migration and invasion capabilities of cells. The results showed that after inhibition of ASAP1, the migration and invasion abilities of A549, NCI-H1299, and PC-9 cells were significantly reduced (Fig. 4A, B). This suggests that ASAP1 plays a promotive role in the migration of lung cancer cells, and its inhibition may slow the metastasis of lung cancer. Matrix metalloproteinase-2 (MMP-2) and matrix metalloproteinase-9 (MMP-9) are enzymes that can degrade the extracellular matrix, and their expression levels are closely related to the aggressiveness and metastatic ability of tumors. After inhibition of ASAP1, the expression levels of MMP-2 and MMP-9 were reduced in A549, NCI-H1299, and PC-9 cells (Fig. 4C, D). This suggests that inhibition of ASAP1 expression can suppress tumor invasion and metastasis by inhibiting the expression of MMP-2 and MMP-9. STAT3 is a signal transduction and transcription activation factor, whose phosphorylated form (p-STAT3) is its activated state, associated with various processes such as tumor growth, invasion, and immune escape. After inhibition of ASAP1, although the total expression of STAT3 did not change, the expression of p-STAT3 significantly decreased (Fig. 4E & Fig. S3). This suggests that ASAP1 may promote the phosphorylation of STAT3, activating the p-STAT3 signaling pathway, thereby promoting the invasion and metastasis of lung cancer.
Inhibition of ASAP1 suppresses migration and invasion of A549, NCI-H1299, and PC-9 cells. A, B Inhibition of ASAP1 reduces the migration and invasion of A549, NCI-H1299, and PC-9 cells in Transwell assays. C Analysis of MMP-2 expression in A549, NCI-H1299, and PC-9 cells following treatment with sh-ASAP1. D Analysis of MMP-9 expression. E Western blot analysis of STAT3 and p-STAT3 in A549 cells treated with sh-ASAP1. Bars show mean ± S.D. n = 3, ***P < 0.001
Anti-metastatic Effects of Inhibition of ASAP1 In Vivo
The number of metastatic tumor nodules is a direct indicator of the metastatic capability of lung cancer. An increase in the number of tumor nodules indicates a strong metastatic ability of lung cancer cells to form more metastatic foci in the lungs. Compared to the control group (shNC), the number of metastatic tumor nodules in the lungs of mice with inhibited ASAP1 (sh-ASAP1) was significantly reduced (Fig. 5A). This suggests that inhibition of ASAP1 can effectively suppress the metastatic capability of lung cancer, reducing the formation of metastatic foci in the lungs. The weight of the spleen can indirectly reflect the impact of tumors on the immune system. The spleen is an important immune organ, and the metastasis and growth of tumors may lead to the activation of the immune system and enlargement of the spleen. Compared to the control group, the spleen weight of mice with inhibited ASAP1 was significantly reduced (Fig. 5B). This may indicate that by inhibiting ASAP1, lung cancer metastasis is reduced, thereby lessening the impact on the immune system and the burden on the spleen. An increase in lung weight is usually associated with pulmonary lesions, increased tumor burden, or the formation of pulmonary metastatic foci. Compared to the control group, the lung weight of mice with inhibited ASAP1 was significantly reduced (Fig. 5C). This result further confirms that inhibition of ASAP1 can effectively suppress the metastasis of lung cancer, reducing the formation of metastatic foci in the lungs, thereby reducing the pulmonary tumor burden.
Regulation of Immune Cells by Inhibition of ASAP1 in a Lung Metastasis Model
Exploring the role of ASAP1 in a lung metastasis model reveals its importance in regulating immune cells. Further, we focused on the impact of ASAP1 inhibition on the regulation of immune cells in the lung metastasis model, specifically including changes in tumor-associated myeloid-derived suppressor cells (MDSCs) and CD8 + CD69+ cells. MDSCs are a heterogeneous group of cells with potent immunosuppressive functions that can inhibit T-cell activity and promote tumor immune evasion. In the tumor microenvironment, an increase in MDSCs is closely related to tumor immune escape, growth, and metastasis. By analyzing CD11b + Gr1+ cells through flow cytometry, the content of MDSCs in the tumor can be quantified. Compared to the control group, inhibition of ASAP1 significantly reduced the content of tumor-associated MDSCs in the lung metastasis model (Fig. 6A). This result suggests that inhibiting ASAP1 can effectively reduce the number of immunosuppressive cells, MDSCs, in the tumor microenvironment, potentially helping to decrease tumor-induced immunosuppression and thereby enhancing the immune system’s ability to attack the tumor. CD8+ cells are a key type of cytotoxic T cells that can recognize and kill tumor cells. CD69 is an early marker of T cell activation, and the presence of CD8 + CD69+ cells indicates that these cells are in an activated state, possessing strong tumor-killing capabilities. Compared to the control group, inhibition of ASAP1 significantly increased the content of CD8 + CD69+ cells. This indicates that inhibition of ASAP1 can increase the number of activated CD8+ cytotoxic T cells in the tumor microenvironment, which may help enhance the immune response against tumor cells, thereby inhibiting tumor growth and metastasis (Fig. 6B).
Regulation of immune cells by ASAP1 inhibition in a lung metastasis model. A ASAP1 inhibition significantly reduces tumor-associated MDSCs (myeloid-derived suppressor cells) in the lung metastasis model. Flow cytometry analysis quantifies CD11b+Gr1+ myeloid cells in tumors treated with sh-ASAP1. B Single-cell suspensions prepared from tumors analyzed by flow cytometry to detect the presence of CD8+CD69+ cells. Bars show mean ± S.D. n = 3, ***P < 0.001
Inhibition of ASAP1 Reduces Tumor Cell Proliferation and Induces Apoptosis in a Lung Metastasis Model
Immunohistochemical analysis was conducted to observe the expression of P-STAT3, Ki-67, Bax, and Bcl-2 in tumor tissues. Compared to the control group (shNC), the expression of P-STAT3 in the tumor tissues of the ASAP1-inhibited group (sh-ASAP1) was significantly reduced (Fig. 7A). This suggests that inhibition of ASAP1 can decrease the activation of P-STAT3, thereby potentially suppressing tumor cell proliferation and survival capabilities. In comparison to the control group, the expression of Ki-67 in the tumor tissues of the ASAP1-inhibited group was significantly reduced (Fig. 7B). This result further confirms that inhibition of ASAP1 can slow down the proliferation rate of tumor cells. Compared to the control group, the expression of Bax in the tumor tissues of the ASAP1-inhibited group was significantly increased (Fig. 7C). This indicates that inhibition of ASAP1 can promote apoptosis in tumor cells. Compared to the control group, the expression of Bcl-2 in the tumor tissues of the ASAP1-inhibited group was significantly reduced (Fig. 7D). This result suggests that inhibition of ASAP1 promotes apoptosis of tumor cells but also reduces their anti-apoptotic capabilities. In addition, the immunohistochemical results show that silencing ASAP1 can significantly reduce the expression of MMP2 and MMP-9. These results further validate the role of ASAP1 in tumor invasion and metastasis.
ASAP1 inhibition reduces tumor cell proliferation and induces apoptosis in a lung metastasis model. To establish a lung metastasis model, 5 × 105 A549 cells were injected intravenously. Immunohistochemical analysis of P-STAT3 (A), Ki-67 (B), Bax (C), Bcl-2 (D), MMP2 (E) and MMP9 (F) expression in tumor tissue. Bars show mean ± S.D. n = 6, *P < 0.05, **P < 0.01, ***P < 0.001
Discussion
In this study, we systematically explored the role and mechanisms of ASAP1 in the metastasis of lung cancer, particularly its process of altering the tumor immune microenvironment by the p-STAT3 signaling pathway. Our experimental results demonstrate that inhibition of ASAP1 significantly reduces the activity of lung cancer cells, induces apoptosis, inhibits cell migration and invasion, and shows notable anti-metastatic effects in vivo. Additionally, we found that inhibiting ASAP1 can regulate immune cells, reduce tumor cell proliferation, and induce tumor apoptosis in lung metastasis models. These findings provide new insights for the treatment of lung cancer.
Our results reveal the crucial role of ASAP1 in the activity of lung cancer cells. By inhibiting ASAP1, the activity of lung cancer cells is significantly reduced, likely due to the regulation of ASAP1 on cell proliferation and survival signaling pathways. This finding is consistent with previous studies, which suggest that ASAP1, as a regulatory protein, can influence tumor cell behavior through various signaling pathways [12,13,14]. Furthermore, we found that inhibition of ASAP1 induces apoptosis in lung cancer cells, particularly through the intrinsic apoptosis pathway. This suggests that ASAP1 may inhibit apoptosis by affecting the expression or activity of apoptosis-related proteins, thereby promoting tumor cell survival. Moreover, our study indicates that inhibiting ASAP1 suppresses migration and invasion of lung cancer cells, which is significant for the metastasis of lung cancer. This effect may be due to ASAP1 regulating the expression or activity of proteins related to cytoskeletal and extracellular matrix interactions, thereby impacting the migratory and invasive capabilities of the cells.
Importantly, our research unveils the mechanism by which ASAP1 alters the tumor immune microenvironment by inhibiting the p-STAT3 signaling pathway. The p-STAT3 signaling pathway plays a key role in regulating tumor immune evasion and promoting tumor growth and metastasis [25, 26]. Our data show that inhibiting ASAP1 reduces the activity of p-STAT3, possibly by affecting upstream kinases or downstream target genes. Through this mechanism, inhibiting ASAP1 can alter the tumor immune microenvironment, enhancing the recognition and elimination of tumor cells by immune cells, thus inhibiting lung cancer metastasis. Our study also finds that inhibiting ASAP1 has a significant anti-metastatic effect in vivo, likely due to its alteration of the tumor immune microenvironment and direct suppression of tumor cell migration and invasion. These results suggest that ASAP1 may be an important therapeutic target for lung cancer, and strategies targeting ASAP1 could have potential clinical applications in inhibiting lung cancer metastasis.
ASAP1 is a protein known to be involved in regulating cytoskeletal reorganization, cell migration, and invasion [19, 27]. The impact of ASAP1 on tumor immunity may operate through several aspects. Firstly, ASAP1 promotes the migration and invasion of tumor cells by regulating cytoskeletal reorganization [20]. This capability may also affect the ability of immune cells, such as T cells and macrophages, to enter the tumor microenvironment. Effective infiltration of immune cells is crucial for tumor immune surveillance and immune response. Furthermore, ASAP1 may indirectly affect tumor immunity by influencing multiple signaling pathways, such as the p-STAT3 signaling pathway [28]. These pathways play roles in regulating the activation, proliferation, and function of immune cells. For example, the STAT3 signaling pathway plays a key role in regulating the immunosuppressive microenvironment of tumors [29, 30]. By affecting the interactions between tumor cells and the surrounding microenvironment, ASAP1 may indirectly alter the behavior of immune cells. For instance, ASAP1 may influence the recruitment and function of immune cells by promoting the secretion of specific cytokines and chemokines by tumor cells. The academic achievement of this study lies in revealing the multiple roles of ASAP1 in the development of lung cancer and elucidating the mechanism by which inhibition of ASAP1 alters the tumor immune microenvironment through the inhibition of the p-STAT3 signaling pathway. This discovery not only enhances our understanding of the complex mechanisms of lung cancer metastasis but also provides a scientific basis for developing new lung cancer treatment strategies targeting ASAP1. Additionally, our study emphasizes the importance of the tumor immune microenvironment in lung cancer metastasis, offering new directions for future research in lung cancer immunotherapy.
Despite the achievements of our study, there are some limitations. Although we have demonstrated that inhibition of ASAP1 can alter the tumor immune microenvironment through the inhibition of the p-STAT3 signaling pathway, the specific molecular mechanisms still require further investigation. Additionally, considering the complexity of the tumor microenvironment, future studies should explore the effects of inhibiting ASAP1 on other immune cells, such as T cells and dendritic cells. Finally, based on the findings of this study, developing small molecule inhibitors or antibodies targeting ASAP1 and evaluating their effectiveness in lung cancer treatment will be an important direction for future research.
Conclusion
In summary, our study provides new insights into the mechanisms of ASAP1 in lung cancer metastasis and reveals the potential mechanism by which inhibition of ASAP1 alters the tumor immune microenvironment through the inhibition of the p-STAT3 signaling pathway. These findings not only enhance our understanding of the complex mechanisms of lung cancer metastasis but also provide a scientific basis for developing new lung cancer treatment strategies. Future research should further explore the interactions between ASAP1 and the p-STAT3 signaling pathway in lung cancer metastasis, as well as potential therapeutic strategies targeting this pathway, in order to provide more effective treatment options for lung cancer patients.
References
Thandra, K. C., Barsouk, A., Saginala, K., Aluru, J. S., & Barsouk, A. (2021). Epidemiology of lung cancer. Contemporary Oncology/Współczesna Onkologia, 25, 45–52.
Travis, W. D. (2020). Lung cancer pathology: current concepts. Clinics in Chest Medicine, 41, 67–85.
Nooreldeen, R., & Bach, H. (2021). Current and future development in lung cancer diagnosis. International Journal of Molecular Sciences, 22, 8661
Bade, B. C., & Cruz, C. S. D. (2020). Lung cancer 2020: epidemiology, etiology, and prevention. Clinics in Chest Medicine, 41, 1–24
Iams, W. T., Porter, J., & Horn, L. (2020). Immunotherapeutic approaches for small-cell lung cancer. Nature Reviews Clinical Oncology, 17, 300–312
Zhao, Y., Guo, S., Deng, J., Shen, J., Du, F., Wu, X., Chen, Y., Li, M., Chen, M., & Li, X. (2022). VEGF/VEGFR-targeted therapy and immunotherapy in non-small cell lung cancer: targeting the tumor microenvironment. International Journal of Biological Sciences, 18, 3845
Wu, F., Fan, J., He, Y., Xiong, A., Yu, J., Li, Y., Zhang, Y., Zhao, W., Zhou, F., & Li, W. (2021). Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nature Communications, 12, 2540
Hsieh, C.-H., Hsieh, H.-C., Shih, F.-H., Wang, P.-W., Yang, L.-X., Shieh, D.-B., & Wang, Y.-C. (2021). An innovative NRF2 nano-modulator induces lung cancer ferroptosis and elicits an immunostimulatory tumor microenvironment. Theranostics, 11, 7072.
Chandra, R., Karalis, J. D., Liu, C., Murimwa, G. Z., Voth Park, J., Heid, C. A., Reznik, S. I., Huang, E., Minna, J. D., & Brekken, R. A. (2021). The colorectal cancer tumor microenvironment and its impact on liver and lung metastasis. Cancers, 13, 6206
Anderson, N. M., & Simon, M. C. (2020). The tumor microenvironment. Current Biology, 30, R921–R925
Neophytou, C. M., Panagi, M., Stylianopoulos, T., & Papageorgis, P. (2021). The role of tumor microenvironment in cancer metastasis: Molecular mechanisms and therapeutic opportunities. Cancers, 13, 2053
Müller, T., Stein, U., Poletti, A., Garzia, L., Rothley, M., Plaumann, D., Thiele, W., Bauer, M., Galasso, A., & Schlag, P. (2010). ASAP1 promotes tumor cell motility and invasiveness, stimulates metastasis formation in vivo, and correlates with poor survival in colorectal cancer patients. Oncogene, 29, 2393–2403
Lin, D., Watahiki, A., Bayani, J., Zhang, F., Liu, L., Ling, V., Sadar, M. D., English, J., Fazli, L., & So, A. (2008). ASAP1, a gene at 8q24, is associated with prostate cancer metastasis. Cancer Research, 68, 4352–4359
Hou, T., Yang, C., Tong, C., Zhang, H., Xiao, J., & Li, J. (2014). Overexpression of ASAP1 is associated with poor prognosis in epithelial ovarian cancer. International Journal of Clinical and Experimental Pathology, 7, 280
Xie, W., Han, Z., Zuo, Z., Xin, D., Chen, H., Huang, J., Zhu, S., Lou, H., Yu, Z., & Chen, C. (2023). ASAP1 activates the IQGAP1/CDC42 pathway to promote tumor progression and chemotherapy resistance in gastric cancer. Cell Death & Disease, 14, 124
Bang, S., Jee, S., Son, H., Cha, H., Sim, J., Kim, Y., Park, H., Myung, J., Kim, H., & Paik, S. (2022). Clinicopathological Implications of ASAP1 Expression in Hepatocellular Carcinoma. Pathology and Oncology Research, 28, 1610635
Park, H., Son, H., Cha, H., Song, K., Bang, S., Jee, S., Kim, H., Myung, J., Shin, S.-J., & Cha, C. (2023). ASAP1 Expression in Invasive Breast Cancer and Its Prognostic Role. International Journal of Molecular Sciences, 24, 14355
He, J., Liu, H., Cai, J., Shen, S., Wang, J., & Liu, H. (2024). ASAP1 Promotes Cholangiocarcinoma Progression via Wnt/β-Catenin Pathway. Journal of Biomedical Nanotechnology, 20, 701–711
Tanna, C. E., Goss, L. B., Ludwig, C. G., & Chen, P.-W. (2019). Arf GAPs as regulators of the actin cytoskeleton—an update. International Journal of Molecular Sciences, 20, 442.
Gasilina, A., Vitali, T., Luo, R., Jian, X., & Randazzo, P. A. (2019). The ArfGAP ASAP1 controls actin stress fiber organization via its N-BAR domain. IScience, 22, 166–180
Tong, M., Wang, J., Jiang, N., Pan, H., & Li, D. (2017). Correlation between p-STAT3 overexpression and prognosis in lung cancer: A systematic review and meta-analysis. PLoS One, 12, e0182282
Liu, Z., Ma, L., Sun, Y., Yu, W., & Wang, X. (2021). Targeting STAT3 signaling overcomes gefitinib resistance in non-small cell lung cancer. Cell Death & Disease, 12, 561
Liu, Y., Zhang, H., Wang, Z., Wu, P., & Gong, W. (2019). 5-Hydroxytryptamine1a receptors on tumour cells induce immune evasion in lung adenocarcinoma patients with depression via autophagy/pSTAT3. European Journal of Cancer, 114, 8–24
Yuan, L., & Ye, J. (2020). Fan D: The B7-H4 gene induces immune escape partly via upregulating the PD-1/STAT3 pathway in non-small cell lung cancer. Human Immunology, 81, 254–261
Groner, B., Lucks, P., & Borghouts, C. (2008). The function of Stat3 in tumor cells and their microenvironment[C]//Seminars in cell & developmental biology. Academic Press, 19, 341–350.
Wang, H. Q., Man, Q. W., Huo, F. Y., Gao, X., Lin, H., Li, S. R., Wang, J., Su, F. C., Cai, L., & Shi, Y. (2022). STAT3 pathway in cancers: Past, present, and future. MedComm, 3, e124.
Randazzo, P. A., Andrade, J., Miura, K., Brown, M. T., Long, Y.-Q., Stauffer, S., Roller, P., & Cooper, J. A. (2000). The Arf GTPase-activating protein ASAP1 regulates the actin cytoskeleton. Proceedings of the National Academy of Sciences, 97, 4011–4016
Priego, N., Zhu, L., Monteiro, C., Mulders, M., Wasilewski, D., Bindeman, W., Doglio, L., Martínez, L., & Martínez-Saez, E. (2018). Ramón y Cajal S: STAT3 labels a subpopulation of reactive astrocytes required for brain metastasis. Nature Medicine, 24, 1024–1035
Lin, W.-H., Chang, Y.-W., Hong, M.-X., Hsu, T.-C., Lee, K.-C., Lin, C., & Lee, J.-L. (2021). STAT3 phosphorylation at Ser727 and Tyr705 differentially regulates the EMT–MET switch and cancer metastasis. Oncogene, 40, 791–805
Saini, U., Naidu, S., ElNaggar, A. C., Bid, H. K., Wallbillich, J. J., Bixel, K., Bolyard, C., Suarez, A. A., Kaur, B., & Kuppusamy, P. (2017). Elevated STAT3 expression in ovarian cancer ascites promotes invasion and metastasis: a potential therapeutic target. Oncogene, 36, 168–181
Author information
Authors and Affiliations
Contributions
J.C. and Y.L. devised the concept and directions of the article. H.Z. collected the literature data. H.Z., Y.L. and Y.W. conducted the literature analysis and wrote the initial draft and response letter of the paper. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Ethics
The experiment met the ethical requirements and was approved by the Ethics Committee of the Fourth Hospital of Hebei Medical University (No. 2022KY252).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, H., Liu, Y., Wu, Y. et al. Inhibition of ASAP1 Modulates the Tumor Immune Microenvironment and Suppresses Lung Cancer Metastasis via the p-STAT3 Signaling Pathway. Cell Biochem Biophys (2024). https://doi.org/10.1007/s12013-024-01349-y
Accepted:
Published:
DOI: https://doi.org/10.1007/s12013-024-01349-y