Abstract
Non-small cell lung cancer (NSCLC) is the most common type of lung cancer. Although significant advances have been achieved in the treatment of NSCLC during the past two decades, the 5-year survival rate of patients with NSCLC remains <20%. Thus, there is an urgent requirement to further understand the molecular mechanisms that promote NSCLC development and to identify novel therapeutic targets. In the present study, the gene expression profiles of patients with NSCLC from The Cancer Genome Atlas database were carefully analyzed and SPINK1 was identified as a tumor-inducing factor. SPINK1 expression level was found to be increased in both NSCLC tissues and cell lines. Moreover, SPINK1 promoted cell proliferation in A549 and H1299 cells. Knockdown of SPINK1 could activate cell autophagy and apoptosis. Mechanistically, SPINK1 was demonstrated to induce the proliferation of NSCLC via activating the MEK/ERK signaling pathway. In conclusion, these findings suggested that SPINK1 may serve as a potential biomarker in NSCLC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer is the leading cause of cancer-related death in the United States and China, with an ~5-year survival rate of ~18% [1]. The most common type of lung cancer is non-small cell lung cancer (NSCLC), which accounts for >80% of all cases [2, 3]. NSCLC can be further divided into three predominant histological subtypes: Adenocarcinoma, squamous cell carcinoma and large cell carcinoma [4]. A total of 70% of patients with lung cancer are diagnosed with advanced-stage disease, which poses more challenges for therapy [5]. Over the past two decades, the application of various targeted therapies, including the EGFR inhibitor afatinib, and the anaplastic lymphoma kinase inhibitor ceritinib, and immunotherapies, such as the immune checkpoint blocker nivolumab, have made remarkable progress in treating patients with advanced NSCLC; however, drug resistance and a poor response prevented the therapeutic efficacy [6]. Therefore, the identification and characterization of novel molecules associated with NSCLC progression are of critical importance to improve both the early diagnosis and treatment of the disease.
Serine peptidase inhibitor Kazal type 1 (SPINK1) is a trypsin inhibitor that was originally isolated from bovine pancreatic tissues [7] and later purified from the urine of patients with ovarian cancer [8, 9]. SPINK1 is expressed in multiple tissues and exerts diverse physiological and pathophysiological functions during the process of multiple diseases. Interestingly, growing evidence demonstrates that SPINK1 plays a role in cancer as a growth factor. Elevated serum SPINK1 was detected and regarded as an independent prognostic factor in renal cancer [10], bladder transitional cell carcinoma [11], metastatic breast cancer [12], hepatocellular carcinoma [13], and ovarian cancer [14, 15]. In vitro, more and more evidence has confirmed that SPINK1 can promote cell proliferation [16], migration [17], and therapeutic resistance [18] while preventing cell apoptosis. Mechanically, previous studies suggested that SPINK1 accelerated malignance mainly through EGFR signaling pathway [16, 19]. However, the effect and underlying mechanism of SPINK1 in the progression of NSCLC are still poorly understood.
In our previous study, Gene Expression Omnibus datasets (GSE19804, GSE18842, GSE27262, and GSE43458) were downloaded and analyzed to screen differentially expressed genes (DEGs) in NSCLC. Among these DEGs, SPINK1 expression level was significantly increased in NSCLC tissues compared with normal tissues, suggesting that SPINK1 may serve as a potential biomarker in NSCLC. Moreover, SPINK1 promoted cell proliferation in A549 and H1299 cells, whereas knockdown of SPINK1 activated cell autophagy and apoptosis. Mechanistically, SPINK1 was demonstrated to induce the proliferation of NSCLC via activating the MEK/ERK signaling pathway. The findings of the present study may provide novel insights for developing new targeted therapy strategies for NSCLC.
Materials and Methods
Microarray Datasets and Online Tools Used for Integrated Bioinformatic Analysis
Gene expression profiles among 59 normal lung tissue cases and 533 NSCLC samples were downloaded from The Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov/).
ELISA
Serum of 20 normal controls, 38 NSCLC patients, and 11 pleural effusion of NSCLC patients in Chongqing Hospital of Traditional Chinese Medicine during 2018.02-2018.06 was collected and SPINK1 concentrations were detected by ELISA (BOSTER, EK1241) according to the reference manual.
Cell Culture
Normal human bronchial epithelial cell line HBE, human NSCLC cell line H1299, SK-MES-1 and H460 were maintained in complete culture medium (RPMI 1640 with 10% fetal bovine serum (FBS), 100 units/mL penicillin, and 100 μg/mL streptomycin). Human NSCLC cell line A549 were routinely cultured with F-12K medium (10% FBS). All cell lines were grown at 37 °C in an atmosphere with 5% CO2. Cell numbers were determined using a Neubauer hemocytometer.
RNA Interference and Exogenous Addition of SPINK1
Small interfering RNAs targeting SPINK1 and the negative control were obtained from TSINGKE (Shanghai, China). The target sequences were as follows:
si-NC: 5′-TTCTCCGAACGTGTCACGT-3′;
si-SPINK1-1: 5′-GCCAGACUUCUAUCCUCAUTT-3′. Cells were seeded in corresponding culture plates and cultured with antibiotics-free medium overnight. Next day the growth medium was removed and cells were transfected with siRNAs (50 nM) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol when cell density was increased to 50–70%. rhSPINK1 was purchased from R&D (#7496-PI-010).
RNA Extraction and Quantitative Real-Time PCR
Total RNA was extracted using IsoPlus reagent (Takara Bio, Japan) following the manufacturer’s protocol and then 1 μg RNA was reverse transcribed with random primers using a PrimeScript™ RT reagent Kit (Takara Bio, Japan). cDNA was amplified using SYBR green master mix (TaKaRa Bio, Japan) on an ABI 7500 Fast Real-Time PCR system (Sigma-Aldrich, St. Louis, MO, USA). GAPDH was amplified as an internal reference. Primers were as follows:
SPINK1-F, TGTGTGT GGGACTGATGGAA;
SPINK1-R, AGGCCCAGATTTTTGAATGA;
GAPDH-F, ACGGCAAATTCAACGGCACAGTCA;
GAPDH-R, TGGGGGCATCGGCAGAAGG.
Viability Assay
The Cell Counting Kit-8 (CCK-8) assay was employed to quantify the vitality of cells. Cells were seeded into 96-well plates at a concentration of 2 × 104 cells/mL and cultured with antibiotics-free medium overnight. The next day cells were transfected with siRNA (50 nM). Then 10 μL CCK8 reagent (Solarbio, China) were added to each well at 0, 24, 48, and 72 h respectively. Cells were incubated for 2.5 h in the dark at 37 °C. The absorbance at 450 nm was measured by Biotek reader (ELx800, USA).
Growth Curve
2 × 104 A549 cells were seeded into 24-well plates and transfected with si-NC and si-SPINK1-1 (50 nM). Total numbers of cells were determined by trypsinization and cell counting using a Neubauer hemocytometer at 0 h, 48 h,72 h, and 96 h post-transfection.
Flow Cytometric Analysis of Cell Cycle Distribution
3 × 105 cells were seeded in six-well plates and treated with siRNAs (50 nM) as described perviously. Cells were collected by trypsinization at 48 h after transfection and washed with cold PBS followed by fixation in 70% ethanol for 12 h at −20 °C. After washing with PBS three times, cells were incubated in PBS with 0.1 mg/mL RNase (Solarbio, China) at 37 °C for 30 min. Then, cells were stained with PI (25 μg/ml, Sigma, USA) in the dark for 10 min. The stained nuclei were counted by flow cytometry and the cell cycle profile was analyzed by the FlowJo software.
Flow Cytometric Analysis of Apoptosis
1.5 × 105 cells were seeded in 12-well plates and transfected with corresponding siRNAs (50 nM) as described previously. Change culture medium with serum-free culture medium supplemented with 0.3% BSA 24 h post-transfection. Total cells were collected by trypsinization at 60 h after transinfection and washed with cold PBS twice. Collected cells were double stained with FITC-Annexin V and PI using FITC Annexin V Apoptosis Detection Kit (BD Biosciences) according to the protocols. The stained cells were analyzed by BD FACSJazz flow cytometer (BD Biosciences), and the data was analyzed using Cell Quest software (BD Biosciences).
Western Blot
Total protein was extracted using radioimmunoprecipitation assay lysis buffer (Beyotime, China) supplemented with 1 mM PMSF (Beyotime, China) and phosphatase inhibitor cocktail A and B (Beyotime, China), quantified using the Enhanced BCA Protein Assay Kit (Beyotime, China), and denatured by boiling with SDS-PAGE Sample Loading Buffer (Beyotime, China) for 10 min. Protein samples (40 µg) were separated by 12% SDS-PAGE, and transferred onto polyvinylidene difluoride (PVDF, 0.20 µm) membranes (Millipore, Bedford, MA). The membranes were blocked with 5% bovine serum albumin in Tris-buffered saline containing 0.1% Tween-20 at 37 °C for 2 h and then incubated with primary antibodies at 4 °C overnight. The primary antibodies were as follows: SPINK1 (sc-37440, 1:500, Santa Cruz, CA), GAPDH (#5174, 1:8000, CST, USA); p-MEK1/2 (#2338, 1:1000, CST, USA); MEK1/2 (#8727, 1:1000, CST, USA); p-ERK (#4370, 1:1000, CST, USA); Erk1/2 (#9102, 1:1000, CST, USA); p-PI3K (#4228, 1:500, CST, USA); PI3K (#4292, 1:800, CST, USA); p-Akt (Ser473)(#4060, 1:500, CST, USA); Akt (#4691, 1:800, CST, USA); LC3 (#12741, 1:1000, CST, USA); P62 (#3912, 1:1000, CST, USA); ATG7 (#8558, 1:800, CST, USA); CDK4 (#12790, 1:1000, CST, USA); Bax (#5023, 1:1000, CST, USA); BCL2 (#196495, Abcam, USA). The membranes were washed for 30 min with TBST and subsequently incubated with the corresponding secondary antibody (#656120, HRP goat anti rabbit, 1:10000, Invitrogen; #A32723, HRP goat anti-mouse, 1:10,000, Invitrogen) for 1 h at 37 °C. Membranes were then washed another three times for 30 min with TBST. Protein bands were visualized using Super Signal electrochemiluminescence (#34580, Thermo Scientific) and quantitated with image Pro Plus 6.0, and the data were normalized to GAPDH.
Statistical Analysis
All data were presented as mean ± SD. GraphPad Prism software (version 5.01) was applied for statistical analysis. Unpaired t tests were used for data with a normal distribution. Differences with p < 0.05 were considered statistically significant.
Results
Convergence of DEGs in NSCLC Tissues Via Integrated Bioinformatic Analysis
To identify the potential role of novel biomarkers related to the pathogenesis and progression of NSCLC, mRNA expression profiles from cancerous and normal lung tissues were downloaded from GEO datasets (GSE19804, GSE18842, GSE27262, and GSE43458) and analyzed by GEO2R respectively. p value < 0.05 and |LogFC| > 1.5 were applied as screening criteria. DEGs identified from the four datasets were as follows: GSE19804: with 129 upregulated and 382 downregulated genes; GSE18842: with 634 upregulated and 902 downregulated genes; GSE27262: with 357 upregulated and 695 downregulated genes; GSE43458: with 54 upregulated and 178 downregulated genes (data not shown). Heatmaps showed 41 upregulated genes (|LogFc| >2) (Fig. 1A) and 64 downregulated genes (|LogFC| > 2.5) (Fig. 1B). Venn Diagram demonstrated the intersections of DEGs obtained from the four datasets, including 23 commonly upregulated genes (Fig. 1C) and 121 commonly downregulated genes (Fig. 1D). These 144 DEGs were further subjected to functional annotation. Results showed that these DEGs were mainly expressed in the plasma membrane, cytoplasm, extracellular, and nucleus of cells (Fig. 1E), acting as cell adhesion molecules, metallopeptidases and receptors (Fig. 1F) involved in a variety of biological processes such as cell communication, signal transduction, growth supporting, and metabolism process (Fig. 1G). In addition, these genes are also abnormally expressed in breast cancer, liver cancer, kidney cancer, head, and neck tumors (Fig. 1H), indicating that these genes are universally disordered in various human tumors and may lead to enhanced activities of proliferation, invasion, and immunologic tolerance of cancer cells.
Identification and functional annotation of genes differently expressed in NSCLC tissues. mRNA profiles were downloaded from GSE19804 dataset and analyzed with GEO2R. Heatmaps showing A 41 upregulated genes (|LogFc| > 2) and B 64 down-regulated genes(|LogFC| > 2.5). Venn Diagrams demonstrating C 23 commonly upregulated genes and (D) 121 commonly downregulated genes from four datasets: GSE19804, GSE18842, GSE27262 and GSE43458. DEGs were subjected to functional annotation, including E cellular components, F molecular functions, G biological process and H sites of expression
SPINK1 Increases in NSCLC Tissues, Serum, and Cell Lines
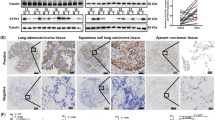
Among the 23 unregulated DEGs, SPINK1 was one of the most significantly overexpressed DEGs by analysis of GEO (Fig. 1A), TCGA (Fig. 2A), GTEx (Fig. 2B) and In Silo Transcriptomics online databases (Fig. 2C). Given that SPINK1 is a secretory protein and the concentration of serum SPINK1 is enhanced in several cancers, serum of 20 normal controls and 38 NSCLC patients, and pleural effusion of 11 NSCLC patients were collected to detect the SPINK1 concentration by ELISA. As shown in Fig. 2D, serum SPINK1 in NSCLC patients was significantly higher than normal controls (1092.9 pg/mL vs 644.5 pg/mL, p < 0.001). Furthermore, the concentration of SPINK1 in pleural effusion was higher than that in serum of NSCLC patients (1928.8 pg/mL vs 1092.9 pg/mL, p < 0.01). In addition, data from the CCLE database were used to examine the relative expression level of SPINK1 in various NSCLC cell lines. It was revealed that the expression level of SPINK1 was relatively high in 10 NSCLC cell lines, particularly in the A549, SK-MES-1, H460, and H1299 cell lines. These results were validated in four NSCLC cell lines (A549, SK-MES-1, H1299, and H460) and one normal lung epithelial cell line (HBE) by RT-qPCR and Western blot(Fig. 2E, F). Altogether, these findings suggested that SPINK1 may serve as a tumor-promoting factor in NSCLC.
Expression levels of SPINK1 in NSCLC tissues and cell lines. mRNA expression levels of SPINK1 in NSCLC tissues compared with normal lung tissues in A TCGA database, B GTEx database and C IST online database. D Concentration of SPINK1 in body fluid was detected by ELISA. E mRNA and F protein levels of SPINK1 in NSCLC cell lines compared with HBE, assessed by qRT-PCR and western blot assays. Data presented as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001
SPINK1 Promotes Proliferation in NSCLC Cells
To investigate the role of SPINK1 in NSCLC progression, SPINK1 was knocked down in A549 and H1299 cells using SPINK1 siRNAs. The transfection efficiency of knockdown (Supplementary Fig. 2A, B) was confirmed by RT-qPCR (Fig. 3A, Supplementary Fig. 1A) and western blot (Fig. 3B, Supplementary Fig. 1B). Subsequently, the effect of SPINK1 on cell proliferation was investigated. As expected, knockdown of SPINK1 significantly inhibited A549 and H1299 cell proliferation and colony formation (Fig. 3C, D, Supplementary Fig. 1C), while rhSPINK1, in turn, remarkably promoted proliferation in H460. Given that SPINK1 is a secreted soluble protein, we speculated that exogenous addition of rhSPINK1 could stimulate the proliferation of H460 with relatively low SPINK1 expression. CCK8 assay results confirmed that rhSPINK1(1 ng/mL) promoted the proliferation of H460 (Fig. 3E). Moreover, pretreatment of SPINK1-knockdowned A549 and H1299 with exogenous rhSPINK1(5 ng/mL) notably rescue cell vitality (Fig. 3F, Supplementary Fig. 1D). Furthermore, cell cycle analysis demonstrated that knockdown of SPINK1 caused cell cycle arrest, with significantly increased percentage of A549 and H1299 cells in the G1-phase and decreased percentage of cells in the S-phase. (Fig. 4A, Supplementary Fig. 2A). Consistently, knockdown of SPINK1 decreased the expression of cell cycle checkpoint protein CDK4 in A549 (Fig. 4B) and H1299 (Supplementary Fig. 2B). Altogether, these findings indicated that SPINK1 promotes proliferation in NSCLC cell lines.
Effect of SPINK1 on proliferation of A549 cell line. A mRNA and B protein expression levels of SPINK1 in A549 post-transfection with si-SPINK1-1(50 nM) and si-NC (50 nM), assessed by qRT-PCR and western blot analysis. C Vitality of A549 was detected by CCK8 assays after endogenous knockdown of SPINK1 at 0 h, 24 h, 48 h, 72 h. D A549 cells were seeded into 24-well plates and transfected with si-NC and si-SPINK1-1 (50 nM). Total numbers of cells were determined by trypsinization and cell counting using a Neubauer hemocytometer. E Vitality of H460 cells after exogenous addition of rhSPINK1 (1 ng/mL), detected by CCK8 assays. F Exogenous addition of rhSPINK1 (5 ng/mL) partially rescued vitality of A549 transfected with si-SPINK1-1(50 nM). Data represent three independent experiments and are presented as mean ± SD (n = 5). *P < 0.05; **P < 0.01; ***P < 0.001
Cell cycle arrest induced by endogenous knockdown of SPINK1. A Cell cycle distribution was detected by flow cytometry in A549 cells transfected with si-NC and si-SPINK1-1 (50 nM). B Cell cycle checkpoint CDK4 was reduced in A549 cells following treatment with si-SPINK1-1, assessed by western blot assay. Data represent three independent experiments and are presented as mean ± SD (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001
Knockdown of SPINK1 Activates Autophagy and Apoptosis in NSCLC Cells
Western blot and Annexin V-DAPI apoptosis assay were performed to investigate the effect of SPINK1 on cell autophagy and apoptosis. As shown in Fig. 5A, Knockdown of SPINK1 in A549 activated autophagy characterized by decreased P62, as well as increased expression of Beclin-1, ATG7, and LC3II/LC3I ratio. Consistently, knockdown of SPINK1 in H1299 induced excessive autophagy (Supplementary Fig. 3A). Meanwhile, the expression level of the pro-apoptotic protein BAX was increased and that of the anti-apoptotic protein BCL2 was decreased following SPINK1 knockdown (Fig. 5B). The early-stage and late-stage apoptotic rate were significantly increased in knockdown-SPINK1 cells compared with control cells (Fig. 3A), suggesting that knockdown of SPINK1 may promote apoptosis in A549 and H1299 cells (Fig. 5C, Supplementary Fig. 3B, C). These results indicated that SPINK1 may inhibit autophagy and apoptosis in NSCLC cells.
Influence of SPINK1 on autophagy and cell apoptosis. A Activation of autophagy following treatment with si-SPINK1-1 (50 nM) in A549 cells, detected by western blot assays. Cell apoptosis was detected by B western blot assays and C cytometry in A549 cells. Data represent three independent experiments and are presented as mean ± SD (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001
SPINK1 Accelerates the Proliferation of NSCLC Via Activation of MEK/ERK Signaling Pathway
It is well established that SPINK1, acting as a pro-survival factor, accelerated the progression of multiple cancers by interacting with EGFR [20,21,22]. In order to clarify signaling pathways downstream of EGFR mediating the carcinogenic effect of SPINK1 in NSCLC, the effect of SPINK1 on the MEK/ERK, PI3K/AKT signaling pathway was investigated. The levels of PI3K, AKT, MEK1/2 and ERK were decreased after si-SPINK1 treatment in A549 cells, among which MEK/ERK was the most significant signaling pathway (Fig. 6A). Consistently, si-SPINK1 treatment in H1299 showed the similar tendency (SFig. 4A). Subsequently, U0126, specific inhibitor of MEK, was used to further examine the association between SPINK1 and the MEK/ERK signaling pathway and verify whether SPINK1-induced proliferation was associated with the MEK/ERK signaling pathway. As demonstrated in Fig. 6B, pretreatment of A549 cells with U0126 resulted in an inhibition of ERK, whereas rhSPINK1 (5 ng/mL) partially reversed the effect of U0126 (Fig. 6B, Supplementary Fig. 4B). In addition, pretreatment with rhSPINK1 significantly enhanced proliferation inhibited by U0126 in A549 and H1299 cells. (Fig. 6C, Supplementary Fig. 4C). Collectively, these findings suggested that SPINK1 may promote proliferation via activation of MEK/ERK signaling pathway.
Signaling pathways involved in biological functions of SPINK1. A Silence of SPINK1 reduced the phosphorylation levels of PI3K, AKT, MEK and ERK in A549 cells, assessed by western blot assays. B Starved A549 cells were pretreated with DMSO or U0126 (10 μM) for 1 h and challenged with PBS or rhSPINK1 in serum-free culture medium for 2 h, phosphorylation levels of MEK/ERK signaling pathway was assessed by western blot assays. C Starved A549 cells were pretreated with DMSO or U0126 (10 μM) for 1 h and challenged with PBS or rhSPINK1 (5 ng/mL) in serum-free culture medium for 48 h, cell vitality was assessed by CCK8 assays. Data represent three independent experiments and are presented as mean ± SD (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001
Discussion
SPINK1 has been reported to be mainly associated with multiple biological functions under various physiological and pathological conditions. As a serpin typically produced by pancreatic acinar cells, SPINK1 can defend against the activation of trypsinogen in the gland vesicle and pancreatic duct [23]. Yamamura et al. reported that SPINK3, the homologous gene of SPINK1 in mouse, may participate in the differentiation and development of mouse embryo [24]. In addition, SPINK1 secreted by cancer cells could co-immmunoprecipitate with serine protease granzyme A (GzmA), thereby suppressing GzmA-mediated apoptosis and establishing a tolerance of cancer cells to the immune surveillance system [25]. More importantly, SPINK1 is aberrantly expressed in multiple human cancers such as ovarian cancer [19], prostatic cancer [26], and so on. Consistently, a recent study reported that tissue SPINK1 is over-expressed in NSCLC patients [27]. Additionally, SPINK1 could be detected in serum and act as independent prognostic factor in prostatic cancer [28] and renal cancer [29]. Previous studies suggested that SPINK1 may serve as a potential tumor promoter in NSCLC. In our study, we confirmed that the concentration of SPINK1 in serum of NSCLC patients was higher than normal controls, and concentration of SPINK1 in pleural effusions was even higher than in serum. These findings may shed new insight into early body fluid diagnosis of NSCLC. Collectively, these results revealed that SPINK1 may serve as a novel tumor promoter in NSCLC.
By performing functional studies for SPINK1, the mechanisms underlying the protumor role of SPINK1 were explored. In eukaryotes, multiple cell cycle checkpoints participate in the regulation of cell cycle progressions such as cyclin-dependent kinases (CDKs), cyclins and CDKs inhibitors. In our study, we indicated that silence of SPINK1 in A549 and H1299 cells resulted in cell cycle arrest, with increased percentage of cells in G1-Phase and decreased percentage of cells in S-Phase. In addition, western blot assays showed decreased expression of CDK4 in SPINK1 silencing groups. Consistent with our results, Haglund et al. reported that upregulation of SPINK1 in BRL-3A cells could increase cell numbers in S-Phase and G2/M Phase [16].
Autophagy is a highly conserved and finely regulated pathway for maintaining cell homeostasis, which can be achieved via inducing lysosomal degradation and recycling of dysfunctional proteins and organelles [30, 31]. The dysregulation of autophagy has been identified as a pathogenic feature of cancer. Previous studies have reported a dual role for autophagy in tumorigenesis. For instance, cancer cells have been demonstrated to induce autophagy under stressful or nutrition‑deprived conditions to promote cell survival [32], but excessive or constitutive activation of autophagy has been indicated to result in cancer cell death. Yamamura et al. reported autophagic cell death of pancreatic acinar cells in SPINK3 deficient mice, with increased ratio of LC3 II/LC3 I and increased formation of autophagosome [33]. Consistent with this report, the results revealed that knockdown of SPINK1 activated autophagy and apoptosis. To some extent when autophagy is no longer able to maintain cell survival, apoptosis will be initiated, with decreased expression of anti-apoptosis protein BCL-2 and increased expression of pro-apoptosis protein BAX [34]. In our models, we speculated that knockdown of SPINK1 conferred sensitivity to starvation and induced excessive autophagy and apoptosis.
Accumulating evidence has suggested that SPINK1 phosphorylates EGFR to exert an oncogenic effect. MEK/ERK and PI3K/AKT [35] signaling pathways were reported as executants downstream of EGFR. Based on these findings, it was hypothesized that SPINK1 may promote proliferation via activation of MEK/ERK signaling pathway. This hypothesis was supported by the decreased level of p-MEK1/2, p-ERK, p-PI3K, and p-AKT upon SPINK1 knockdown, while the phosphorylation level of P38 was not affected. Notably, compared with the PI3K/AKT signaling pathway, the phosphorylation of MEK/ERK decreased significantly after rhSPINK1 treatment. In addition, high-dose MEK inhibitor U0126 inhibited the MEK/ERK signaling pathway and cell proliferation, suggesting that SPINK1 partially accelerated NSCLC progression through the MEK/ERK signaling pathway.
In conclusion, the present study validated the role of SPINK1 as a tumor promoter in NSCLC. Knockdown of SPINK1 in NSCLC cell lines could inhibit cell proliferation, induce cell cycle arrest, and activate cell autophagy and apoptosis. Furthermore, SPINK1 promotes progression of NSCLC partially through MEK/ERK signaling pathway. To sum up, our study revealed a novel mechanism by which SPINK1 may promote tumor progression, and hence provided a new target for the treatment of NSCLC.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
References
Jemal, A., Siegel, R., & Ward, E., et al. (2007). Cancer statistics, 2007. CA: A Cancer Journal for Clinicians, 57, 43–66.
Zheng, X., Cheng, M., & Fu, B., et al. (2015). Targeting LUNX inhibits non-small cell lung cancer growth and metastasis. Cancer Research, 75, 1080–1090.
Molina, J. R., Yang, P., & Cassivi, S. D., et al. (2008). Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clinic Proceedings, 83, 584–594.
Herbst, R. S., Heymach, J. V., & Lippman, S. M. (2008). Lung cancer. New England Journal of Medicine, 359, 1367–1380.
Lemjabbar-Alaoui, H., Hassan, O. U., & Yang, Y. W., et al. (2015). Lung cancer: Biology and treatment options. Biochim Biophys Acta, 1856, 189–210.
Herbst, R. S., Morgensztern, D., & Boshoff, C. (2018). The biology and management of non-small cell lung cancer. Nature, 553, 446–54.
Kazal, L. A., Spicer, D. S., & Brahinsky, R. A. (1948). Isolation of a crystalline trypsin inhibitor-anticoagulant protein from pancreas. Journal of the American Chemical Society, 70, 3034–3040.
Huhtala, M. L., Pesonen, K., & Kalkkinen, N., et al. (1982). Purification and characterization of a tumor-associated trypsin inhibitor from the urine of a patient with ovarian cancer. The Journal of Biological Chemistry, 257, 13713–13716.
Stenman, U. H., Huhtala, M. L., & Koistinen, R., et al. (1982). Immunochemical demonstration of an ovarian cancer-associated urinary peptide. International Journal of Cancer, 30, 53–57.
Lukkonen, A., Lintula, S., & von Boguslawski, K., et al. (1999). Tumor-associated trypsin inhibitor in normal and malignant renal tissue and in serum of renal-cell carcinoma patients. International Journal of Cancer, 83, 486–490.
Kelloniemi, E., Rintala, E., & Finne, P., et al. (2003). Tumor-associated trypsin inhibitor as a prognostic factor during follow-up of bladder cancer. Urology, 62, 249–253.
El-mezayen, H. A., Metwally, F. M., & Darwish, H. (2014). A novel discriminant score based on tumor-associated trypsin inhibitor for accurate diagnosis of metastasis in patients with breast cancer. Tumour Biology: the Journal of the International Society for Oncodevelopmental Biology and Medicine, 35, 2759–2767.
Lyytinen, I., Lempinen, M., & Nordin, A., et al. (2013). Prognostic significance of tumor-associated trypsin inhibitor (TATI) and human chorionic gonadotropin-β (hCGβ) in patients with hepatocellular carcinoma. Scandinavian Journal of Gastroenterology, 48, 1066–1073.
Paju, A., Vartiainen, J., & Haglund, C., et al. (2004). Expression of trypsinogen-1, trypsinogen-2, and tumor-associated trypsin inhibitor in ovarian cancer: prognostic study on tissue and serum. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research, 10, 4761–4768.
Venesmaa, P., Stenman, U. H., & Forss, M., et al. (1998). Pre-operative serum level of tumour-associated trypsin inhibitor and residual tumour size as prognostic indicators in Stage III epithelial ovarian cancer. British jJournal of Obstetrics and Gynaecology, 105, 508–11.
Chang, C., Zhao, W., & Luo, Y., et al. (2017). Serine peptidase inhibitor Kazal type I (SPINK1) promotes BRL-3A cell proliferation via p38, ERK, and JNK pathways. Cell Biochemistry and Function, 35, 339–48.
Marchbank, T., Mahmood, A., & Playford, R. J. (2013). Pancreatic secretory trypsin inhibitor causes autocrine-mediated migration and invasion in bladder cancer and phosphorylates the EGF receptor, Akt2 and Akt3, and ERK1 and ERK2. American Journal of Physiology Renal Physiology, 305, F382–F9.
Chen, F., Long, Q., & Fu, D., et al. (2018). Targeting SPINK1 in the damaged tumour microenvironment alleviates therapeutic resistance. Nature Communications, 9, 4315.
Mehner, C., Oberg, A. L., & Kalli, K. R., et al. (2015). Serine protease inhibitor Kazal type 1 (SPINK1) drives proliferation and anoikis resistance in a subset of ovarian cancers. Oncotarget, 6, 35737–35754.
Venesmaa, P., Stenman, U. H., & Forss, M., et al. (1998). Pre-operative serum level of tumour-associated trypsin inhibitor and residual tumour size as prognostic indicators in Stage III epithelial ovarian cancer. British Journal of Obstetrics and Gynaecology, 105, 508–511.
Chang, C., Zhao, W., & Luo, Y., et al. (2017). Serine peptidase inhibitor Kazal type I (SPINK1) promotes BRL-3A cell proliferation via p38, ERK, and JNK pathways. Cell Biochemistry & Function, 35, 339–48.
Ying, H. Y., Gong, C. J., & Feng, Y., et al. (2017). Serine protease inhibitor Kazal type 1 (SPINK1) downregulates E-cadherin and induces EMT of hepatoma cells to promote hepatocellular carcinoma metastasis via the MEK/ERK signaling pathway. Journal of Digestive Diseases, 18, 349–58.
Ohmuraya, M., Hirota, M., & Araki, K., et al. (2006). Enhanced trypsin activity in pancreatic acinar cells deficient for serine protease inhibitor kazal type 3. Pancreas, 33, 104–106.
Wang, J., Ohmuraya, M., & Hirota, M., et al. (2008). Expression pattern of serine protease inhibitor kazal type 3 (Spink3) during mouse embryonic development. Histochemistry and Cell Biology, 130, 387–397.
Lu, F., Lamontagne, J., & Sun, A., et al. (2011). Role of the inflammatory protein serine protease inhibitor Kazal in preventing cytolytic granule granzyme A-mediated apoptosis. Immunology, 134, 398–408.
Paju, A., Hotakainen, K., & Cao, Y., et al. (2007). Increased expression of tumor-associated trypsin inhibitor, TATI, in prostate cancer and in androgen-independent 22Rv1 cells. European Urology, 52, 1670–1679.
Xu, L., Lu, C., & Huang, Y., et al. (2018). SPINK1 promotes cell growth and metastasis of lung adenocarcinoma and acts as a novel prognostic biomarker. BMB Reports, 51, 648–53.
Tomlins, S. A., Rhodes, D. R., & Yu, J., et al. (2008). The role of SPINK1 in ETS rearrangement-negative prostate cancers. Cancer Cell, 13, 519–528.
Räsänen, K., Itkonen, O., & Koistinen, H., et al. (2016). Emerging Roles of SPINK1 in Cancer. Clinical Chemistry, 62, 449–457.
Dikic, I., Johansen, T., & Kirkin, V. (2010). Selective autophagy in cancer development and therapy. Cancer Research, 70, 3431–3434.
Janku, F., McConkey, D. J., & Hong, D. S., et al. (2011). Autophagy as a target for anticancer therapy. Nature Reviews Clinical Oncology, 8, 528–539.
Guo, X. L., Li, D., & Sun, K., et al. (2013). Inhibition of autophagy enhances anticancer effects of bevacizumab in hepatocarcinoma. Journal of Molecular Medicine, 91, 473–483.
Ohmuraya, M., Hirota, M., & Araki, M., et al. (2005). Autophagic cell death of pancreatic acinar cells in serine protease inhibitor Kazal type 3-deficient mice. Gastroenterology, 129, 696–705.
Yang, D., Liu, X., & Zhang, R., et al. (2011). Increased apoptosis and different regulation of pro-apoptosis protein bax and anti-apoptosis protein bcl-2 in the olfactory bulb of a rat model of depression. Neuroscience Letters, 504, 18–22.
Ozaki, N., Ohmuraya, M., & Hirota, M., et al. (2009). Serine protease inhibitor Kazal type 1 promotes proliferation of pancreatic cancer cells through the epidermal growth factor receptor. Molecular Cancer Research, 7, 1572–1581.
Acknowledgements
This study is supported by National Natural Science Foundation grants of China (No. csfc81373151) and Natural Science Foundation Project of CQCSTC (No. cstc2018jcyjAX0257). We thank Zhiqiang Ding for the correction of the English usage of the manuscript.
Author information
Authors and Affiliations
Contributions
D.Y.L. and Y.H. conceived of and designed the research. X.Z., Q.H., D.Y.L., Y.T.Z., and R.S.A. performed the experiments and analyzed the data. X.Z. and Y.H. wrote the manuscript. Z.M.W. and C.F.J. interpreted the data and corrected the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationship that could be construed as a potential conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, D., Zhang, X., Ding, Z. et al. Identification and Exploration of Serine Peptidase Inhibitor Kazal Type I (SPINK1) as a Potential Biomarker Correlated with the Progression of Non-Small Cell Lung Cancer. Cell Biochem Biophys 80, 807–818 (2022). https://doi.org/10.1007/s12013-022-01098-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-022-01098-w