Abstract
Under normal physiological conditions, IGF-1 (insulin-like growth factor-1) has important biological effects. However, many studies have found that IGF-1 is closely related to the occurrence and development of breast cancer. But up to now, the cellular properties of IGF-1 have not been systematically explored in breast cancer cell. It is well-known that the cellular properties and behaviors of IGF-1/IGF-1R are closely related to its biological functions. In the current study, we used the breast cancer cell line as a model to explore the biological characteristics of IGF-1/IGF-1R, and found that IGF-1/IGF-1R can be internalized into the cytoplasm. In addition, we also found that IGF-1R can also enter cell nuclei under the mediation of IGF-1. Further research found that the nuclear-localized IGF-1R has important potential biological effects, which is closely associated to the proliferation of breast cancer cell, this may be achieved by regulating IGF-1R-mediated intracellular signaling. The current research has laid the foundation for investigating the relationship between IGF-1/IGF-1R system and the occurrence and development of breast cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A series of studies have shown that IGF-1 (insulin-like growth factor-1) has many important biological functions and roles [1, 2]. IGF-1R is almost expressed in all tissues, therefore IGF-1 has the potential biological effect on all tissues and organs, such as promoting cell differentiation, regulating cell proliferation, and promoting protein and DNA synthesis [3, 4]. IGF-1 needs to interact with IGF-1R to play its biological role. Structurally, IGF-1R is a tetramer composed of two α subunits and two β subunits linked by disulfide bonds. The α subunit of the extracellular domain contains the ligand binding site; the β subunit consists of a transmembrane domain and the intracellular domain, in which the intracellular domain has tyrosine kinase (TK) activity. When IGF-1 binds to its receptor (IGF-1R) expressed on the cell membrane, it can cause IGF-1R autophosphorylation, which in turn activates a series of signal cascades, such as phosphatidylinositol 3-kinase (PI3K)-Akt and mitogen-activated protein kinase pathways. The activated signal transduction molecules are then transported to the nucleus, where they initiate gene transcription [4,5,6,7].
Although under normal physiological conditions, IGF-1 mainly secreted by the liver has important biological effects [7], studies have showed that IGF-1/IGF-1R system is closely related to the occurrence and development of many types of malignant tumors [8]. It has been reported that IGF-1 is involved in tumor proliferation and metastasis [9]. In addition, studies have shown that IGF-1R is highly expressed in many types of tumor tissues [10]. Therefore, the IGF-1R can be used as a potential target for anti-tumor therapy.
The cellular properties of IGF-1/IGF-1R are closely related to its biological functions [11, 12]. Studies have shown that IGF-1R is closely associated with breast cancer [13,14,15], but the cellular properties of IGF-1/IGF-1R have not been systematically studied in breast cancer cells. For this reason, in the current study, we used the breast cancer cell line as a model to explore the cellular characteristics of IGF-1/IGF-1R, and found that IGF-1/IGF-1R could be internalized into the cytoplasm. More importantly, we also found that IGF-1R could also transport into the cell nuclei under IGF-1 induction. Further study indicated that the nuclear-localized IGF-1R has important potential biological functions. Taken together, the current research has laid the foundation for exploring the relationship between IGF-1/IGF-1R system and the occurrence and development of breast cancer.
Materials and Methods
Antibodies and Reagents
IGF-1 was purchased from Sigma (St. Louis, USA). p-AKT, p-ERK1/2, total AKT, and total ERK1/2 antibodies were purchased from Cell Signaling Technology. BCA protein assay kit was purchased from Thermo Scientific. MTT assay kit was purchased from Life Technologies. Anti-IGF-1Rβ, anti-EEA1, anti-Rab5, anti-Rab11, anti-clathrin, anti-caveolin, and Nup 358 antibodies were purchased from Abcam. Fetal bovine serum (FBS) and DMEM medium were purchased from Invitrogen (USA). Cell lysate and PVDF membrane were purchased from Biyuntian (Shanghai, China). Low-fluorescence PVDF membrane was purchased from Bio-Rad (DMEM). Cell Counting Kit 8 (WST-8/CCK8) was purchased from Abcam (ab228554). Unless otherwise stated, reagents were purchased from Sigma-Aldrich.
Cell Culture
Human breast cancer cell line MCF-7 and T47D were purchased from ATCC, the cells were cultured in DMEM (Dulbecco’s Modified Eagle Medium) with 10% FBS at 37 °C in a humidified atmosphere of 5% CO2.
Western-blot Analysis
Cell protein was extracted using RIPA lysate. Protein concentration was determined using BCA kit according to manufacturer’s instructions. The protein samples were subjected to SDS-polyacrylamide gel electrophoresis for separation of proteins. The protein on the gel was transferred to the PVDF membrane. After washing PVDF membrane three times with TBST, the PVDF membrane was blocked with 5% BSA for 2 h at 37 °C. After washing the PVDF membrane twice, primary antibodies were added and incubated at 4 °C for 12 h. Then, after the PVDF membranes were washed three times, the PVDF membrane was incubated with the secondary antibody at 37 °C for 2 h. After washing, the membranes were detected using a fluorescence imaging system (Bio-Rad).
Immunofluorescence
MCF-7 and T47D cells were seeded on coverslips at a density of 2 × 105 per well. When MCF-7 and T47D cells grew to about 30–50% confluence. After washing three times with PBS, the cells were fixed with 4% PFA for 30 min at room temperature. After washing the cells twice, the cells were permeabilized with 0.4% Triton X-100 for 30 min. The cells were washed and blocked with 10% donkey serum at 37 °C for 1 h. After washing the cell 3 times with PBS, the cell samples were incubated with the primary antibody overnight at 4 °C. The cells were washed twice with PBS, and then incubated with the secondary antibody for 1 h at 37 °C. DAPI was used to stain the nucleus. The cell sample was detected with a confocal laser scanning microscope (CLSM) (Olympus, FV3000).
ELISA Assay
After MCF-7 and T47D cells were stimulated with IGF-1, the cytoplasmic and nuclear proteins were extracted using the Subcellular Protein Fraction Kit (Thermo Fisher). The samples were tested using the ELISA kit according to the manufacturer’s instructions.
Cell Proliferation Assay
The CCK-8 kit was used to detect cell proliferation. MCF-7 and T47D cells were added to a 96-well culture plate and incubated at 37 °C for 24–72 h. After the cells were washed twice, IGF-1 was added and incubated for 24 h at 37 °C. Then, the cells were washed and 10 μL of CCK solution was added, and then incubated at 37 °C for 120 min. An ELISA reader (Bio-Rad, USA) was used to determine the OD value at 450 nm.
Analysis of Cell Cycle by Flow Cytometry
After IGF-1 treatment, MCF-7 and T47D cells were digested from the cell culture plate and prepared as a single cell suspension. After the cells were centrifuged, the supernatant was discarded. The cell samples were then treated with 70% alcohol at 4 °C for 0.5 h. After the cell samples were centrifuged again, the supernatant was discarded. The cell pellet was treated with RNase A (20 μg/mL) for 0.5 h. The cell sample was mixed with PI and incubated at 37 °C for 0.5 h. The cell cycle was then tested a BD Accuri C6 flow cytometer.
Cell Transfection
For cell transfection with siRNAs, MCF-7 and T47D cells (2 × 105 cells/mL) were used in this study. SiRNA against importin-β and Nup 358 were transfected into MCF-7 and T47D cells using lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). Western blot was used to determine siRNA knockdown efficiency and specificity.
Analyze Cell Proliferation Using Cell-IQ
MCF-7 and T47D cells were seeded into a 48-well plate (2 × 104 cells per well), 0.5 mL medium with 10% FBS in DMEM medium was added. The cell was cultured in a humidified atmosphere of 5% CO2 and 95% air. When the cells were in logarithmic growth phase, different concentration of IGF-1 was added. The cell proliferation was monitored by the Cell-IQ system. Cell-IQ software were used to analyze cell proliferation.
RT-PCR
MCF-7 or T47D cells (2 × 105/well) were seeded into a 12-well cell culture plates. MCF-7 or T47D cells in a logarithmic growth phase were trypsinized and counted. RNA was extracted and purified from MCF-7 or T47D cells using the RNeasy Mini kit following the manufacturer’s protocol. total RNA was immediately reverse-transcribed into first-strand cDNA with ReverTra Ace (TOYOBO, Osaka, Japan). RT-PCR was done using the following primer: IGF-1R primers forward: 5′-GGAGTTGTATTTGCCATCACCAGGG-3′, reverse: 5′-ATGCGCGGGCAAATTTGATCCCATA-3′.
Analysis of Intracellular Signaling by Flow Cytometry
The cells (MCF-7 or T47D) were starved for 10 h. The cells were then stimulated with IGF-1. After IGF-1 treatment, the cells were collected by centrifugation (1000 rpm, 8 min). The cells were washed with ice-cold PBS, and fixed in 70% ethanol for 2 h at 4 °C. the cells were then blocked with 5% BSA for 2 h, the cells were incubated with primary antibodies overnight, followed by incubation with a fluorophore-conjugated secondary antibody. After centrifugation, the cell samples were determined by Flow cytometry (Becton Dickinson).
Statistical Analysis
All data are expressed as mean ± standard deviation (SD), and analyzed using SPSS 20.0 statistical software (SPSS Inc., IL). The p values were calculated using one-way analysis of variance or Student t test. These data obey normal distribution, and normality was tested by the Kolmogorov–Smirnov test. P < 0.05 was considered to indicate a statistically significant difference.
Results
Analysis of IGF-1R Expression in MCF-7 and T47D Cells
The expression of IGF-1R was analyzed using a CLSM. As shown in Fig. 1A, IGF-1R was mainly localized on the cell membrane in MCF-7 or T47D cells (Supplementary Fig. 1). Additionally, we also analyzed the expression of IGF-1R using flow cytometry, the results indicated that MCF-7 and T47D also expressed high levels of IGF-1R (Fig. 1B). Furthermore, we also detected interaction between IGF-1R and IGF-1R on the MCF-7 and T47D cells (Supplementary Fig. 1).
A Evaluation of IGF-1R expression in breast cancer cell lines (MCF-7 and T47D). The cells were seeded on coverslips. After the cells grew to about 30–50% confluence, the cells were fixed with 4% PFA, permeabilized with 0.4% Triton X-100, and blocked with 10% donkey serum at 37 °C for 1 h. After washing, the cell samples were incubated with the anti-IGF-1R antibody (1:1000 dilutions) overnight at 4 °C. The cells were washed twice with PBS, and then incubated with the secondary antibody (1:2000 dilutions) for 1 h at 37 °C. The cell sample was detected with a confocal laser scanning microscope (Olympus, FV3000). B Detection of the IGF-1R expression by flow cytometry. The cells were collected by centrifugation. After washing, the cells were fixed in 70% ethanol for 2 h at 4 °C, and blocked with 5% BSA for 2 h, the cells were then incubated with anti-IGF-1R (1:1000 dilutions) overnight, followed by incubation with a fluorophore-conjugated secondary antibody (1:500 dilutions) at 37 °C for 1 h. After centrifugation and washing, the cell samples were determined by Flow cytometry (Becton Dickinson). C IGF-1 interacted with IGF-1R in MCF-7 and T47D cells. The cells were cultured in serum-free medium for 5 h. The cells were then stimulated with IGF-1 (50 ng/mL) for 5 min. The cells were then fixed, permeabilized, and blocked with 10% donkey serum at 37 °C for 1 h. The cells were incubated with anti-IGF-1 (1:500 dilutions) and anti-IGF-1R antibody (1:1000 dilutions) at 37 °C for 1 h. After washing, the cells were incubated with the fluorescently labeled secondary antibody for 1 h at 37 °C. The cell sample was detected with a confocal laser scanning microscope (Olympus, FV3000). Each experiment was repeated at least three times. Data are expressed as mean ± SD. An asterisk (*) indicates a significant difference (p < 0.05)
Intracellular Trafficking of IGF-1/IGF-1R in MCF-7 and T47D Cells
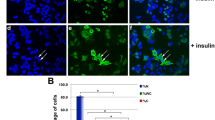
CLSM was used to evaluate the cellular behavior of IGF-1 on the MCF-7 cell model. The cells were incubated with IGF-1 (50 ng/mL) for different time points (1–90 min). The results showed that fluorescently labeled IGF-1 was internalized into MCF-7 cells in a time-dependent manner (Fig. 2A). It can be seen that IGF-1 was mainly localized on the cell membrane after treatment with IGF-1 for 1 min. The cytoplasmic fluorescence signal gradually increased with time increasing (3–30 min). The intracellular fluorescence signal reached an almost maximal level 30–60 min after IGF-1 treatment and subsided rapidly within 60–90 min.
A Time course of IGF-1 internalization of in MCF-7 cells. The cells were cultured in serum-free medium for 5 h. The cells were then incubated with fluorescently labeled IGF-1 (50 ng/mL) for 1–90 min. After washing, the cells were fixed. The cell samples then analyzed with a confocal laser scanning microscope (Olympus, FV3000) excitation wavelength of 488 nm and emission wavelength of 521 nm. B Time course of IGF-1 internalization of in T47D cells. The cells were cultured in serum-free medium for 5 h. After washing, the cells were then stimulated with fluorescently labeled IGF-1 (50 ng/mL) for 1–90 min. After washing, the cells were fixed. The cell samples then detected with a confocal laser scanning microscope (Olympus, FV3000) at excitation wavelength of 488 nm and emission wavelength of 521 nm. Each experiment was repeated at least three times
Interestingly, CLSM observation showed that IGF-1 could transport into the nucleus in the MCF-7 cell model (Fig. 2A). Additionally, we also analyzed IGF’s intercellular trafficking in T47D cells, and the results showed that IGF-1 also transported into cell nuclei of T47D (Fig. 2B).
The Subcellular Localization of IGF-1R Under IGF-1 Stimulation
Here, the intracellular trafficking of IGF-1R was analyzed under IGF-1 stimulation on the MCF-7 cell model. It was found that IGF-1R can be internalized into the MCF-7 cells in a time-dependent manner after IGF-1 treatment, and IGF-1R was also transported to the cell nuclei. As shown in Fig. 3A, IGF-1R was mainly localized on the cell membrane without IGF-1 stimulation, and IGF-1R rapidly internalized into cell nuclei after treatment with IGF-1 for 5 min. The intracellular fluorescence signal gradually increased with time increasing (3–30 min). The intracellular fluorescence signal reached an almost maximal level 30–60 min after IGF-1 treatment and subsided rapidly within 60–90 min. In addition, we also analyzed the intracellular trafficking of IGF-1R in T47D cells (Supplementary Fig. 2). The results form ELISA also confirmed these findings (Fig. 3B). These results imply that nuclear-localized IGF-1R may have important biological functions.
A Detection of subcellular localization under IGF-1 treatment in MCF-7 cell. The cells were seeded on coverslips. After the cells grew to about 30–50% confluence, the cells were stimulated with IGF-1 (50 ng/mL) for the indicated time points, after which, the cells were fixed, permeabilized, and blocked at 37 °C for 1 h. After washing, the cell samples were incubated with the anti-IGF-1R antibody (1:1000 dilutions) overnight at 4 °C. After washing, the cells were incubated with the fluorescently labeled secondary antibody (1:2000 dilutions) for 1 h at 37 °C. The cell sample was detected with a confocal laser scanning microscope (Olympus, FV3000). B the cells were stimulated with IGF-1 (50 ng/mL) for the indicated time points (1–60 min). Subcellular fractions were extracted using a subcellular fractionation kit. The IGF-1R levels were then analyzed by ELISA kit according to the manufacturer’s instructions. Each experiment was repeated at least three times. Data are expressed as mean ± SD
Clathrin was Involved in IGF-1R’s Endocytosis
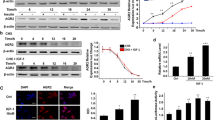
The endocytosis is the first step of IGF-1R’s internalization. Here, to study the potential endocytosis mechanism of IGF-1R, Co-localization analysis was done. It is well-known that clathrin is an important molecule that mainly mediates the endocytosis of growth factors and cytokines. Therefore, we analyzed whether clathrin is involved in IGF-1R’s endocytosis in breast cancer cells by co-localization analyses, and the results indicated that the co-localization signal between clathrin and IGF-1R could be detected (Fig. 4A), indicating that clathrin may be involved in the endocytosis of IGF-1R in breast cancer cells. To further confirm the effect of clathrin in the process of IGF-1R’s endocytosis, clathrin inhibitor (30 μM) was used to pretreat the cells for 0.5 h, and the results showed that clathrin inhibitor could significantly inhibit the endocytosis of IGF-1R (Fig. 4B). In addition, clathrin was knocked down, and the results also showed that IGF-1R’s endocytosis was significantly blocked (Fig. 4B).
A Detection of interaction between clathrin and IGF-1R by co-localization analysis. The cells were cultured in serum-free medium for 6 h. the cells were treated with IGF-1 (50 ng/mL) for 10 min. After washing, the cells were fixed, permeabilized, and blocked at 37 °C for 1 h. After three washes, the cell samples were incubated with the anti-IGF-1R antibody (1:1000 dilutions) or anti-clathrin antibody (1:1000 dilutions) overnight at 4 °C, respectively. After washing, the cells were incubated with the fluorescently labeled secondary antibody for 1 h at 37 °C. The cell samples were detected with a confocal laser scanning microscope (Olympus, FV3000). B Analysis of the effect of clathrin on IGF-1R’s endocytosis. The cells were cultured in serum-free medium for 6 h. the cells were then preincubated clathrin inhibitor (30 μM) for 0.5 h before the cells were stimulated with IGF-1 (50 ng/mL) for 15 min, and then IGF-1R was detected by CLSM (additionally, the effect of clathrin on IGF-1R’s endocytosis was detected by knocking down expression of clathrin). Each experiment was repeated at least three times. Data are expressed as mean ± SD. An asterisk (*) indicates a significant difference (p < 0.05)
The Internalized IGF-1R Transported into Different Types of Endosomes
It is well-known that endosomes play an important role in the intracellular transport of cytokine receptors. To analyze which types of endosomes are involved in the intracellular transport of IGF-1R, indirect immunofluorescence assays were performed, and the results showed that IGF-1R entered into the EEA1-positive early endosomes. In addition, IGF-1R also entered into Rab11, and Rab9-positive endosomes (Fig. 5). These findings provide an explanation for the different subcellular locations of IGF-1R.
IGF-1R entered into different types of endosomes by IFA assays. The cells were stimulated with IGF-1 (50 ng/mL) for 15 min, after which, the cells were fixed, permeabilized, and blocked at 37 °C for 1 h. After three washes, the cells were incubated with the anti-IGF-1R antibody (1:1000 dilutions), anti-EEA1 antibody (1:1000 dilutions), anti-Rab5 antibody (1:1500 dilutions), anti-Rab9 antibody (1:2000 dilutions) overnight at 4 °C, respectively. After washing, the cells were incubated with the fluorescently labeled secondary antibody for 1 h at 37 °C. The cell samples were detected with a confocal laser scanning microscope (Olympus, FV3000)
Tyrosine Phosphorylation of IGF-1R is Required for IGF-1R’s Nuclear Transport
We investigated whether the phosphorylation of IGF-1R is involved in its nuclear localization. IGF-1R phosphorylation inhibitor (Linsitinib, 100 nmol/L) was used to pretreat the cells for 2 h, and it was found that the nuclear transport of IGF-1R was significantly inhibited (Fig. 6A). However, its cytoplasmic internalization was not significantly affected. These experimental results indicate that the phosphorylation of IGF-1R is required for its nuclear localization.
A IGF-1R tyrosine phosphorylation was required for IGF-1R’s nuclear localization. IGF-1R phosphorylation inhibitor (Linsitinib, 100 nmol/L) were used to pretreat the cells for 2 h before the cells were treated with IGF-1 (50 ng/mL) for 0.5 h. After IGF-1 treatment, the cells were fixed, permeabilized, and blocked. The cells were incubated with the anti-IGF-1R antibody (1:1000 dilutions). After washing, the cells were incubated with the fluorescently labeled secondary antibody for 1 h at 37 °C. The cell samples were detected with a confocal laser scanning microscope (Olympus, FV3000). B NUP 358 is required for IGF-1R nuclear localization. NUP 358 were knock down by siRNA method. The cells were then stimulated with IGF-1 (50 ng/mL) for 30 min. After washing, the cells were fixed, permeabilized, and blocked. The cells were incubated with the anti-IGF-1R antibody (1:1000 dilutions). The cell samples were then detected by CLSM. Data are expressed as mean ± SD (50 cells were analyzed each treatment). An asterisk (*) indicates a significant difference (p < 0.05)
In addition, crossing the nuclear membrane is one of the key steps for IGF-1R’s nuclear translocation. Studies have shown that NUP 358 is involved in the nuclear translocation of IGF-1R in other types of tumor cell lines [16]. Here, to detect whether NUP 358 is required for IGF- 1R’s nuclear localization in breast cancer cell line (MCF-7), the co-localization analysis was performed, and the results indicated that co-localization signals between IGF-1R and NUP 358 could be detected (Fig. 6A). To further demonstrate that NUP 358 was involved in the nuclear transport of IGF-1R, the expression of NUP 358 was knocked down by siRNA method, we can see that the nuclear localization of IGF-1R was significantly reduced. This further confirms that NUP 358 plays important role in the process of nuclear localization of IGF-1R (Fig. 6B).
IGF-1R’s Nuclear Localization is Closely Related to the Cell Proliferation
To study the biological functions of nuclear-localized IGF-1R, we established a nonnuclear-localized IGF-1R cell model by knocking down Nup 358 (MCF-7 cell). It can be seen that knockdown of NUP 358 did not affect the proliferation of MCF-7 cells and did not lead to cell apoptosis (Fig. 7A, B), but knockdown of NUP 358 significantly inhibited the nuclear localization of IGF-1R (Fig. 7C). We then evaluated the effect of IGF-1R’s nuclear localization on cell proliferation. As shown in Fig. 7C, the reduction of IGF-1R’s nuclear localization significantly weakened the cell’s proliferation ability (Fig. 7D). In order to further evaluate the role of nuclear-localized IGF-1R. In addition, the cell cycle also was also changed (Fig. 7E). Additionally, the expression of PCNA (cell proliferation marker) was also significantly reduced compared to the control (Fig. 7F).
A Knockdown of NUP 358 expression does not affect the cell proliferation of MCF-7 cells. MCF-7 cells were added to a 96-well culture plate and incubated at 37 °C. IGF-1 (50 ng/mL) was then added and incubated for 24 h at 37 °C. After washing, 10 μL of CCK solution was added, and incubated at 37 °C for 2 h. An ELISA reader was used to determine the OD value at 450 nm. B Knockdown of NUP 358 expression did not cause breast cancer cell apoptosis. The cell apoptosis was analyzed by flow cytometry using Annexin V-FITC/PI staining kit according to manufacturer’s instruction. C NUP 358 Knockdown significantly weakened the nuclear localization of IGF-1R. D The reduction of IGF-1R nuclear localization significantly leaded to the cell proliferation decrease. E The IGF-1R’s nuclear localization is associated with the cell cycle. After IGF-1 (50 ng/mL) treatment for 30 min, the cells were digested from the cell culture plate and prepared as a single cell suspension. After centrifugation, the supernatant was discarded. The cell samples were then treated with 70% alcohol at 4 °C for 30 min. After washing, the cell pellet was treated with RNase A (20 µg/mL) for 30 min. The cell cycle was then determined by a BD Accuri C6 flow cytometer. F Decreased nuclear localization of IGF-1R downregulates PCNA expression. Western blot was used to detect PCNA expression. Each experiment was repeated at least three times. Data are expressed as mean ± SD. An asterisk (*) indicates a significant difference (p < 0.05)
Nuclear-localized IGF-1R Increases the Phosphorylation of Signaling Molecules
We further analyzed the potential molecular mechanism by which nuclear-localized IGF-1R promotes cell proliferation. We study this issue from the perspective of signaling transduction. As indicated in Fig. 8, the results showed that the activation of p-ERK1/2 was significantly prolonged and increased compared to the nonnuclear localized IGF-1R group, these results indicated that nuclear-localized IGF-1R could be associated with IGF-1R-mediated signaling transduction. This may be one of the potential mechanisms by which nuclear localization of IGF-1R can promote breast cell proliferation. In addition, the results of ELISA analysis also confirmed this observation (Supplementary Fig. 3).
The nuclear localization of IGF-1R increased the activation of intracellular signaling molecules. The cells were starved for 10 h. The cells were then stimulated with IGF-1 (50 ng/mL) for the indicated time points. After IGF-1 stimulation, the cells were collected by centrifugation (1000 rpm, 8 min). The cells were washed and fixed in 70% ethanol for 2 h at 4 °C. After washing, the cells were then blocked with 5% BSA for 2 h, the cells were incubated with primary antibodies (anti-p-ERK1/2) overnight, followed by incubation with a fluorophore-conjugated secondary antibody. After centrifugation, the cell samples were determined by Flow cytometry (Becton Dickinson). Each experiment was repeated at least three times. Data are expressed as mean ± SD. An asterisk (*) indicates a significant difference (p < 0.05)
Discussion
More than 60 years ago, Salmon and Daughaday first described IGF-1 [17]. Subsequently, a series of intensive studies were carried out around the IGF-1 molecule. Although under normal physiological conditions, IGF-1/IGF-1R can promote cell proliferation, tissue growth and development [1]. However, a series of recent studies have shown that IGF-1/IGF-1R is closely related to the occurrence and development of many types of tumors [17,18,19]. IGF-1R usually is over-expressed in malignant tumor cells and low expression in normal tissues [10]. Studies have shown that the occurrence and development of breast cancer is also closely related to IGF/IGF-1R system. Furthermore, it has been reported that elevated nuclear IGF-1R levels are negatively correlated with the survival rate of cancer patients. The cellular behavior of IGF-1 is closely related to its biological activity, and the biological characteristics of IGF-1R have not been revealed yet on breast cancer cells. Therefore, in the current study, breast cancer cell lines were used as a model to investigate the cell behavior on the IGF-1/IGF-1R. We found that IGF-1R was internalized into cell cytoplasm, and some were transported to cell nuclei of MCF-7 and T47D cells. Further research showed that the nuclear-localized IGF-1R is closely related to the proliferation of breast cancer cells via regulating ERK1/2 signaling (Fig. 9).
The endocytosis of IGF-1R is the first step in the process of IGF-1R’s nuclear localization. In the current study, we found that clathrin-mediated endocytosis plays an important role in the process of IGF-1R’s endocytosis. Moreover, inhibition of clathrin could significantly reduce the IGF-1R’s internalization. Previous studies have shown that the endocytic pathway of IGF-1R is mediated by clathrin-dependent endocytosis or caveolin-mediated endocytosis [16]. Of course, there is also non-clathrin and caveolin-dependent endocytic pathways. In addition, we also initially analyzed which types of endosomes are involved in the cytoplasmic transport of IGF-1R, the results showed that IGF-1R entered into the EEA1-positive early endosomes. In addition, IGF-1R also entered into Rab11, and Rab9-positive endosomes. These findings provide an explanation for the different subcellular locations of IGF-1R.
The macromolecules shuttling between the nucleus and the cytoplasm is mediated by the nuclear pore complex [20, 21]. Therefore, how IGF-1R crosses the nuclear membrane is an extremely important scientific question. Recently, Packham et al. found that the entry of IGF-1R into the nucleus requires the participation of NUP 358 under the mediation of importin-β. In addition, they also found that p150Glued also plays an important role in the nuclear localization of IGF-1R. In the current study, we also found that NUP 358 (also termed as Ran/BP2) was involved in the nuclear localization of IGF-1R [22].
In addition, IGF-1R is a TK receptor, which has high homology with insulin receptor (IR). Since the breast cancer cell line used in the current study expressed both IGF-1R and IR. To eliminate the potential impact of IR, we analyzed whether IGF-1 can bind to IR in MCF-7 cells. The results show that IGF-1’s affinity for IR is very weak, which is similar to previous reports [23].
What are the important physiological functions of nuclear localization of IGF-1R? We found that nuclear-localized IGF-1R can enhance the activation of ERK1/2 (phosphorylation). This may be one of the important mechanisms by which nuclear-localized IGF-1R can promote the proliferation of breast cancer cells. IGF-1R can activate many signal transduction pathways, among which, the activated ERK1/2 can promote cell growth. Of course, this may not be the only mechanism by which IGF-1R functions in the nucleus. Nuclear-localized IGF-1R may have a series of important biological activities. These scientific issues need further exploration in the future.
In summary, we studied the cell behavior of IGF-1/IGF-1R in detail in the breast cancer cell lines (MCF-7 and T47D cells). In the current study, we found that IGF-1R not only can internalize into the cytoplasm, but also can transported into the cell nuclei. Further studies indicated that the nuclear localization of IGF-1R is closely related to cell proliferation of breast cancer. Current work suggests that the development of inhibitors of nuclear transport of IGF-1R may have anti-breast cancer potential.
References
Feng, Z., & Levine, A. J. (2010). The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends in Cell Biology, 20(7), 427–434.
Ascenzi, F., Barberi, L., Dobrowolny, G., Villa Nova Bacurau, A., Nicoletti, C., Rizzuto, E., & Musarò, A. (2019). Effects of IGF-1 isoforms on muscle growth and sarcopenia. Aging Cell, 18(3), e12954.
Rigiracciolo, D. C., Nohata, N., Lappano, R., Cirillo, F., Talia, M., Scordamaglia, D., & Maggiolini, M. (2020). IGF-1/IGF-1R/FAK/YAP transduction signaling prompts growth effects in triple-negative breast cancer (TNBC) cells. Cells, 9(4), 1010.
Forbes, B. E., Blyth, A. J., & Wit, J. M. (2020). Disorders of IGFs and IGF-1R signaling pathways. Molecular and Cellular Endocrinology, 518, 1110354
Delafontaine, P., Song, Y. H., & Li, Y. (2004). Expression, regulation, and function of IGF-1, IGF-1R, and IGF-1 binding proteins in blood vessels. Arteriosclerosis, Thrombosis, and Vascular Biology, 24(3), 435–444.
Troncoso, R., Ibarra, C., Vicencio, J. M., Jaimovich, E., & Lavandero, S. (2014). New insights into IGF-1 signaling in the heart. Trends in Endocrinology and Metabolism, 25(3), 128–137.
Shuang, T., Fu, M., Yang, G., Wu, L., & Wang, R. (2018). The interaction of IGF-1/IGF-1R and hydrogen sulfide on the proliferation of mouse primary vascular smooth muscle cells. Biochemical Pharmacology, 149, 143–152.
Baserga, R., Peruzzi, F., & Reiss, K. (2003). The IGF-1 receptor in cancer biology. International Journal of Cancer, 107(6), 873–877.
AsghariHanjani, N., & Vafa, M. (2019). The role of IGF-1 in obesity, cardiovascular disease, and cancer. Medical Journal of the Islamic Republic of Iran, 33, 56.
Jentzsch, T., Robl, B., Husmann, M., Bode-Lesniewska, B., & Fuchs, B. (2014). Worse prognosis of breast cancer patients expressing IGF-1 on a tissue microarray. Anticancer Research, 34(8), 3881–3889.
Tang, W., Feng, X., Zhang, S., Ren, Z., Liu, Y., Yang, B., & Ge, N. (2015). Caveolin-1 confers resistance of hepatoma cells to anoikis by activating IGF-1 pathway. Cellular Physiology and Biochemistry, 36(3), 1223–1236.
Foti, M., Ahmed Moukil, M., Dudognon, P., & Carpentier, J. L. (2004). Insulin and IGF-1 receptor trafficking and signalling. In Biology of IGF-1: its interaction with insulin in health and malignant states: Novartis Foundation Symposium 262 (Vol. 262, pp. 125–147). Chichester, UK: John Wiley & Sons, Ltd.
Armakolas, N., Armakolas, A., Antonopoulos, A., Dimakakos, A., Stathaki, M., & Koutsilieris, M. (2016). The role of the IGF-1 Ec in myoskeletal system and breast cancer pathophysiology. Critical Reviews in Oncology/Hematology, 108, 137–145.
Burrow, S., Andrulis, I. L., Pollak, M., & Bell, R. S. (1998). Expression of insulin-like growth factor receptor, IGF-1, and IGF-2 in primary and metastatic breast cancer. Journal of Surgical Oncology, 69(1), 21–27.
Zhang, W., Lee, J. C., Kumar, S., & Gowen, M. (1999). ERK Pathway Mediates the Activation of Cdk2 in IGF-1–Induced Proliferation of Human Breast cancer MG-63 Cells. Journal of Bone and Mineral Research, 14(4), 528–535.
Crudden, C., Song, D., Cismas, S., Trocmé, E., Pasca, S., Calin, G. A., & Girnita, L. (2019). Below the surface: IGF-1R therapeutic targeting and its endocytic journey. Cells, 8(10), 1223.
Anisimov, V. N., & Bartke, A. (2013). The key role of growth hormone–insulin–IGF-1 signaling in aging and cancer. Critical Reviews in Oncology/Hematology, 87(3), 201–223.
Tentori, L., & Graziani, G. (2007). Doping with growth hormone/IGF-1, anabolic steroids or erythropoietin: is there a cancer risk? Pharmacological Research, 55(5), 359–369.
Tentori, L., & Graziani, G. (2007). Doping with growth hormone/IGF-1, anabolic steroids or erythropoietin: is there a cancer risk? Pharmacological Research, 55.5, 359–369.
Pemberton, L. F., Blobel, G., & Rosenblum, J. S. (1998). Transport routes through the nuclear pore complex. Current Opinion in Cell Biology, 10(3), 392–399.
Cronshaw, J. M., Krutchinsky, A. N., Zhang, W., Chait, B. T., & Matunis, M. J. (2002). Proteomic analysis of the mammalian nuclear pore complex. The Journal of Cell Biology, 158(5), 915–927.
Packham, S., Warsito, D., Lin, Y., Sadi, S., Karlsson, R., Sehat, B., & Larsson, O. (2015). Nuclear translocation of IGF-1R via p150 Glued and an importin-β/RanBP2-dependent pathway in cancer cells. Oncogene, 34(17), 2227–2238.
Moloney, A. M., Griffin, R. J., Timmons, S., OConnor, R., Ravid, R., O, & Neill, C. (2010). Defects in IGF-1 receptor, insulin receptor and IRS-1/2 in Alzheimer’s disease indicate possible resistance to IGF-1 and insulin signalling. Neurobiology of Aging, 31(2), 224–243.
Acknowledgements
This work was supported by Jilin Scientific and Technological Development Program [grant no. 20190905003SF], [grant no. 20190701060GH]; China Postdoctoral Science Foundation Grant [grant no. 2019M651216].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The author declares no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Guo, B., Lv, Z., Cui, C. et al. IGF-1R Transported to the Cell Nuclei to Regulate the Proliferation of Breast Cancer Cells. Cell Biochem Biophys 79, 801–813 (2021). https://doi.org/10.1007/s12013-021-00989-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-021-00989-8