Abstract
Circular RNAs (circRNAs) have been discovered to serve as vital regulators in atherosclerosis (AS). However, the role and mechanism of circ_0002331 in AS process are still unclear. Human umbilical vein endothelial cells (HUVECs) were treated with ox-LDL to establish an in vitro model for AS. The expression levels of circ_0002331, Cyclin D2 (CCND2) and ELAVL1 were analyzed by quantitative real-time PCR. Cell proliferation, apoptosis, migration, invasion and angiogenesis were assessed by EdU assay, flow cytometry, transwell assay and tube formation assay. The protein levels of CCND2, ELAVL1, and autophagy-related markers were detected using western blot analysis. IL-8 level was analyzed by ELISA. The relationship between ELAVL1 and circ_0002331 or CCND2 was analyzed by RIP assay and RNA pull-down assay. Moreover, FISH assay was used to analyze the co-localization of ELAVL1 and CCND2 in HUVECs. Our data showed that circ_0002331 was obviously downregulated in AS patients and ox-LDL-induced HUVECs. Overexpression of circ_0002331 could promote proliferation, migration, invasion and angiogenesis, while inhibit apoptosis, autophagy and inflammation in ox-LDL-induced HUVECs. Furthermore, CCND2 was positively regulated by circ_0002331, and circ_0002331 could bind with ELAVL1 to promote CCND2 mRNA stability. Besides, CCND2 overexpression suppressed ox-LDL-induced HUVECs dysfunction, and its knockdown also reversed the regulation of circ_0002331 on ox-LDL-induced HUVECs dysfunction. In conclusion, circ_0002331 might be a potential target for AS treatment, which could improve ox-LDL-induced dysfunction of HUVECs via regulating CCND2 by binding with ELAVL1.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Atherosclerosis (AS), a chronic inflammatory disease that occurs in arterial wall, is the leading pathological basis of multiple-site vascular diseases [1, 2]. Additionally, the mortality rate of vascular diseases has already surpassed carcinoma in recent years [3]. Endothelial cells are important parts of vasculature and play important roles in maintaining vascular homeostasis [4]. Besides, ox-LDL has been identified to be involved in many AS relevant cellular processes [5, 6]. Therefore, exploring the molecular mechanisms that affect ox-LDL-induced endothelial cell dysfunction are necessary to discover new potential target for AS treatment.
Circular RNAs (circRNAs) are non-coding RNAs with stable circular structure formed by back-splicing [7]. Importantly, circRNAs have been reported to mediate disease process by regulating multiple cellular activities, including proliferation, oxidative stress, and autophagy [8]. At present, many circRNAs are confirmed to participate in AS process. For example, circ_CHMP5 might be a target for AS treatment, which was upregulated in AS patients and could inhibit proliferation and angiogenesis in ox-LDL-induced human umbilical vein endothelial cells (HUVECs) [9]. On the contrary, circ_0026218 could suppress ox-LDL-induced HUVECs apoptosis, oxidative stress and inflammation, so it played a negative role in AS process [10]. Therefore, the search for effective circRNAs may be of great significance for AS treatment.
Through circRNA microarray analysis, Li et al. found 943 differentially expressed circRNAs in HUVECs treated with or without ox-LDL, among which circ_0002331 was significantly downregulated in ox-LDL-induced HUVECs [11]. However, the role of circ_0002331 is AS process is still unclear. Our study aimed to explore the role of circ_0002331 in ox-LDL-induced HUVECs dysfunction and to reveal its underlying mechanism. This study showed that circ_0002331 could improve ox-LDL-induced dysfunction in HUVECs. In addition, we found that circ_0002331 interacted with RNA-binding protein ELAVL1 to regulate the mRNA stability of Cyclin D2 (CCND2). These results suggest that circ_0002331 may be a molecular target for AS therapy.
Materials and Methods
Specimen Collection
This research was approved by the Ethics Committee of Lishui People's Hospital. Written informed consents were acquired from all participants. A total of 30 AS patients (44–72 years old; 18 males and 12 females) and 30 healthy normal controls (39–70 years old; 18 males and 12 females) were recruited from the Lishui People's Hospital. In AS group (n = 30), AS patients with ≥ 50% stenosis of at least one of the major coronaries were recruited (Supplementary Fig. 1). The clinical pathological features of AS patients are shown in Table 1. In addition, patients with any malignant tumor, trauma, acute and chronic infection, liver or kidney disease, cerebrovascular accident, or recent surgical history were excluded. In normal control group (n = 30), age-, sex-, and ethnicity-matched healthy volunteers were included. The blood was harvested from each participant and the serum was prepared by centrifugation. This research was approved by the Ethics Committee of Lishui People's Hospital. Written informed consents were acquired from all participants.
Cell Lines and Cell Culture
HUVECs (Biovector NTCC, Beijing China) were propagated in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) containing 10% FBS, 1% Penicillin–Streptomycin, 0.1 mg/mL heparin and 0.05 mg/mL ECGs under standard culture conditions (5% CO2, 37 °C). For ox-LDL stimulation, HUVECs were incubated with different concentration of ox-LDL (Invitrogen, Carlsbad, CA, USA) for 24 h or incubated with 100 mg/L of ox-LDL for different time. HUVECs were transfected with the small interfering RNA (siRNA, 50 nM) and pcDNA overexpression vector (4.0 μg) of circ_0002331, CCND2, ELAVL1, as well as corresponding negative controls (si-NC and vector) by Lipofectamine 3000 (Invitrogen).
Quantitative Real-Time PCR (qRT-PCR)
Total RNAs isolated by TRIzol (Invitrogen) were reverse-transcribed into cDNA using cDNA synthesis kit (Takara, Dalian, China). After that, qRT-PCR was conducted using the SYBR Green (Takara) in PCR detection system following thermocycling conditions: 95 °C for 1 min, 40 cycles of 95 °C for 15 s, 55 °C for 30 s and 72 °C for 30 s. Expression was calculated using 2−ΔΔCt method and normalized to GAPDH. The sequences of primers: circ_0002331 (F 5’-CTCCCAACCTCATGGTCAGT-3’; R 5’-CTCATCCTCCTCGTCCTCTG-3’); CCND2 (F 5’-CTGTCTCTGATCCGCAAGCAT-3’; R 5’-GGTGGGTACATGGCAAACTTAAA-3’); ELAVL1 (F 5’-GGGTGACATCGGGAGAACG-3’; R 5’-CTGAACAGGCTTCGTAACTCAT-3’); GAPDH (F 5’-CTGGGCTACACTGAGCACC-3’; R 5’-AAGTGGTCGTTGAGGGCAATG-3’). For subcellular localization assay, RNA was isolated from the cytoplasmic and nuclear of HUVECs using Cytoplasmic and Nuclear RNA Purification Kit (Amyjet scientific, Wuhan, China). Then, circ_0002331 expression was measured by qRT-PCR with GAPDH as a cytoplasmic control and U6 as a nuclear control.
Cell Proliferation Assay
According to EdU Kit (Ribobio, Guangzhou, China), HUVECs were seeded into a 96-well plate (2 × 103 cells per well) and then incubated with EdU diluent for 2 h. After fixing with 4% paraformaldehyde for 15 min, cells were treated with glycine and TritonX-100. Then, cells were stained with Apollo and DAPI solution for 30 min. Fluorescence images were captured under a microscope, and EdU-positive cells were analyzed using Image J software.
Cell Apoptosis Assay
After treatment and transfection, HUVECs were digested with trypsin. The single cell suspension (5 × 105 cells) was added into the binding buffers containing Annexin V-FITC and PI (Beyotime, Shanghai, China) for 15 min. The apoptotic cells were analyzed under an FACS Calibur flow cytometer.
Western Blot Assay
Total proteins were extracted by RIPA buffer (Beyotime). After quantified by a BCA kit (Beyotime), proteins were loaded onto SDS-PAGE gel and then shifted onto PVDF membranes. After incubated with the primary antibodies at 4 °C overnight, membranes were reacted with the secondary antibodies (1:2000 dilution; Boster, Wuhan, China) at room temperature for 1 h. The protein was visualized by ECL reagent (Beyotime). The primary antibodies were: anti-Bcl-2 (1:1500 dilution; Boster), anti-Cleavsed-caspase 3 (1:1500 dilution; Boster), anti-Bax (1:1500 dilution; Boster), anti-LC3I/LC3II (1:1500 dilution; Boster), anti-Beclin-1 (1:1500 dilution; Boster), anti-p62 (1:1500 dilution; Boster), anti-CCND2 (1:1500 dilution; Boster), anti-p-eNOS (1:1000 dilution; Invitrogen), anti-eNOS (1:1000 dilution; Invitrogen) and anti-GAPDH (1:2000 dilution; Boster).
Migration and Invasion Assays
HUVECs (1 × 105 cells/well for migration and 4 × 105 cells/well for invasion) were seeded into the upper chamber of 24-well transwell chamber (Corning, Franklin Lakes, NJ, USA) with serum-free medium, while the medium contained 10% FBS was added into lower chamber. After 24 h, migrated cells on the basal side of the membrane were fixed by 4% formaldehyde and stained by crystal violet. Each well was photographed under the microscope. Transwell chamber was coated with Matrigel (Corning) for invasion assay, and other steps were same as that in cell migration assay.
Tube Formation Assay
HUVECs were seeded into 96-well plates (1 × 104 cells/well) pre-coated with Matrigel (Corning). Following incubation for 24 h, the number of branches was counted under a microscope with Image-Pro Plus software.
ELISA
The culture medium of HUVECs was collected for the measurement of IL-8 level according to the instructions of Human IL-8 ELISA Kit (R&D Systems, Minneapolis, MN, USA).
RIP and RNA Pull-Down Assays
RIP assay was conducted with an RIP kit (Millipore, Bedford, MA, USA). HUVECs were lysed by RIP buffer and then interacted with anti-ELAVL1 or anti-IgG embracing magnetic beads. The immunoprecipitated RNAs were extracted for qRT-PCR assay.
HUVECs were incubated with Bio-circ_0002331 probe and Bio-NC probe (RiboBio). After 48 h, whole-cell lysates from HUVECs was pulled down by streptavidin-coupled magnetic beads (Millipore). The pull-down complexes were collected for qRT-PCR assay to detect the ELAVL1 enrichment.
FISH Assay
HUVECs were fixed in paraformaldehyde, permeabilized in Triton X-100, and incubated with the corresponding FISH probes (Genechem, Shanghai, China) in the hybridization solution. After staining with DAPI, the cells were imaged under a laser confocal microscope.
Statistical Analysis
All experiments were performed in triplicate, with each independent experiment set 3 times. All analyses were conducted by GraphPad Prism 7. Results were exhibited as mean ± SD. Student’s t-test or ANOVA was used to analyze significance of difference for groups. P-value less than 0.05 was defined to be statistically significant.
Results
Circ_0002331 was Downregulated in AS Serum Samples and ox-LDL-Induced HUVECs
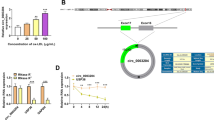
Circ_0002331 is derived from the back-splicing of FBXW8 gene (Fig. 1A). Circ_0002331 only could be amplified by cDNA but no products were observed in gDNA (Fig. 1B), confirming that circ_0002331 was generated by back-splicing. Circ_0002331 was obviously decreased in the serum of AS patients compared to normal control group (Fig. 1C). Interestingly, ROC assay showed that the AUC of circ_0002331 was 0.8867 (Fig. 1D), indicating that circ_0002331 might be a diagnostic biomarker for AS patients. Besides, circ_0002331 was mainly located in the cytoplasm of cells (Fig. 1E). Moreover, circ_0002331 was lowly expressed in ox-LDL-induced HUVECs in a concentration-dependent manner and time-dependent manner (Fig. 1F, G). These results showed that circ_0002331 might play an important role in AS progression.
Circ_0002331 expression in AS patients and ox-LDL-induced HUVECs. (A) The basic information of circ_0002331. (B) The characteristic of circ_0002331 was verified by agarose gel electrophoresis. (C) The level of circ_0002331 was determined by qRT-PCR in serum specimens (n = 30). (D) The clinical diagnostic value of circ_0002331 was analyzed by ROC curves in AS patients (n = 30). (E) Circ_0002331 expression in the cytoplasmic and nuclear of cells was detected by qRT-PCR. (F,G) Circ_0002331 level was measured by qRT-PCR in ox-LDL-induced HUVECs. *P < 0.05, **P < 0.01, ***P < 0.001
Circ_0002331 Overexpression Promoted ox-LDL-Induced HUVECs Cell Functions
Then, gain-of-functional experiments were used to identify the roles of circ_0002331 in ox-LDL-induced HUVECs. As shown in Fig. 2A, circ_0002331 was significantly overexpressed in HUVECs transfected with circ_0002331 overexpression vector. Circ_0002331 upregulation enhanced the EdU positive cell rate and reduced the apoptosis in ox-LDL-induced HUVECs (Fig. 2B–D). Treatment with ox-LDL decreased Bcl-2 level, increased Cleaved-caspase 3 level and enhanced Bax level in HUVECs, which was overturned by circ_0002331 overexpression (Fig. 2E). The suppressive effects of ox-LDL on the migration and invasion of HUVECs was abolished by circ_0002331 overexpression (Fig. 2F, G). Considering autophagy was closely tied to endothelial dysfunction, we also measured the expression of autophagy-related proteins, and found that ox-LDL increased LC3II/LC3I and Beclin-1 levels while decreased p62 level in HUVECs, which was rescued by circ_0002331 overexpression (Fig. 2H). Besides, eNOS is a subtype of NOS that regulate the production of NO, which has an important influence on AS [12]. It was reported that ox-LDL exerts its cytotoxicity by reducing eNOS expression to promote endothelial cells apoptosis, thereby accelerating the progression of AS [12, 13]. Next, we further investigated the expression level of eNOS in ox-LDL-induced HUVECs. As displayed in Fig. 2I, ox-LDL significantly inhibited the phosphorylation of eNOS, while overexpression of circ_0002331 partly overturned this effect. Dr. JA Berliner and colleagues reported that minimally ox-LDL (MM-LDL) and ox-PAPC can activate human aortic endothelial cells to produce monocyte chemotactic activators and IL-8 [14, 15]. Here, we also measured angiogenesis and IL-8 levels. Our data showed that ox-LDL reduced the number of branches and promoted IL-8 level in HUVECs, and these effects were abolished by circ_0002331 overexpression (Fig. 2J, K). Therefore, these findings implied that circ_0002331 promoted proliferation, migration, invasion, and angiogenesis, while repressed apoptosis, autophagy, and inflammation in ox-LDL-induced HUVECs.
Effects of circ_0002331 on ox-LDL-induced HUVECs functions. (A) The level of circ_0002331 was measured by qRT-PCR in HUVECs transfected with circ_0002331 or vector. (B-K) HUVECs were divided into 4 groups: con, ox-LDL, ox-LDL + vector, and ox-LDL + circ_0002331 groups. EdU assay (B) and flow cytometry (C,D) were used to assess cell proliferation and apoptosis. (E) The expression of Bcl-2, Cleavsed-caspase 3, and Bax was quantified by WB. (F,G) Transwell assay was employed to measure the migration and invasion of HUVECs. (H,I) WB was used to show the protein expression levels of LC3I/LC3II, Beclin-1, p62, and p-eNOS/eNOS. (J) Branch number was analyzed by tube formation assay. (K) IL-8 level was examined by ELISA. *P < 0.05, **P < 0.01, ***P < 0.001
CCND2 was Positively Regulated by Circ_0002331 in HUVECs
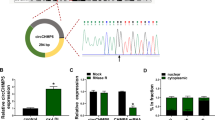
In the serum of AS patients, CCND2 had decreased expression and was positively correlated with circ_0002331 expression (Fig. 3A, B). The results of WB indicated that CCND2 was lowly expressed in the serum of AS patients and ox-LDL-induced HUVECs (Fig. 3C, D). After transfection with si-circ_0002331 into HUVECs, circ_0002331 expression was markedly decreased, indicating the successful transfection (Fig. 3E). Importantly, CCND2 protein expression could be promoted by circ_0002331 overexpression and reduced by circ_0002331 knockdown (Fig. 3F). All data revealed that circ_0002331 positively regulated CCND2 expression in HUVECs.
CCND2 was positively regulated by circ_0002331. (A) The serum level of CCND2 was determined by qRT-PCR in serum specimens (n = 30). (B) Pearson’s correlation analysis was used to analyze relationship between CCND2 and circ_0002331 in serum specimens (n = 30). (C,D) CCND2 protein expression was detected by WB analysis in serum specimens and ox-LDL-induced HUVECs. (E) The transfection efficiency of si-circ_0002331 was confirmed by qRT-PCR. (F) CCND2 protein expression was determined by WB assay in HUVECs transfected with circ_0002331 overexpression vector and si-circ_0002331. **P < 0.01, ***P < 0.001
Circ_0002331 Regulated CCND2 mRNA Stability by ELAVL1
Basing on the Circinteractome prediction, circ_0002331 is found to have binding sites with ELAVL1 (Fig. 4A). Then, RIP assay suggested that circ_0002331 was enriched in anti-ELAVL1 (Fig. 4B), and RNA pull-down assay uncovered that ELAVL1 enrichment was increased in Bio-circ_0002331 probe group (Fig. 4C). These data indicated that circ_0002331 could bind with ELAVL1. Besides, Starbase software predicted that CCND2 could bind to ELAVL1 (Fig. 4D), and RIP assay showed that CCND2 enrichment was markedly enhanced in anti-ELAVL1 (Fig. 4E). Besides, the co-localization of ELAVL1 and CCND2 in cells was observed by FISH assay (Fig. 4F). Furthermore, ELAVL1 protein expression was silenced by si-ELAVL1 and promoted by ELAVL1 overexpression vector (Fig. 4G). CCND2 protein expression could be reduced by ELAVL1 knockdown and increased by ELAVL1 overexpression (Fig. 4H). In addition, actinomycin D results showed that circ_0002331 knockdown inhibited the mRNA stability of CCND2, and this effect could be reversed by overexpression of ELAVL1 (Fig. 4I). The promotion effect of circ_0002331 on CCND2 protein expression could also be abolished by ELAVL1 knockdown (Fig. 4J). Thus, we believed that circ_0002331 positively regulated CCND2 mRNA stability by recruiting ELAVL1.
Circ_0002331 regulated CCND2 mRNA stability by ELAVL1. (A) Circinteractome predicted the circ_0002331 binding site on ELAVL1. RIP assay (B) and RNA pull down assay (C) were used to assess the interaction between circ_0002331 and ELAVL1. (D) Starbase predicted the CCND2 binding site on ELAVL1. (E) The association between CCND2 and ELAVL1 was analyzed by RIP assay. (F) FISH assay was used to analyze the co-localization of ELAVL1 and CCND2 in HUVECs. (G) The transfection efficiencies of si-ELAVL1 and ELAVL1 overexpression vector were confirmed by WB analysis. (H) CCND2 protein expression was detected by WB assay in HUVECs transfected with si-ELAVL1 and ELAVL1 overexpression vector. (I) Actinomycin D assay was used to analyze the mRNA stability of CCND2 in HUVECs transfected with si-circ_0002331 and ELAVL1 overexpression vector. (J) CCND2 protein expression was measured by WB assay in HUVECs transfected with circ_0002331 overexpression vector and si-ELAVL1. ***P < 0.001
CCND2 Overexpression Suppressed ox-LDL-Induced HUVECs Apoptosis, Autophagy and Inflammation
Then, the effect of CCND2 on ox-LDL-induced HUVECs functions was investigated. WB assay indicated that CCND2 was overexpressed in HUVECs transfected with CCND2 overexpression vector than that in control group (Fig. 5A). Upregulation of CCND2 reversed the ox-LDL-induced inhibitory effect on EdU positive cell rate (Fig. 5B). CCND2 overexpression reduced apoptosis rate, Cleaved-caspase 3 and Bax levels, while increased Bcl-2 level in ox-LDL-induced HUVECs (Fig. 5C, D). Also, migration and invasion were inhibited in ox-LDL-induced HUVECs, which was rescued by CCND2 overexpression (Fig. 5E, G). Moreover, CCND2 abolished ox-LDL-dependent autophagy by decreasing the levels of LC3II/LC3I and Beclin-1 and increasing the level of p62 (Fig. 5H). In addition, CCND2 overexpression also relieved ox-LDL-induced dephosphorylation of eNOS (Fig. 5I). Besides, upregulation of CCND2 enhanced the number of branches and reduced IL-8 level in ox-LDL-induced HUVECs (Fig. 5J, K). All data suggested that CCND2 could relieve ox-LDL-induced dysfunctions in HUVECs.
Effects of CCND2 overexpression on ox-LDL-induced HUVECs functions. (A) The expression of CCND2 was evaluated by WB in HUVECs transfected with vector or CCND2. (B-K) HUVECs were divided into 4 groups: con, ox-LDL, ox-LDL + vector, and ox-LDL + CCND2. Cell proliferation and apoptosis were analyzed by EdU (B) and flow cytometry (C,D). (E) WB was used for examining Bcl-2, Cleavsed-caspase 3, and Bax expression. (F,G) Transwell assay was carried out to assess migration and invasion of HUVECs. (H,I) The protein levels of LC3I/LC3II, Beclin-1, p62, and p-eNOS/eNOS were analyzed by WB assay. (J) Branch number was tested by tube formation assay. (K) IL-8 level was analyzed by ELISA. **P < 0.01, ***P < 0.001
Circ_0002331 Regulated ox-LDL-Induced HUVECs Functions by CCND2
To determine whether circ_0002331 regulated ox-LDL-induced HUVECs functions through CCND2, ox-LDL-induced HUVECs were co-transfected with circ_0002331 overexpression vector and si-CCND2. Under ox-LDL condition, the enhancement effect of circ_0002331 overexpression on HUVECs proliferation was weakened by CCND2 knockdown (Fig. 6A). Circ_0002331 upregulation inhibited cell apoptosis, Cleaved-caspase 3 and Bax levels, and enhanced Bcl-2 level in ox-LDL-induced HUVECs, which were reversed by silencing CCND2 (Fig. 6B, C). The migration and invasion of HUVECs were increased in ox-LDL-induced HUVECs after upregulation of circ_0002331, which was rescued by CCND2 knockdown (Fig. 6D, F). The autophagy inhibition was found in ox-LDL-induced HUVECs when overexpression of circ_0002331, which was abolished by CCND2 silencing (Fig. 6G). Besides, the promotion effect of circ_0002331 on eNOS phosphorylation and branch number, as well as the inhibition effect on IL-8 level, were partly reversed by CCND2 knockdown (Fig. 6H, J). These data showed that circ_0002331 suppressed ox-LDL-induced HUVECs dysfunction by regulating CCND2 expression.
Circ_0002331 regulated ox-LDL-induced HUVECs functions by CCND2. HUVECs were treated with ox-LDL and transfected with vector, circ_0002331, circ_0002331 + si-NC, or circ_00023314 + si-CCND2. EdU assay (A) and flow cytometry (B) were used to detect cell proliferation and apoptosis. (C) WB was used to test expression of Bcl-2, Cleavsed-caspase 3, and Bax. (D-F) Migration and invasion of HUVECs were examined by transwell. (G-H) The expression levels of LC3I/LC3II, Beclin-1, p62, and p-eNOS/eNOS were analyzed by WB. (I) Branch number was detected by tube formation assay. (J) IL-8 level was measured by ELISA. **P < 0.01, ***P < 0.001
Discussion
Although the pathogenesis of AS is multifaceted and unclear, but it is universally acknowledged that endothelial cells injury is the initiating phase for AS progression [16, 17]. According to previous reports, ox-LDL is a commonly accepted rick factor for AS, and ox-LDL after oxidative modification can promote AS more severely [5, 6]. Consistent with previous researches [18, 19], our study was performed in ox-LDL-treated HUVECs. In our current research, circ_0002331 was downregulated in AS serum and ox-LDL-induced HUVECs. Furthermore, circ_0002331 overexpression promoted proliferation, migration, invasion and angiogenesis, while inhibited apoptosis, autophagy and inflammation in ox-LDL-induced HUVECs, supporting the protective role of circ_0002331 in AS.
CCND2 belongs to the cyclin D-type family protein, which acts biological function via mediating the cyclin-dependent kinase (CDK) signaling [20]. Dysregulation of CCND2 is found to be associated with cell proliferation, indicating that CCND2 may be a key pro-proliferative cyclin [21, 22]. A previous study confirmed the role of CCND2 in endothelial cells, implying the potential association between CCND2 and vascular diseases [23]. Huang et al. revealed that CCND2 decreased ox-LDL-triggered anti-proliferative effects in HUVECs [24]. Analogously, Wu et al. discovered that CCND2 could suppress apoptosis and promote proliferation in HUVECs [25]. Consistent with these reported, our study confirmed that CCND2 overexpression enhanced proliferation, migration, invasion and angiogenesis, while suppressed apoptosis, autophagy and inflammation in ox-LDL-induced HUVECs. Besides, we found that CCND2 expression was positively regulated by circ_0002331, and its knockdown abolished the effect of circ_0002331 on ox-LDL-induced HUVECs dysfunction. These results provide evidence that circ_0002331 regulates AS process through CCND2.
Several aberrantly expressed circRNAs, including circ_0002331, have been identified in AS. At present, most researches focus on investigating circRNA-mediated regulatory networks in the progression of AS. However, there are few reports on how circRNAs decrease/increase in cells. Current research suggests that some RNA-binding proteins can bind to circRNAs flanking intron sequences, thereby participating in the regulation of the generation of circRNAs [26]. By searching CircInteractome database, we found that the flanking regions of circ_0002231 contained the RNA-binding sites of ELAVL1. Importantly, the binding sites between ELAVL1 and CCND2 were discovered by Starbase software. Further analysis showed that circ_0002331 promoted CCND2 mRNA stability by interacting with ELAVL1. This is a new finding for in our study.
Collectively, our results showed that circ_0002331 protected HUVECs from ox-LDL-induced dysfunction through ELAVL1/CCND2 axis. Therefore, circ_0002331/ELAVL1/CCND2 axis might provide significant theoretical foundation for developing novel therapies for AS in the future. However, we have only validated this at the molecular and cellular level, and animal studies in vivo are needed to further confirm our findings. In addition, activated endothelial cell induced leukocyte recruitment and adhesion, which exerted a pivotal role in AS development [27]. Thus, the next phase of our project is to investigate the correlation between leukocyte-endothelial cell interaction and circ_0002331/ELAVL1/CCND2 axis in AS development.
Conclusion
To sum up, our data revealed that circ_0002331 overexpression alleviated ox-LDL-induced HUVECs apoptosis, autophagy and inflammation. Mechanistically, circ_0002331 could interact with ELAVL1 to regulate CCND2 mRNA stability, thereby protecting HUVECs from ox-LDL-induced dysfunction. Our finding highlights circ_0002331 as a new potential molecular targets for AS therapy.
Data Availability
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
References
Stratton, M. S., Farina, F. M., & Elia, L. (2019). Epigenetics and vascular diseases. Journal of Molecular and Cellular Cardiology, 133, 148–163. https://doi.org/10.1016/j.yjmcc.2019.06.010
Mathur, K. S. (1970). Vascular diseases. Journal of the Association of Physicians of India, 18, 31–42.
Lu, M., Wu, L., Shi, P., Kang, S., Shi, L., & Wu, Y. (2004). Hypertension and subclinical carotid atherosclerosis in a suburban general population in China. Journal of Hypertension, 22, 1699–1706. https://doi.org/10.1097/00004872-200409000-00013
Sturtzel, C. (2017). Endothelial Cells. Advances in Experimental Medicine and Biology, 1003, 71–91. https://doi.org/10.1007/978-3-319-57613-8_4
Kattoor, A. J., Kanuri, S. H., & Mehta, J. L. (2019). Role of Ox-LDL and LOX-1 in atherogenesis. Current Medicinal Chemistry, 26, 1693–1700. https://doi.org/10.2174/0929867325666180508100950
Di Pietro, N., Formoso, G., & Pandolfi, A. (2016). Physiology and pathophysiology of oxLDL uptake by vascular wall cells in atherosclerosis. Vascular Pharmacology, 84, 1–7. https://doi.org/10.1016/j.vph.2016.05.013
Kristensen, L. S., Andersen, M. S., Stagsted, L. V. W., Ebbesen, K. K., Hansen, T. B., & Kjems, J. (2019). The biogenesis, biology and characterization of circular RNAs. Nature Reviews Genetics, 20, 675–691. https://doi.org/10.1038/s41576-019-0158-7
Cao, Q., Guo, Z., Du, S., Ling, H., & Song, C. (2020). Circular RNAs in the pathogenesis of atherosclerosis. Life Sciences, 255, 117837. https://doi.org/10.1016/j.lfs.2020.117837
Wang, Y., Liu, P., Chen, X., & Yang, W. (2023). Circ_CHMP5 aggravates oxidized low-density lipoprotein-induced damage to human umbilical vein endothelial cells through miR-516b-5p/TGFbetaR2 axis. Clinical Hemorheology and Microcirculation. https://doi.org/10.3233/CH-231722
Yang, L., Chen, W., Li, B., Hu, Y., Lu, H., Zhang, P., et al. (2023). Circular RNA circ_0026218 suppressed atherosclerosis progression via miR-338-3p/SIRT6 axis. BioMed Research International. https://doi.org/10.1155/2023/5647758
Li, C. Y., Ma, L., & Yu, B. (2017). Circular RNA hsa_circ_0003575 regulates oxLDL induced vascular endothelial cells proliferation and angiogenesis. Biomedicine & Pharmacotherapy, 95, 1514–1519. https://doi.org/10.1016/j.biopha.2017.09.064
Hong, F. F., Liang, X. Y., Liu, W., Lv, S., He, S. J., Kuang, H. B., et al. (2019). Roles of eNOS in atherosclerosis treatment. Inflammation Research: Official journal of the European Histamine Research Society, 68, 429–441. https://doi.org/10.1007/s00011-019-01229-9
Gliozzi, M., Scicchitano, M., Bosco, F., Musolino, V., Carresi, C., Scarano, F., et al. (2019). Modulation of nitric oxide synthases by oxidized LDLs: Role in vascular inflammation and atherosclerosis development. International Journal of Molecular Sciences. https://doi.org/10.3390/ijms20133294
Lee, H., Shi, W., Tontonoz, P., Wang, S., Subbanagounder, G., Hedrick, C. C., et al. (2000). Role for peroxisome proliferator-activated receptor alpha in oxidized phospholipid-induced synthesis of monocyte chemotactic protein-1 and interleukin-8 by endothelial cells. Circulation Research, 87, 516–521. https://doi.org/10.1161/01.res.87.6.516
Yeh, M., Leitinger, N., de Martin, R., Onai, N., Matsushima, K., Vora, D. K., et al. (2001). Increased transcription of IL-8 in endothelial cells is differentially regulated by TNF-alpha and oxidized phospholipids. Arteriosclerosis, Thrombosis, and Vascular Biology, 21, 1585–1591. https://doi.org/10.1161/hq1001.097027
Konukoglu, D., & Uzun, H. (2017). Endothelial dysfunction and hypertension. Advances in Experimental Medicine and Biology, 956, 511–540. https://doi.org/10.1007/5584_2016_90
Gimbrone, M. A., Jr., & Garcia-Cardena, G. (2016). Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circulation Research, 118, 620–636. https://doi.org/10.1161/CIRCRESAHA.115.306301
Zhong, X., Zhang, L., Li, Y., Li, P., Li, J., & Cheng, G. (2018). Kaempferol alleviates ox-LDL-induced apoptosis by up-regulation of miR-26a-5p via inhibiting TLR4/NF-kappaB pathway in human endothelial cells. Biomedicine & Pharmacotherapy, 108, 1783–1789. https://doi.org/10.1016/j.biopha.2018.09.175
Chen, X., Lin, J., Hu, T., Ren, Z., Li, L., Hameed, I., et al. (2019). Galectin-3 exacerbates ox-LDL-mediated endothelial injury by inducing inflammation via integrin beta1-RhoA-JNK signaling activation. Journal of Cellular Physiology, 234, 10990–11000. https://doi.org/10.1002/jcp.27910
Peters, G. (1994). The D-type cyclins and their role in tumorigenesis. Journal of Cell Science Supplement, 18, 89–96. https://doi.org/10.1242/jcs.1994.supplement_18.13
Di, W., Li, Q., Shen, W., Guo, H., & Zhao, S. (2017). The long non-coding RNA HOTAIR promotes thyroid cancer cell growth, invasion and migration through the miR-1-CCND2 axis. American Journal of Cancer Research, 7, 1298–1309.
Li, L., Sarver, A. L., Alamgir, S., & Subramanian, S. (2012). Downregulation of microRNAs miR-1, -206 and -29 stabilizes PAX3 and CCND2 expression in rhabdomyosarcoma. Laboratory Investigation, 92, 571–583. https://doi.org/10.1038/labinvest.2012.10
Hu, Y., Jin, G., Li, B., Chen, Y., Zhong, L., Chen, G., et al. (2019). Suppression of miRNA let-7i-5p promotes cardiomyocyte proliferation and repairs heart function post injury by targetting CCND2 and E2F2. Clinical Science (London, England), 133, 425–441. https://doi.org/10.1042/CS20181002
Huang, R., Hu, Z., Cao, Y., Li, H., Zhang, H., Su, W., et al. (2019). MiR-652-3p inhibition enhances endothelial repair and reduces atherosclerosis by promoting Cyclin D2 expression. eBioMedicine, 40, 685–694. https://doi.org/10.1016/j.ebiom.2019.01.032
Wu, R., Tang, S., Wang, M., Xu, X., Yao, C., & Wang, S. (2016). MicroRNA-497 induces apoptosis and suppresses proliferation via the Bcl-2/Bax-Caspase9-Caspase3 pathway and cyclin D2 protein in HUVECs. PLoS ONE, 11, e0167052. https://doi.org/10.1371/journal.pone.0167052
Wei, Y., Lu, C., Zhou, P., Zhao, L., Lyu, X., Yin, J., et al. (2021). EIF4A3-induced circular RNA ASAP1 promotes tumorigenesis and temozolomide resistance of glioblastoma via NRAS/MEK1/ERK1-2 signaling. Neuro-oncology, 23, 611–624. https://doi.org/10.1093/neuonc/noaa214
Bozic, M., Álvarez, Á., de Pablo, C., Sanchez-Niño, M. D., Ortiz, A., Dolcet, X., et al. (2015). Impaired vitamin D signaling in endothelial cell leads to an enhanced leukocyte-endothelium interplay: Implications for atherosclerosis development. PLoS ONE, 10, e0136863. https://doi.org/10.1371/journal.pone.0136863
Acknowledgements
Not applicable.
Funding
No funding was received.
Author information
Authors and Affiliations
Contributions
All authors made substantial contribution to conception and design, acquisition of the data, or analysis and interpretation of the data; take part in drafting the article or revising it critically for important intellectual content; gave final approval of the revision to be published; and agree to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Ethics Approval and Consent to Participate
The present study was approved by the ethical review committee of Lishui People's Hospital. Written informed consent was obtained from all enrolled patients.
Consent for Publication
Patients agree to participate in this work.
Additional information
Handling Editor: Amie Lund.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, F., Yu, X. Circ_0002331 Interacts with ELAVL1 to Improve ox-LDL-Induced Vascular Endothelial Cell Dysfunction via Regulating CCND2 mRNA Stability. Cardiovasc Toxicol 24, 625–636 (2024). https://doi.org/10.1007/s12012-024-09865-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12012-024-09865-2