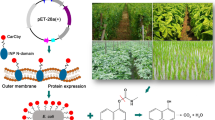
Abstract
Enzymatic degradation of polyethylene terephthalate (PET) is attracting attention as a new technology because of its mild reaction conditions. However, the cost of purified enzymes is a major challenge for the practical application of this technology. In this study, we attempted to display the surface of the PET-degrading enzyme, PETase, onto Escherichia coli using the membrane anchor, PgsA, from Bacillus subtilis to omit the need for purification of the enzyme. Immunofluorescence staining confirmed that PETase was successfully displayed on the surface of E. coli cells when a fusion of PgsA and PETase was expressed. The surface-displaying E. coli was able to degrade 94.6% of 1 mM bis(2-hydroxyethyl) terephthalate in 60 min, and the PET films were also degraded in trace amounts. These results indicate that PgsA can be used to present active PETase on the cell surface of E. coli. This technique is expected to be applied for efficient PET degradation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Polyethylene terephthalate (PET) is a plastic widely used in industry and daily life [1]. However, its high durability results in the accumulation of discarded PET in the environment for hundreds of years. This is a concern because of its negative impact on ecosystems and human health [2,3,4].
Several recycling methods have been developed for the recovery of PET. Mechanical and chemical recycling are representative methods [5, 6]. Mechanical recycling is a method in which PET is crushed, dissolved, and remolded. It is considered to be cheaper than chemical recycling [7]. However, the drawback is that the product properties deteriorate with each cycle [8]. In chemical recycling, PET is chemically depolymerized into monomers, which are then repolymerized. Therefore, PET can be recycled with minimal quality loss [9]. However, chemical recycling is more expensive than mechanical recycling and thus, it offers fewer economic benefits [10]. In addition, the chemical decomposition of PET requires high temperature, high-pressure conditions, and large amounts of energy [11].
PET degradation by microorganism-derived enzymes proceeds under mild conditions, such as 30 to 70 °C and normal pressure. Therefore, this strategy has attracted attention as a new option for environment-friendly PET recycling. To date, several enzymes have been reported to be involved in the degradation of PET, such as the cutinase HiC from Humilica insolens [12], the cutinase LCC from leaf and branch compost [13], and hydrolase TfH from Thermobifida fusca [14].
In 2016, the bacterium Ideonella sakaiensis 201-F6 was shown to grow on PET as a major energy and carbon source in recycling plants in Japan [15]. PETase, a PET-degrading enzyme secreted by this bacterium, shows higher PET degradation efficiency and substrate specificity than other PET-degrading enzymes at room temperature. PETase hydrolyzes PET and releases mono(hydroxyethyl)terephthalate (MHET) and terephthalate (TPA). In recent years, protein engineering modifications of this enzyme have been widely used to improve the enzymatic activity and thermal stability of PET [16, 17]. Notable mutant enzymes include DuraPETase [18] and FAST-PETase [19]. Both mutant PETases have extremely high PET degradation activity and thermal stability compared to wild-type PETase, and therefore, the implementation of enzymatic PET degradation is becoming a reality.
When such enzymes are used industrially, they are generally prepared using microorganisms such as recombinant E. coli. The desired enzyme can be obtained through cultivation of recombinant Escherichia coli, cell disruption, and enzyme purification. However, the cost of enzyme purification is known to be very high [20]. In addition, because the enzyme is water-soluble, it can only be used once for the required reaction and then it becomes waste. To implement PET degradation using enzymes, it is desirable to address these issues.
Techniques have been developed to display target enzymes on the cell surface using membrane anchors to eliminate the enzyme purification process [21, 22]. When fused with the target enzyme, membrane anchors display the target enzyme and express enzyme activity at the cell surface [23]. Therefore, these cells can be used as immobilized catalysts. Cells can be easily separated from the generated monomers by centrifugation or filtration, without the need for cell disruption. Furthermore, the separated cells can be reused [24].
Various membrane-anchor-based surface display systems have been developed for E. coli. Outer membrane proteins [25], ice nucleation proteins [26], and autotransporters [27] are used as anchors. In this study, we used the PgsA protein from Bacillus subtilis as the anchor protein. This protein is part of the enzyme complex that synthesizes poly-γ-glutamic acid (PGA) in B. subtilis [28]. Narita et al. successfully fused PgsA with α-amylase (AmyA) from Streptococcus bovis 148 and lipase B (CALB) from Candida antarctica to display these enzymes in an active form on the cell surface of E. coli [29]. Gallus et al. developed a new cell surface display system using the post-translational fusion of target proteins and membrane anchors using the SpyCatcher/SpyTag system [30]. This system has been reported to successfully display heme- and diflavin-containing cytochrome P450 BM3 monooxygenase from Bacillus megaterium in E. coli, with higher levels of presentation than conventional genetic fusion using a plasmid [31].
We heterologously expressed PETase and PgsA in E. coli via genetic and post-translational fusion. In both cases, we successfully expressed the fusion protein and confirmed that the active form of the PETase was present on the cell surface of E. coli. E. coli expressing PETase by genetic fusion was able to degrade the PET intermediate bis(2-Hydroxyethyl) terephthalate (BHET) more efficiently than E. coli expressing PETase intracellularly or in a crude enzyme solution. It was also confirmed that E. coli can degrade PET films, albeit in small amounts, indicating that this is a promising new approach to PET degradation, although further improvement of the degradation efficiency is necessary.
Materials and Methods
Strains and Growth Conditions
E. coli DH5α was used to generate the plasmids. Cells were transformed with plasmids for cloning using heat shock. The cells were incubated overnight at 37 °C in Luria–Bertani medium (20 g/L LB broth, Lenox; Nacalai Tesque, Kyoto, Japan) containing 100 μg/mL ampicillin.
E. coli BL21(DE3) cells were used for protein expression analysis. The cells were transfected with each plasmid by electroporation. They were then incubated in LB medium containing 100 μg/mL ampicillin overnight at 37 °C. The transformants were incubated overnight at 37 °C, 180 rpm in test tubes containing 4 mL of LB liquid medium with 100 μg/mL ampicillin. The precultures were inoculated into flasks containing 100 mL of LB liquid medium with 100 μg/mL ampicillin and incubated at 37 °C, 150 rpm until the optical density at 600 nm (OD600) reached 0.5–0.8. Protein expression was then induced with 0.5 mM isoIsopropyl-β-d-thiogalactopyranoside, and the cultures were incubated at 20 °C and 150 rpm for 20 h. After the induction of expression, the cells were harvested by centrifugation (12,000 rpm, 3 min, 4 °C), washed with phosphate-buffered saline (PBS, pH 7.4), 50 mM Tris–HCl buffer (pH 8.0) with 100 mM NaCl or 50 mM Tris–HCl buffer (pH 9.0), and resuspended in the respective solutions.
Plasmid Construction and Transformation
The plasmids used in this study are listed in Table S1 and the primers are listed in Table S2. Polymerase chain reaction (PCR) was performed using KOD Plus or KOD One master mixes (Toyobo Co., Ltd., Osaka, Japan). The vectors and inserts were ligated using NEBuilder (New England Biolabs Inc., Ipswich, MA, USA) according to the manufacturer’s protocol.
Primers 1 and 2 were used to amplify PgsA using the pHLA vector as the template [29]. The SpyCatcher gene with a glycine-serine (GS) linker was amplified by PCR using primers 3 and 4, and a synthetic gene (synthesized by Integrated DNA Technologies Inc., Coralville, IA, USA) was used as a template. The amplified fragment was inserted into the NcoI site of the pETDuet-1 vector (pDuet), and the resulting plasmid was named pDuet_PgsA-SC.
The PETase gene with a Myc Tag was amplified using primers 5, 6, and 7 with a synthetic gene (Eurofins Genomics Inc., Tokyo, Japan) as the template. The amplified fragment was inserted into the NdeI site of the pETDuet-1 vector, and the resulting plasmid was named pDuet_PETase-Myc.
Primers 1 and 8 were used to amplify PgsA, using pDuet_PgsA-SC as the template. The PETase gene with a Myc tag was also amplified using primers 9 and 10 with pDuet_PETase-Myc as the template. These amplified fragments were inserted into the NcoI site of the pETDuet-1 vector, and the resulting plasmid was named pDuet_PgsA-PETase-Myc.
Primers 5, 11, 12, and 13 were used to amplify the PETase genes with SpyTag and MycTag, using synthetic genes as templates. The amplified fragment was inserted into the NdeI site of pDuet_PgsA-SC, and the resulting plasmid was named pDuet_PgsA-SC_PETase-ST.
Using primers 14 and 15 and the synthetic gene as a template, the PETase gene with a His tag was amplified by PCR. After treating the pETDuet-1 vector with the restriction enzymes NcoI and AvrII, the amplified fragment was inserted, and the resulting plasmid was named pDuet_PETase-His.
Sodium Dodecyl Sulfate–Polyacrylamide Gel Electrophoresis
Cells were resuspended in 50 mM Tris–HCl Buffer (pH 7.5) and adjusted to an OD600 of 5.0. The cell suspension was collected in microtubes and sonicated using a BIORUPTOR UCD-250 instrument (Tosho Electric Co., Ltd., Tokyo, Japan). Sodium dodecyl sulfate (SDS) treatment was performed by mixing equal amounts of SDS sample buffer with the protein solution after sonication and heating at 95 °C for 5 min. Proteins were separated by SDS–polyacrylamide gel electrophoresis (PAGE) (4.5% or 15% [w/v] acrylamide) and stained with Coomassie Brilliant Blue.
Crude Enzyme Preparation
After the induction of protein expression, BL21(DE3) cells harboring pDuet_PETase-His (BL21/pDuet_PETase) were resuspended in 50 mM Tris–HCl buffer (pH 8.0) containing 100 mM NaCl and adjusted to an OD600 of 10. The cell suspension was collected in microtubes and sonicated using a BIORUPTOR UCD-250 instrument (Tosho Electric Co., Ltd.). The resulting solution was centrifuged (13,000 rpm, 10 min, 4 °C), and the supernatant was used as a PETase crude enzyme solution.
Immunofluorescence Analysis
After culture, cell suspensions of each strain were resuspended in PBS to an OD600 of 5.0 and 2 mg/L rabbit anti-myc affinity-purified antibody was added to the solution and incubated at 25 °C, 800 rpm for 1 h. After centrifugation (13,000 rpm, 1 min, 25 °C) and removal of the supernatant, the cells were washed with PBS. To the washed cells, 4 mg/L Alexa Flour® anti-rabbit IgG was added, and the cells were incubated at 25 °C, 800 rpm for 1 h. The cells were centrifuged (13,000 rpm, 1 min, 25 °C) to remove the supernatant and then washed with PBS. After resuspending the cells in PBS, fluorescence intensity was measured at an excitation wavelength of 485 nm and an emission wavelength of 528 nm using a Synergy LC plate reader (BioTek, Winooski, VT, USA).
BHET Degradation Reaction
Enzymatic reactions were performed with cells or crude enzymes (OD600 = 2.5) in 50 mM Tris–HCl Buffer (pH 8.0) containing 100 mM NaCl. BHET (dissolved in dimethyl sulfoxide) was added to a final concentration of 1 mM, and the samples were incubated at 30 °C for 1 h in 96-well plates. The reaction was stopped by the addition of 16.7% acetonitrile.
Evaluation of Reusability
Reusability was evaluated using suspensions of BL21(DE3) cells harboring pDuet_PgsA-PETase-Myc (OD600 = 2.5) in 50 mM Tris–HCl buffer (pH 8.0) containing 100 mM NaCl. Then, 1 mM BHET (dissolved in dimethyl sulfoxide) was added and incubated in microtubes at 30 °C, 800 rpm for 15 min. The reaction was stopped by centrifugation (13,000 rpm, 1 min, 4 °C), and the supernatant was diluted with 16.7% acetonitrile. After removing the remaining supernatant, the cells were washed with 50 mM Tris–HCl buffer (pH 8.0) containing 100 mM NaCl. The cells were then resuspended in 50 mM Tris–HCl buffer (pH 8.0) containing 100 mM NaCl. Then, 1 mM BHET (dissolved in dimethyl sulfoxide) was added, and the samples were incubated in a microtube at 30 °C, 800 rpm for 15 min. This procedure was repeated five times.
PET Degradation Reaction
A sheet of PET film (15 mm × 15 mm, 0.2-mm thick; RP Topla, Co., Ltd., Osaka, Japan) was immersed in 4 mL of the cell suspensions of each strain (OD600 = 5.0) in 50 mM Tris–HCl buffer (pH 9.0) and incubated at 30 °C for 48 h. The solution was collected, and 16.7% acetonitrile was added to stop the reaction.
Product Analysis
The concentration of BHET, MHET, and TPA were determined by high-performance liquid chromatography (Shimadzu Co. Kyoto, Japan; solvent delivery system, LC-20AD; column, 5C18-AR-II (Nacalai Tesque, Inc., Kyoto, Japan); column temperature, 35 °C; detector, SPD-10A). The peak of each compound was detected in the order of TPA, MHET, and BHET.
Results and Discussion
Preparation of PETase-Displaying E. coli
Fusion of a membrane anchor with an enzyme of interest allows the enzyme to be displayed on the cell surface. In this study, we used PgsA, a membrane protein from Bacillus subtilis, as a membrane anchor, when expressed by fusing an enzyme to the C-terminus of PgsA, it acts as a membrane anchor in E. coli [29]. Genetic fusion between an anchor and an enzyme is employed in cell-surface display systems. Generally, genetic fusions may lead to unfavorable domain-domain interactions and misfolding, resulting in reduced display efficiency and loss of enzyme function. Therefore, we attempted to use a post-translational fusion system with SpyCatcher/SpyTag [30]. Using this method, the membrane anchor fused to SpyCatcher and the enzyme fused to SpyTag are expressed separately, and the two are then combined by the covalent binding of SpyCatcher and SpyTag. This method is expected to improve display efficiency and enzyme activity by reducing unfavorable domain interactions and misfolding because the membrane anchor and enzyme are folded separately and independently. Therefore, we used two expression systems, one with genetic fusion at the plasmid stage (pDuet_PgsA-PETase-Myc) (Fig. 1a) and the other with post-translational modification using the SpyCatcher/SpyTsg system (pDuet_PgsA-SC_PETase-ST-Myc) (Fig. 1b). In the former, PETase and PgsA were fused via a flexible GS linker. In the latter, PETase was fused to SpyTag and PgsA to SpyCatcher via a flexible GS linker. Schematic illustrations showing protein translation and membrane localization of the respective transformants are shown in Fig. 1c and d.
Schematic diagram of the cell surface display of PETase. a Plasmid expressing PETase on the cell surface by genetic fusion: pDuet_PgsA-PETase-Myc. b Plasmid expressing PETase on the cell surface by post-translational fusion using the SpyCatcher/SpyTag system: pDuet_PgsA-SpyC_PETase-SpyT-Myc. c Image of genetic fusion of PgsA and PETase on BL21/pDuet_PgsA-PETase. d Image of post-translational fusion of PgsA-SpyCatcher and PETase-SpyTag on BL21/pDuet_PgsA-SpyC_PETase-ST
To assess protein expression, the lysates of each cell line were analyzed using SDS-PAGE. As shown in Fig. 2, a band corresponding to PgsA-PETase-Myc (lane 2; 71.9 kDa) was observed. Although the PETase-SpyT-Myc (30.7 kDa) band was observed in lane 3, conjugation between PETase-SpyT-Myc and PgsA-SpyC was also observed (PgsA-SpyC-SpyT-Myc-PETase; 85.9 kDa). This result indicates that the SpyCatcher component of PgsA-SC and the SpyTag component of PETase-ST-Myc formed a covalent bond, resulting in the fusion protein, PgsA-SpyC-SpyT-Myc-PETase. The expression level of PgsA-SpyC-SpyT-Myc-PETase was higher than the expression level of PgsA-PETase-Myc. Therefore, expression levels of the fusion proteins were higher when post-translational fusion was performed using SpyCatcher/SpyTag than when PETase and PgsA were fused to a plasmid. This is presumably because folding occurs more smoothly when the individual proteins are expressed separately.
Evaluation of Cell Surface Expression of PETase
Surface exposure of the fusion protein was analyzed by immunofluorescence staining using a rabbit anti-Myc primary antibody that binds to the Myc tag attached to the C-terminus of PETase and an Alexa488-conjugated anti-rabbit secondary antibody. Figure 3 shows the fluorescence quantification results obtained using a microplate reader. BL21(DE3) harboring pDuet_PgsA-PETase-Myc (BL21/pDuet_PgsA-PETase) and BL21(DE3) harboring pDuet_PgsA-SC_PETase-ST-Myc (BL21/pDuet_PgsA-SC_PETase-ST) exhibited higher fluorescence intensities than BL21(DE3) harboring pETDuet-1 (BL21/pDuet). This result indicated that both strains had PETase on the cell surface. Additionally, the fluorescence intensity was higher for BL21/pDuet_PgsA-PETase than BL21/pDuet_PgsA-SC_PETase-ST. Although the SDS-PAGE results (Fig. 1) showed that the expression level was higher for the post-translational fusion product than the genetic fusion product, the amount of displayed PETase was greater with the genetic fusion than with the post-translational fusion using SpyCatcher/SpyTag. This is probably because the proportion of fusion proteins transported to the outer membrane of the total fusion protein expressed was lower in the case of post-translational fusion.
BHET Degradation by PETase-Displaying E. coli
Two types of surface-displaying strains were employed to degrade BHET. PET degradation is much slower than BHET degradation; therefore, we used BHET as a substrate to ensure that the surface-displayed PETase remained active initially. BHET is hydrolyzed to MHET by PETase. An intracellular PETase expression strain (BL21/pDuet_PETase) and a crude enzyme solution were also evaluated for BHET degradation. Figure 4 shows that BL21/pDuet_PETase degraded only 7% of the BHET in 60 min, whereas BL21/pDuet_PgsA-PETase degraded 94.6% and BL21/pDuet_PgsA-SC_PETase-ST degraded 24.4% of the BHET. Both strains displaying PETase degraded more BHET than the strain expressing PETase intracellularly, indicating that the degradation of poorly membrane-permeable BHET favored the surface-display system. The results of BHET degradation and immunofluorescence staining indicated that the use of PgsA enabled the display of the active form of PETase on the cell surface of E. coli. The extent of BHET degradation was greater in BL21/pDuet_PgsA-PETase than in BL21/pDuet_PgsA-SC_PETase-ST cells. This result was consistent with the immunofluorescence staining results, suggesting that the strategy of genetic fusion with the PgsA anchor was suitable for this study.
Results of bis(2-Hydroxyethyl) terephthalate (BHET) degradation to mono(hydroxyethyl)terephthalate (MHET) by BL21/pDuet_PgsA-PETase (closed green squares), BL21/pDuet_PgsA-SC_PETase-ST (open green squares), BL21/pDuet_PETase (closed blue circles), crude enzyme (closed blue triangles), and BL21/pDuet (closed gray circles). Data are presented as the average of triplicate independent experiments, and error bars represent the standard deviation
The crude enzyme solution degraded 56.5% of the BHET in 60 min. As shown in Fig. S1, no soluble PETase band was observed, suggesting that the expression level of PETase was low. Previous studies have also reported low expression levels of soluble PETase [32]. In addition, intracellular PETase has almost no activity in Fig. 4. Hence, the activity of BL21/pDuet_PgsA-PETase was derived from PgsA-PETase localized on the cell surface. It is thought that the activity of displayed-PETase was higher than that of the crude enzyme solution because the amount of PgsA-PETase on the cell surface of BL21/pDuet_PgsA-PETase was greater than the soluble PETase expressed by BL21/pDuet_PETase. Considering that the amount of BHET degraded by BL21/pDuet_PgsA-PETase was only 1.7 times the amount degraded by the crude enzyme solution, the expression level of PETase on the cell surface of BL21/pDuet_PgsA-PETase was expected to be relatively low. In Fig. S1, a band corresponding to PgsA-PETase was clearly observed in lane 2. This suggests that most of the identified fusion proteins may not be transported to the outer membrane, but remain in the cytoplasm. PgsA has no signal peptide targeting it to the periplasmic space [33]. Therefore, passage through the inner membrane and insertion were presumed to occur via spontaneous insertion into the membrane by the hydrophobic portion of PgsA. Thus, improving the transportation of fusion proteins by adding a signal peptide may facilitate BHET degradation.
The reusability of BL21/pDuet_PgsA-PETase was investigated using the BHET degradation reaction. The BHET degradation reaction was performed five times in microtubes. Between cycles, the cells were separated by centrifugation and washed to remove unreacted BHET. As shown in Fig. 5, 55% of the BHET-degrading activity was maintained in one reaction, and 36% was maintained in two reactions. The reason for the decrease in BHET degradation with each cycle may be the inevitable loss of cells per cycle or desorption of fusion proteins during the reaction and washing steps.
PET Degradation Reaction
BL21/pDuet_PgsA-PETase was used for PET film degradation. Figure 6 shows the amount of MHET and TPA released. Although no PET monomer was detected in the reaction solution of BL21/pDuet, 59 nmol of PET monomer was detected in the reaction solution of BL21/pDuet_PgsA-PETase. This confirmed that the PET film was degraded by surface-displayed PETase. However, the relatively low level of degradation may be due to the low presentation of PETase on the cell surface and the insufficient contact angle between the cells and the PET film. Further studies are required to improve the contact between the PET film and cells, as well as the activity of PETase.
Conclusions
In this study, the PET-degrading enzyme, PETase, was displayed on the cell surface of E. coli using the membrane anchor PgsA from B. subtilis. The fusion of PgsA and PETase was performed using two strategies: genetic fusion and post-translational fusion. Immunofluorescence staining showed that both the strains successfully displayed PETase on their cell surfaces. Strains expressing PgsA-PETase generated by genetic fusion showed higher BHET-degrading activity than the strains with intracellular expression or crude enzyme solution. This strain also degraded PET films. These results indicate that the surface display of PETase by PgsA is a promising approach for PET degradation.
Availability of Data and Materials
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Yan, L., Hou, L., Sun, S., & Wu, P. (2020). Dynamic diffusion of disperse dye in a polyethylene terephthalate film from an infrared spectroscopic perspective. Industrial and Engineering Chemistry Research, 59, 7398–7404. https://doi.org/10.1021/acs.iecr.9b07110
Leng, Z., Padhan, R. K., & Sreeram, A. (2018). Production of a sustainable paving material through chemical recycling of waste PET into crumb rubber modified asphalt. Journal of Cleaner Production, 180, 682–688. https://doi.org/10.1016/j.jclepro.2018.01.171
Zhang, J., Wang, L., & Kannan, K. (2019). Polyethylene terephthalate and polycarbonate microplastics in pet food and feces from the United States. Environmental Science & Technology, 53, 12035–12042. https://doi.org/10.1021/acs.est.9b03912
Lebreton, L. C. M., Zwet, J. V. D., Damsteeg, J. W., Slat, B., Andrady, A., & Reisser, J. (2017). River plastic emissions to the world’s oceans. Nature Communications, 8, 15611. https://doi.org/10.1038/ncomms15611
Shamsaei, M., Aghayan, I., & Kazemi, K. A. (2017). Experimentalinvestigation of using cross-linked polyethylene waste as aggregate in roller compacted concrete pavement. Journal of Cleaner Production, 165, 290–297. https://doi.org/10.1016/j.jclepro.2017.07.109
Lopez-Fonseca, R., Duque-Ingunza, I., Rivas, B. D., Arnaiz, S., & Gutierrez-Ortiz, J. I. (2010). Chemical recycling of post-consumer PET wastes by glycolysis in the presence of metal salts. Polymer Degradation and Stability, 95, 1022–1028. https://doi.org/10.1016/j.polymdegradstab.2010.03.007
Badia, J. D., Vilaplana, F., Karlsson, S., & Ribes-Greus, A. (2009). Thermal analysis as a quality tool for assessing the influence of thermo-mechanical degradation on recycled poly (ethylene terephthalate). Polymer Testing, 28, 169–175. https://doi.org/10.1016/j.polymertesting.2008.11.010
Bartolome, L., Imran, M., Cho, B. G., Al-Masry, W. A., & Kim, D. H. (2012). Recent developments in the chemical recycling of PET. Material Recycling-Trends and Perspectives. https://doi.org/10.5772/33800
Geyer, B., Lorenz, G., & Kandelbauer, A. (2016). Recycling of poly(ethylene terephthalate)-A review focusing on chemical methods. Express Polymer Letters, 10, 559–586. https://doi.org/10.3144/expresspolymlett.2016.53
Geyer, B., Rohner, S., Lorenz, G., & Kandelbauer, A. (2014). Designing oligomeric ethylene terephtalate building blocks by chemical recycling of polyethylene terephtalate. Journal of Applied Polymer Science, 131, 39786–39786. https://doi.org/10.1002/app.39786
Kamber, N. E., Tsujii, Y., Keets, K., Waymouth, R. M., Pratt, R. C., Nyce, G. W., & Hedrick, J. L. (2010). The depolymerization of poly (ethylene terephthalate)(PET) using N-heterocyclic carbenes from ionic liquids. Journal of Chemical Education, 87, 519–521. https://doi.org/10.1021/ed800152c
Ronkvist, A. M., Xie, W., Lu, W., & Gross, R. A. (2009). Cutinase-catalyzed hydrolysis of poly(ethylene terephthalate). Macromolecules, 42, 5128–5138. https://doi.org/10.1021/ma9005318
Sulaiman, S., Yamato, S., Kanaya, E., Kim, J. J., Koga, Y., Takano, K., & Kanaya, S. (2012). Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Applied and Environment Microbiology, 78, 1556–1562. https://doi.org/10.1128/AEM.06725-11
Muller, R. J., Schrader, H., Profe, J., Dresler, K., & Deckwer, W. D. (2005). Enzymatic degradation of poly(ethylene terephthalate): Rapid hydrolyse using a hydrolase from T. fusca. Macromolecular Rapid Communications, 26, 1400–1405. https://doi.org/10.1002/marc.200500410
Yoshida, S., Hiraga, K., Takehana, T., Taniguchi, I., Yamaji, H., Maeda, Y., Toyohara, K., Miyamoto, K., Kimura, Y., & Oda, K. (2016). A bacterium that degrades and assimilates poly(ethylene terephthalate). Science, 351, 1196–1199. https://doi.org/10.1126/science.aad6359
Ma, Y., Yao, M., Li, B., Ding, M., He, B., Chen, S., Zhou, X., & Yuan, Y. (2018). Enhanced poly (ethylene terephthalate) hydrolase activity by protein engineering. Engneering, 4, 888–893. https://doi.org/10.1016/j.eng.2018.09.007
Pirillo, V., Orlando, M., Tessaro, D., Pollegioni, L., & Molla, G. (2021). An efficient protein evolution workflow for the improvement of bacterial PET hydrolyzing enzymes. International Journal of Molecular Sciences, 23, 264. https://doi.org/10.3390/ijms23010264
Cui, Y., Chen, Y., Liu, X., Dong, S., Tian, Y., Qiao, Y., Mitra, R., Han, J., Li, C., Han, X., Liu, W., Chen, Q., Wei, W., Wang, X., Du, W., Tang, S., Xiang, H., Liu, H., Liang, Y., … Wu, B. (2021). Computational redesign of a PETase for plastic biodegradation under ambient condition by the GRAPE strategy. ACS Catalysis, 11, 1340–1350. https://doi.org/10.1021/acscatal.0c05126
Lu, H., Diaz, D. J., Czarnecki, N. J., Zhu, C., Kim, W., Shroff, R., Acosta, D. J., Alexander, B. R., Cole, H. O., Zhang, Y., Lynd, N. A., Ellington, A. D., & Alper, H. S. (2022). Machine learning-aided engineering of hydrolases for PET depolymerization. Nature, 604, 662–667. https://doi.org/10.1038/s41586-022-04599-z
Tufvesson, P., Lima-Ramos, J., Nordblad, M., & Woodley, J. M. (2011). Guidelines and cost analysis for catalyst production in biocatalytic processes. Organic Process Research & Development, 15, 266–274. https://doi.org/10.1021/op1002165
Heinisch, T., Schwizer, F., Garabedian, B., Csibra, E., Jeschek, M., Vallapurackal, J., Vallapurackal, J., Pinheiro, V. B., Marliere, P., Panke, S., & Ward, T. R. (2018). E. coli surface display of streptavidin for directed evolution of an allylic deallylase. Chemical Science, 9, 5383–5388. https://doi.org/10.1039/c8sc00484f
Quehl, P., Hollender, J., Schuurmann, J., Brossette, T., Mass, R., Jose, J. (2016). Co-expression of active human cytochrome P450 1A2 and cytochrome P450 reductase on the cell surface of Escherichia coli. Microbial Cell Factories, 15. https://doi.org/10.1186/s12934-016-0427-5
Bloois, E. V., Winter, R. T., Kolmar, H., & Fraaije, M. W. (2011). Decorating microbes: Surface display of proteins on Escherichia coli. Trends in Biotechnology, 29, 79–86. https://doi.org/10.1016/j.tibtech.2010.11.003
Jose, J., Maas, R. M., & Teese, M. G. (2012). Autodisplay of enzymes-Molecular basis and perspectives. Journal of Biotechnology, 161, 92–103. https://doi.org/10.1016/j.jbiotec.2012.04.001
Verhoeven, G. S., Alexeeva, S., Dogterom, M., & Blaauwen, T. (2009). Differential bacterial surface display of peptides by the transmembrane domain of ompA. PLoS One, 4, e6739. https://doi.org/10.1371/journal.pone.0006739
Samuelson, P., Gunneriusson, E., Nygren, P. A., & Stahl, S. (2002). Display of proteins on bacteria. Journal of Biotechnology, 96, 129–154. https://doi.org/10.1016/S0168-1656(02)00043-3
Jose, J., & Meyer, T. F. (2007). The autodisplay story, from discovery to biotechnical and biomedical applications. Microbiology and Molecular Biology Reviews, 71, 600–619. https://doi.org/10.1128/MMBR.00011-07
Ashiuchi, M., Nawa, C., Kamei, T., Song, J. J., Hong, S. P., Sung, M. H., Soda, K., Yagi, T., & Misono, H. (2001). Physiological and biochemical characteristics of poly γ-glutamate synthetase complex of Bacillus subtilis. European Journal of Biochemistry, 268, 5321–5328. https://doi.org/10.1046/j.0014-2956.2001.02475.x
Narita, J., Okano, K., Tateno, T., Tanino, T., Sewaki, T., Sung, M., Fukuda, H., & Kondo, A. (2006). Display of active enzymes on the cell surface of Escherichia coli using pgsA anchor protein and their application to bioconversion. Applied Microbiology and Biotechnology, 70, 564–572. https://doi.org/10.1007/s00253-005-0111-x
Gallus, S., Peschke, T., Paulsen, M., Burgahn, T., Niemeyer, C. M., & Rabe, K. S. (2020). Surface display of complex enzymes by in situ spycatcher-spytag interaction. ChemBioChem, 21, 2126–2131. https://doi.org/10.1002/cbic.202000102
Gallus, S., Mittmann, E., & Rabe, K. S. (2022). A modular system for the rapid comparison of different membrane anchors for surface display on Escherichia coli. ChemBioChem, 23, e202100472. https://doi.org/10.1002/cbic.202100472
Ko, M. K., Kim, M. J., Yi, J., Kang, J., Bae, J. H., Sohn, J. H., & Sung, B. H. (2021). A novel protein fusion partner, carbohydrate-binding module family 66, to enhance heterologous protein expression in Escherichia coli. Microbial Cell Factories, 20, 232. https://doi.org/10.1186/s12934-021-01725-w
Nascimento, B. M., & Nair, N. U. (2020). Characterization of a membrane enzymatic complex for heterologous production of poly-γ-glutamate in E. coli. Metabolic Engineering Communications, 11, e00144. https://doi.org/10.1016/j.mec.2020.e00144
Acknowledgements
We would like to thank Editage (http://www.editage.com) for English language editing.
Funding
This study was supported by a Grant from the Fuji Seal Foundation.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by TY and TM. The first draft of the manuscript was written by TY and TM commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical Approval
This study was approved by Osaka Metropolitan University and carried out according to the guidelines of the committee at Osaka Metropolitan University.
Consent to Participate
All authors have their consent to participate.
Consent for Publication
All authors have their consent to publish their work.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key Points
• PETase was successfully displayed on the E. coli surface via PgsA anchor.
• Genetic fusion between PETase and PgsA exhibited its highest activity.
• Displayed PETase efficiently degraded BHET rather than using the crude enzyme.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yamashita, T., Matsumoto, T., Yamada, R. et al. Display of PETase on the Cell Surface of Escherichia coli Using the Anchor Protein PgsA. Appl Biochem Biotechnol 196, 5471–5483 (2024). https://doi.org/10.1007/s12010-023-04837-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-023-04837-8