Abstract
The conventional process for converting starch to glucose is energy-intensive. To lower the cost of this process, a novel strain of Paenibacillus phyllosphaerae CS-148 was isolated and identified, which could directly hydrolyze raw starch into glucose and accumulate glucose in the fermentation broth. The effects of different organic and inorganic nitrogen sources, the culture temperature, the initial pH, and the agitation speed on the yield of glucose were optimized through the one-factor-at-a-time method. Nine factors were screened by Plackett–Burman design, and three factors (raw corncob starch, yeast extract and (NH4)2SO4) had significant effects on glucose yield. Three significant factors were further optimized using Box-Behnken design. Under the optimized fermentation conditions (raw corncob starch 40.4 g/L, yeast extract 4.27 g/L, (NH4)2SO4 4.39 g/L, KH2PO4 2 g/L, MgSO4`7H2O 2 g/L, FeSO4`7H2O 0.02 g/L, NaCl 2 g/L, KCl 0.5 g/L, inoculums volume 4%, temperature 35 °C, agitation rate 150 rpm, and initial pH 7.0), the maximum glucose yield reached 17.32 ± 0.46 g/L, which is 1.33-fold compared to that by initial fermentation conditions. The maximum conversion rate and glucose productivity were 0.43 ± 0.01 g glucose/g raw corn starch and 0.22 ± 0.01 g/(L·h), respectively. These results implied that P. phyllosphaerae CS-148 could be used in the food industry or fermentation industry at a low cost.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Starch is the most important storage polysaccharide in plants, its hydrolysis products, such as glucose, maltose, and other syrups are utilized extensively in the food, pharmaceutical, and fermentation industries [1]. However, starch granules show some resistance to chemicals and enzymes hydrolysis because of their polycrystalline structure [2]. Therefore, the conventional enzymatic saccharification of starch entails three steps: gelatinization, liquefaction, and saccharification [3]. Starch slurry is first gelatinized at about 100 °C to destroy the crystalline structure; then, the gelatinized starch is liquefied at about 95 °C by thermophilic α-amylases. Subsequently, the liquefied solution is hydrolyzed using glucoamylases at 50–60 °C to produce glucose syrup [4, 5]. This is a high-energy process, resulting in high production costs. Therefore, researchers are exploring various techniques to reduce energy consumption and production costs. Among them, the direct hydrolysis of starch using raw starch digesting α-amylase (RSDA) or microorganisms may be a good choice.
RSDA can directly hydrolyze raw starch granules into glucose without gelatinization [6]. It can simplify the conventional starch hydrolysis process and lower energy consumption [7]. Thus, it has garnered significant attention in recent years. Many researchers have reported that RSDA could be produced by various strains, such as Penicillium oxalicum [8], Bacillus licheniformis [9], Aspergillus fumigatus [2], and Aspergillus niger [10]. Many tesearches have been conducted into improving the activity of RSDA, such as screening new strains, optimizing the fermentation conditions, mutation breeding, and molecular cloning [8, 11,12,13,14]. However, the production of RSDA activity is still relatively low, and the fermentation and purification of RSDA also further increase the cost of enzyme application. Thus, more research effort must be made.
In this paper, a novel Paenibacillus phyllosphaerae CS-148 strain was screened, which can directly hydrolyze raw starch into glucose, however, the glucose was not consumed by the microorganism. Next, fermentation conditions were optimized. To the best of the authors’ knowledge, this is the first time reported a microorganism which can directly hydrolyze raw starch and accumulate glucose in fermentation broth. The purpose of this study was to determine a novel technique to reduce the energy consumption and cost of starch processing.
Materials and Methods
Isolation and Identification of Microorganisms
Soil samples were collected from different flour mills in Huaian City, China. Soil samples (1 g) were mixed with 9-mL sterile water and the suspensions were serially diluted, the supernatants (0.2 mL) with appropriate dilutions were then spread onto raw starch agar plates, which containing (g/L): corn starch 20, (NH4)2SO4 2,K2HPO4 1, MgSO4`7H2O 0.5, NaCl 1, KCl 0.5, FeSO4`7H2O 0.01, and agar 15.The pH of the medium was natural (~7.2). Corn starch was sterilized using dry heat sterilization at 110 °C for 3 h, and added to sterile nutrient agar at 50–60 °C. After incubation at 32 °C for 3–5 days, the strains that can hydrolyze raw starch were screened for the formation of clearing zones around the colonies, and these screened strains were further confirmed by liquid culture. The liquid medium was the same as the raw starch agar medium without agar. Fermentation was performed in a 250-mL Erlenmeyer flask containing 50 mL of liquid medium. The cultivation was maintained at 32 °C for 4 days with shaking at 150 rpm, where the inoculum volume was 4%. After centrifugation at 10,000 rpm for 5 min, the concentration of glucose in fermentation broths was detected using a Biosensor Analyzer.
The strain that could produce the highest concentration of glucose in the fermentation broth was named CS-148. The physiological and biochemical characteristics of CS-148 were investigated using previously described approaches [15, 16]. The genomic DNA of CS-148 was extracted using a TIANGEN Bacterial Genomic Extraction Kit, and the partial 16S rDNA was amplified using the PCR master mix kit (Sangon Biotech Shanghai, China). The PCR primers were 27F (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-GGTTACCTTGTTACGACTT-3’). The amplified 16S rDNA fragment was sent to Sangon Biotech Shanghai for sequencing. Further the obtained partial 16S rRNA sequence was used for homologous comparison using BLAST at the NCBI website to identify the strain. The phylogenetic tree was constructed by the neighbor-joining method of MEGA XI, using the maximum composite likelihood method to compute the evolutionary distances.
Optimization of Culture Conditions
The different fermentation parameters, including organic nitrogen sources (peptone, tryptone, yeast extract, beef extract, and corn steep liquor, with the pH adjusted to 7), inorganic nitrogen sources (NH4Cl, (NH4)2SO4, NaNO3, NH4NO3, with the pH adjusted to 7), cultural temperature (26, 29, 32, 35 and 38 °C), initial pH (5,6,7,8, and 9) and agitation speed (100, 150, 200, and 250 rpm) were initially screened using the one-factor-at-a-time(OFAT) method to investigate the effect of the glucose yield, while the concentrations of other ingredients were the same as the raw starch agar medium, such as corn starch, K2HPO4, MgSO4`7H2O, NaCl, KCl, and FeSO4`7H2O.
Screening of Fermentation Parameters
Plackett–Burman design (PBD) can identify significant factors from various parameters using few experimental runs [17]. The effects of nine factors on glucose production were screened by PBD: the inoculum amount, along with the concentration of raw corn starch, yeast extract, (NH4)2SO4, KH2PO4, MgSO4`7H2O, NaCl, KCl, and FeSO4`7H2O. Each factor was set at two levels, coded by (+1) and (−1), as shown in Table 1.
Optimization by Box–Behnken design (BBD)
According to the results of PBD, three significant factors (raw corncob starch concentration, yeast extract concentration, and ammonium sulfate concentration) were screened from the nine factors and were further optimized using BBD. The coded forms of the significant parameters and their actual values are shown in Table 2. A total of 17 runs were performed. The correlation relationship between the variables and response was fitted by a second-order polynomial model, and can be expressed as follows:
where Y is the predicted response (glucose yield); β0 is the model constant-coefficient; X1, X2, and X3 are the concentration of raw corn starch, yeast extract and (NH4)2SO4 (g/L), respectively. β1, β2, and β3 are linear coefficients, β11, β22, and β33 are the quadratic coefficients, β12, β13, and β23 are the interaction coefficients.
Quantitative Determination of Glucose and Fermentation Parameters
The concentration of glucose was assayed following the method proposed by Zhao [18] using a Biosensor Analyzer (SBA-40E, Biology Institute of Shandong Academy of Sciences, China). The samples were centrifuged at 10,000 rpm for 5 min, and 25 μL of supernatant was injected into the Biosensor Analyzer to measure the glucose concentration.
The glucose yield was defined as the concentration of glucose in the fermentation broths. The conversion rate of raw starch was calculated as the ratio of produced glucose (g) to total mass of the raw corn starch (g). Glucose productivity was calculated as the ratio of glucose concentration (g/L) in the fermentation broth to the fermentation time (h).
Statistical Analysis
The data was analyzed by one-way ANOVA, and the differences of the means (n=3) were determined using Tukey’s test (p < 0.05). The PBD and BBD experiments were designed and analyzed in the Design Expert (v. 8.0.6) software (State-Ease Inc. Minneapolis, USA).
Scanning Electron Microscopy of Starch Granules
The fermentation broth was centrifuged at 4000 rpm for 5 min, and the precipitates of hydrolysis starch granules were dried via lyophilization. Then, starch granules were gold-sputtered and observed using an S-3400N scanning electron microscope (SEM) (Hitachi Limited, Tokyo, Japan) under vacuum conditions using a 10 kV accelerating voltage [19].
Results and Discussion
Isolation and Identification of Microorganisms
The strains from the soil were screened on the raw starch plate, eight colonies were screened out by observing the zone of clearance due to the hydrolysis of raw starch. Then, the strains were further screened using a liquid culture. Among them, the CS-148 fermentation broth produced the highest concentration of glucose; therefore, it was subjected to the experiment. The colony morphology of this strain on the raw starch plate is shown in Fig. 1, and the morphological and biochemical characteristics of CS-148 are shown in Table 3. It can be seen that the colonies of CS-148 were round, smooth, convex, transparent, sticky, and formed transparent zones on raw starch agar plates. Furthermore, CS-148 cells were rod-shaped, and Gram staining was variable. Rivas et al. [16] also reported similar results regarding the strain of P. phyllosphaerae PALXIL04T. Many members of Paenibacillus are known to be Gram variable [20,21,22,23], which may be caused by differences in the chemical and structural characteristics of the bacterial cell walls of those strains. The gram-positive bacteria had a thick, relatively impermeable wall, and Gram-negative bacteria had a thin peptidoglycan layer plus lipid–protein bilayer wall. Therefore, they have intermediate structure cell walls with variable staining characteristics [24]. Furthermore, as shown in Table 3, strain CS-148 could utilize sucrose, maltose, glycerol, and inositol, but could not utilize glucose, fructose, galactose, mannose, xylose, and citrate. The 16S rDNA sequence of the strain was submitted in the NCBI GenBank (GenBank accession number: OQ608826) and compared with the data contained therein, which revealed that the screened strain was phylogenetically related to Paenibacillus phyllosphaerae. As shown in Fig. 2, the strain CS-148 and P. phyllosphaerae strain SM26 were grouped together in one branch and had the highest sequence similarity. Thus, the strain was named as P. phyllosphaerae CS-148.
Effects of Different Culture Parameters on the Yield of Glucose
The effects of different organic and inorganic nitrogen sources, cultural temperature, initial pH, and agitation speed on the yield of glucose were examined, and the results are presented in Fig. 3.
As shown in Fig. 3, different cultural conditions have varied effects on the yield of glucose by P. phyllosphaerae CS-148. Fig. 3a shows that there were significant differences (p<0.05) in glucose yields among different organic nitrogen sources. Yeast extract and tryptone could be good organic nitrogen sources for P. phyllosphaerae CS-148 fermentation. However, when yeast extract was used as the organic nitrogen source, fermentation could achieve higher glucose yields (13.03 ± 0.76 g/L) at lower costs. Therefore, the best organic nitrogen source was determined as yeast extract. The high glucose production compared with other organic nitrogen sources is possibly due to significant bio-accessible nutrients for P. phyllosphaerae CS-148 in yeast extract, such as amino acids, nucleic acids, structural proteins, and vitamins. Many researchers have reported that the optimal organic nitrogen source was yeast extract for Paenibacillus spp. [25, 26]. As shown Fig. 3b, the glucose yields using (NH4)2SO4 are significantly higher than those using the other three inorganic nitrogen sources. (NH4)2SO4 is considered to be a suitable inorganic nitrogen sources, while (NH4)2SO4 is a good industrial nitrogen source because of its low cost. It is widely known that environmental variables, such as temperature, pH, and agitation speed, have significant effects on cell growth and metabolite synthesis. The effects of temperature on the yield of glucose by P. phyllosphaerae CS-148 can be seen in Fig. 3c, where the maximum yield of glucose was obtained at 35 °C, at 13.40 ± 0.53 g/L. These results are consistent with a study by Priest [23], which claimed that the optimum growth temperature is generally at 28–40 °C for the genus of Paenibacillus. According to Fig. 3d, the yield of glucose increased with increases in pH from 5 to 7, and significantly decreased with an increase in pH from 7 to 9. The highest yield of glucose was at pH 7, reaching 13.37 ± 0.67 g/L. Similarly, previous reports have suggested that the optimum pH of Paenibacillus spp. is 7 [23]. The pH has a significant impact on cellular metabolism, because different pH values can change the membrane charge, membrane permeability, and ionization of nutrients [27]. When the pH value is higher and lower than the optimal pH, the yields of glucose were significantly decreased. Furthermore, shaking is an important parameter that allows submerged fermentation to overcome mass transfer resistances [28]. Fig. 3e illustrates the effect of agitation speed on the glucose yield. When the agitation speed was increased from 100 to 150 rpm, the glucose yield significantly increased from 11.20 ± 0.56 g/L to 13.17± 0.47 g/L. It was also found that there is no significant difference in the glucose yield of fermentation between the agitation speed of 150 and 200 rpm. Hence, the agitation speed of 150 rpm was selected as the optimal agitation speed for fermentation. When the agitation speed further increased to 200 rpm, the yield of glucose decreased to 12.03 ± 0.83 g/L. This might be caused by shear damage to the cells at high agitation speeds [29].
Screening of Significant Parameters Using PBD
The levels of nine factors for the PBD were selected according to previous experiments, and the experimental results for the PBD are listed in Table 4 and Table 5.
Based on the analysis of PBD (Table 5), the model had a low p-value (0.0245), indicating its statistical significance. The R2 value of the equation was 0.9945, and the predicted R2 of 0.8020 is in agreement with the adjusted R2 of 0.9697, with a difference of less than 0.2. These results confirmed that the design was a good fit for the model, and the model could be used to predict the effects of the factors on glucose yield by P. phyllosphaerae CS-148. The predictive model is represented as follows: The yield of glucose (g/L) = 14.63+1.17X1+1.21X2+0.53X3+0.11X4+0.057X5+0.17X6+0.09X7-0.12X8+0.16X9.
As shown in Table 4, three factors, namely raw corn starch, yeast extract, and (NH4)2SO4, were found to be the significant parameters affecting glucose yield , their p-values were less than 0.05.
Determining the Optimum Levels of Significant Factors Using BBD
The combined effect of three important factors for glucose yield was further studied by BBD, and the design matrix with responses is listed in Table 6. Analysis of the variance of BBD is shown in Table 7.
According to Table 7, this regression model was found to be significant, and the lack of fit was not significant, which indicated that the model was well-fitted and can be used to predict the effects of the variables on glucose yield. The value of the determination coefficient was 0.9867, which further confirmed that the model is satisfactory. The model terms X2 (yeast extract, p=0.0013), X3 (ammonium sulfate, p=0.0053), X12 (raw corn starch, p<0.0001), X22 (p<0.0001), and X32 (p=0.0001) were found to be significant model terms for the yield of glucose. The quadratic response surface model was fitted to the following equations: The yield of glucose (g/L)=17.71+0.145*X1+0.46*X2+0.355*X3−0.13*X1*X2+0.04*X1*X3+0.03*X2*X3−1.66*X12−1.69*X22−0.936*X32.
The interactions between the two factors are shown in 3D curves and contour plots (Fig. 4). Fig. 4a shows the interactive effect of raw corncob starch and yeast extract on glucose yield by keeping ammonium sulfate at the center point. The yield of glucose increased as the concentration of raw corncob starch increased from 30 to 40.4 g/L, and yeast extract from 2 to 4.27 g/L. However, the circular contour lines suggested that the interaction between raw corn starch and yeast extract was weak, which is also consistent with the results in Table 7. In other interactions, the relative effect of raw corncob starch and (NH4)2SO4, yeast extract, and (NH4)2SO4 were also studied and are shown in Fig. 4b, c, the results were similar to the interaction between raw corn starch and yeast extract. All of the 3D response surface graphs exhibited a convex shape, and the maximum glucose yield could be well-defined. From the quadratic response surface model, the optimized parameters were raw corncob starch 40.0 g/L, yeast extract 4.3 g/L, and (NH4)2SO4 4.4 g/L; furthermore, the theoretical yield of glucose was 17.78 g/L.
Verification Test and SEM of Starch Granules
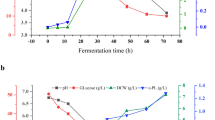
The validation experiments were carried out three times in a 250-mL Erlenmeyer flask containing 50 mL of the culture under the optimized fermentation conditions (raw corncob starch 40.0 g/L, yeast extract 4.3 g/L, (NH4)2SO4 4.4 g/L, K2HPO4 2 g/L, MgSO4`7H2O 2 g/L, FeSO4`7H2O 0.02 g/L, NaCl 2 g/L, KCl 0.5 g/L, inoculum volume 4%, temperature 35 °C, agitation rate 150 rpm, and initial pH 7.0). The glucose yields, conversion rates and glucose productivities were analyzed every 24 h. As shown in Fig. 5, after 24 h of fermentation, a low concentration of glucose was observed, which was 3.80 ± 0.25g/L, and the conversion rate was 0.10 ± 0.01 g glucose/g raw corn starch. However, the glucose productivity was relatively high, at 0.16 ± 0.01 g/(L·h). The highest yield of glucose, 17.32 ± 0.46 g/L, was obtained at 96 h of fermentation, which decreased subsequently. The change in the conversion rate was the same as in the glucose yield; its highest value was 0.43 ± 0.01 g glucose/g raw corn starch. This relatively low conversion rate was due to some raw starch not being hydrolyzed into glucose. Another reason is that some raw starch may be hydrolyzed into oligosaccharides, which cannot be detected by the Biosensor Analyzer. The glucose productivity generally increased with fermentation time, the maximum value was recorded at 72 h, at 0.22 ± 0.01 g/(L·h). However, when the yield of glucose reached its maximum, the glucose productivity decreased to 0.18 ± 0.01 g/(L·h). The results show that the experimental values were closely related to the predicted values, proving that the model is adequate.
In addition, the surface of raw and hydrolyzed corncob starch granules was observed by SEM. The surface morphological changes are shown in Fig. 5, where it can be seen that the surface of the raw starch granules was smooth, while the surface of starch granules became pitted after 24 h of fermentation, and many large holes appeared after 48 h. As the fermentation duration increased, larger cavities formed in the starch granules and, then, the granules cracked and ruptured. These results further confirmed that the strain of P. phyllosphaerae CS-148 can directly hydrolyze raw starch and accumulate glucose.
Conclusion
In this study, a novel P. phyllosphaerae CS-148 strain was isolated and identified, which could directly hydrolyze raw starch into glucose. However the glucose in the fermentation broth was not consumed by the microorganism. Then, the effects of the fermentation conditions on the glucose yields by P. phyllosphaerae CS-148 were investigated. The effects of different organic and inorganic nitrogen sources, the cultural temperature, the initial pH, the agitation speed, and the fermentation duration on the yield of glucose were optimized using the OFAT method. The screening results of PBD showed that the raw corncob starch, yeast extract, and (NH4)2SO4 concentration significantly affected the glucose yields. Thus, the optimum concentrations of the selected variables were calculated using the Box-Behnken design. The maximum glucose yield reached 17.32 ± 0.46 g/L under the optimized fermentation conditions, which is 1.33-fold that by initial fermentation conditions. The maximum conversion rate and glucose productivity were 0.43 ± 0.01 g glucose/g raw corn starch and 0.22 ± 0.01 g/(L·h), respectively. These results suggest that P. phyllosphaerae CS-148 has great potential for applications in the food industry or fermentation industry. However, more research effort should focus on improving the conversion rate of raw starch and understanding the starch hydrolysis mechanism for industrial application.
Data Availability
All the data generated or analyzed during this study are included in this published article.
References
Zhang, L., Zhong, L., Wang, J., Zhao, Y., Zhang, Y., Zheng, Y., Dong, W., Ye, X., Huang, Y., Li, Z., & Cui, Z. (2021). Efficient hydrolysis of raw starch by a maltohexaose-forming α-amylase from Corallococcus sp EGB. LWT, 152, 112361.
Song, W., Tong, Y., Li, Y., Tao, J., Li, J., Zhou, J., & Liu, S. (2021). Expression and characterization of a raw-starch glucoamylase from Aspergillus fumigatus. Process Biochemistry, 111, 97–104.
Fang, W., Xue, S., Deng, P., Zhang, X., Wang, X., Xiao, Y., & Fang, Z. (2019). AmyZ1: a novel α-amylase from marine bacterium Pontibacillus sp. ZY with high activity toward raw starches. Biotechnology for Biofuels, 12, 95.
Lin, H. J., Xian, L., Zhang, Q. J., Luo, X. M., Xu, Q. S., Yang, Q., Duan, C. J., Liu, J. L., Tang, J. L., & Feng, J. X. (2011). Production of raw cassava starch-degrading enzyme by Penicillium and its use in conversion of raw cassava flour to ethanol. Journal of Industrial Microbiology & Biotechnology, 38, 733–742.
Goyal, N., Gupta, J. K., & Soni, S. K. (2005). A novel raw starch digesting thermostable α-amylase from Bacillus sp. I-3 and its use in the direct hydrolysis of raw potato starch. Enzyme and Microbial Technology, 37, 723–734.
Nwagu, T., Lin, H. J., Xian, L., Zhang, Q. J., Luo, X. M., Xu, Q. S., Yang, Q., Duan, C. J., Liu, J. L., Tang, J. L., & Feng, J. X. (2012). Adsorption and stabilization of a raw sarch digesting amylase on micro bead silica Gel 300 A. British Biotechnology Journal, 2, 85–101.
Shofiyah, S. S., Yuliani, D., Widya, N., Sarian, F. D., Puspasari, F., Radjasa, O. K., & Ihsanawati, & Natalia D. (2020). Isolation, expression, and characterization of raw starch degrading α-amylase from a marine lake Bacillus megaterium NL3. Heliyon, 6, e05796.
Gu, L. H., Tan, M. Z., Li, S. H., Zhang, T., Zhang, Q. Q., Li, C. X., Luo, X. M., Feng, J. X., & Zhao, S. (2020). ARTP/EMS-combined multiple mutagenesis efficiently improved production of raw starch-degrading enzymes in Penicillium oxalicum and characterization of the enzyme-hyperproducing mutant. Biotechnology for Biofuels, 13, 187.
Lomthong, T., Chotineeranat, S., & Kitpreechavanich, V. (2015). Production and characterization of raw starch degrading enzyme from a newly isolated thermophilic filamentous bacterium, Laceyella sacchari LP175. Starch - Stärke, 67, 255–266.
Sun, H., & Peng, M. (2017). Improvement of glucoamylase production for raw-starch digestion in Aspergillus niger F-01 by maltose stearic acid ester. Biotechnology Letters, 39, 561–566.
Xie, F., Quan, S., Liu, D., Ma, H., Li, F., Zhou, F., & Chen, G. (2014). Purification and characterization of a novel α-amylase from a newly isolated Bacillus methylotrophicus strain P11-2. Process Biochemistry, 49, 47–53.
Finore, I., Kasavi, C., Poli, A., Romano, I., Oner, E. T., Kirdar, B., Dipasquale, L., Nicolaus, B., & Lama, L. (2011). Purification, biochemical characterization and gene sequencing of a thermostable raw starch digesting α-amylase from Geobacillus thermoleovorans subsp. stromboliensis subsp. nov. World Journal of Microbiology and Biotechnology, 27, 2425–2433.
Duque, S. M. M., Dizon, E. I., Merca, F. E., & Flores, D. M. (2016). Optimization of raw-starch-digesting amylase (RSDA) production medium for Enterococcus faecium DMF78. International Food Research Journal, 23, 1280–1288.
Tang, S., Xu, T., Peng, J., Zhou, K., Zhu, Y., Zhou, W., Cheng, H., & Zhou, H. (2020). Overexpression of an endogenous raw starch digesting mesophilic α-amylase gene in Bacillus amyloliquefaciens Z3 by in vitro methylation protocol. Journal of the Science of Food and Agriculture, 100, 3013–3023.
Buchanan, R. E., & Gibbons, N. E. (1984). Bergey’s manual of determinative bacteriology (8th ed.). Beijing, China: Science Press.
Rivas, R., Mateos, P. F., Martínez-Molina, E., & Velázquez, E. (2005). Paenibacillus phyllosphaerae sp. nov., a xylanolytic bacterium isolated from the phyllosphere of Phoenix dactylifera. International Journal of Systematic and Evolutionary Microbiology, 55, 743–746.
Bhaturiwala, R., Bagban, M., Mansuri, A., & Modi, H. (2022). Successive approach of medium optimization using one-factor-at-a-time and response surface methodology for improved β-mannanase production from Streptomyces sp. Bioresource technology, 18, 101087.
Zhao, H., Xia, J., Wang, J., Yan, X., Wang, C., Lei, T., Xian, M., & Zhang, H. (2018). Production of bacterial cellulose using polysaccharide fermentation wastewater as inexpensive nutrient sources. Biotechnology and Biotechnological Equipment, 32, 350–356.
Govindaraju, I., Chakraborty, I., Baruah, V. J., Sarmah, B., Mahato, K. K., & Mazumder, N. (2021). Structure and morphological properties of starch macromolecule using biophysical techniques. Starch - Stärke, 73, 2000030.
Roldán, D. M., Costa, A., Králová, S., Busse, H.-J., Amarelle, V., Fabiano, E., & Menes, R. J. (2022). Paenibacillus farraposensis sp. nov., isolated from a root nodule of Arachis villosa. International Journal of Systematic and Evolutionary Microbiology, 72, 005294.
Sáez-Nieto, J. A., Medina-Pascual, M. J., Carrasco, G., Garrido, N., Fernandez-Torres, M. A., Villalón, P., & Valdezate, S. (2017). Paenibacillus spp. isolated from human and environmental samples in Spain: Detection of 11 new species. New Microbes and New Infections, 19, 19–27.
Aw, Y.-K., Ong, K.-S., Lee, L.-H., Cheow, Y.-L., Yule, C. M., & Lee, S.-M. (2016). Newly isolated Paenibacillus tyrfis sp. nov., from Malaysian tropical Peat swamp soil with broad spectrum antimicrobial activity. Frontiers in Microbiology, 7, 219.
Priest, F. G. (2015). Paenibacillus. In S. D. M. E. Trujillo, P. DeVos, B. Hedlund, P. Kämpfer, F. A. Rainey, & W. B. Whitman (Eds.), Bergey’s Manual of Systematics of Archaea and Bacteria (pp. 1–40). New York: Wiley.
Beveridge, T. J. (2001). Use of the Gram stain in microbiology. Biotechnic & Histochemistry, 76, 111–118.
Liang, T. W., & Wang, S. L. (2015). Recent advances in exopolysaccharides from Paenibacillus spp.: Production, isolation, structure, and bioactivities. Marine Drugs, 13, 1847–1863.
Häßler, T., Schieder, D., Pfaller, R., Faulstich, M., & Sieber, V. (2012). Enhanced fed-batch fermentation of 2,3-butanediol by Paenibacillus polymyxa DSM 365. Bioresource Technology, 124, 237–244.
Zhang, X., Li, H., Kang, X., Lim, S., & Li, F. (2021). Isolation, identification and optimization of fermentation conditions against Sclerotinia sclerotiorum strains in high salt Doenjang. Food Science and Human Wellness, 10, 205–213.
Rahman, S. S. A., Pasupathi, S., & Karuppiah, S. (2022). Conventional optimization and characterization of microbial dextran using treated sugarcane molasses. International Journal of Biological Macromolecules, 220, 775–787.
Choudhury, A. R., Sharma, N., & Prasad, G. S. (2012). Deoiledjatropha seed cake is a useful nutrient for pullulan production. Microbial Cell Factories, 11, 39.
Funding
This work was supported by the Major Basic Research Project of the Natural Science Foundation of the Jiangsu Higher Education Institutions (18KJA550001).
Author information
Authors and Affiliations
Contributions
GY: funding acquisition, conceptualization, supervision, investigation, writing—original draft; YZ: supervision, writing—review and editing; CJ: methodology; JW: funding acquisition; LZ: resources; WW: data curation
Corresponding author
Ethics declarations
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
All authors have their consent to publish their work.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yan, G., Zhou, Y., Wu, J. et al. Novel Strain of Paenibacillus phyllosphaerae CS-148 for the Direct Hydrolysis of Raw Starch into Glucose: Isolation and Fermentation Optimization. Appl Biochem Biotechnol 196, 4125–4139 (2024). https://doi.org/10.1007/s12010-023-04750-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-023-04750-0