Abstract
Atherosclerosis (AS) is one of the most common and important vascular diseases. It is believed that the abnormal expression of circular RNAs (circRNAs) plays an important role in AS. Hence, we investigate the function and mechanism of circ-C16orf62 in AS development.
In this study, oxidized low-density lipoprotein (ox-LDL)-treated human macrophages (THP-1) were used as pathological conditions of AS in vitro. The expression of circ-C16orf62, miR-377 and Ras-related protein (RAB22A) mRNA was detected by real-time quantitative polymerase chain reaction (RT-qPCR) or western blot. Cell viability or cell apoptosis was assessed by cell counting kit-8 (CCK-8) assay or flow cytometry assay. The releases of proinflammatory factors were investigated using enzyme-linked immunosorbent assay (ELISA). The production of malondialdehyde (MDA) and superoxide dismutase (SOD) was examined to assess oxidative stress. Total cholesterol (T-CHO) level was detected, and cholesterol efflux level was tested using a liquid scintillation counter. The putative relationship between miR-377 and circ-C16orf62 or RAB22A was verified by dual-luciferase reporter assay and RNA immunoprecipitation (RIP) assay.
circ-C16orf62 expression was elevated in AS serum samples and ox-LDL-treated THP-1 cells. Apoptosis, inflammation, oxidative stress and cholesterol accumulation induced by ox-LDL were suppressed by circ-C16orf62 knockdown. Circ-C16orf62 could bind to miR-377 and thus increased the expression level of RAB22A. Rescued experiments showed that circ-C16orf62 knockdown alleviated ox-LDL-induced THP-1 cell injuries by increasing miR-377 expression, and miR-377 overexpression lessened ox-LDL-induced THP-1 cell injuries by degrading RAB22A level.
In conclusion, circ-C16orf62 played a crucial role in the regulation of apoptosis, inflammation, oxidative stress and cholesterol accumulation in ox-LDL-treated human macrophages via mediating the miR-377/RAB22A axis, hinting that circ-C16orf62 might be involved in AS progression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Atherosclerosis (AS) is one of the common chronic inflammatory diseases, leading to a variety of cardiovascular diseases, such as acute myocardial infarction, coronary heart disease and so on [1]. Oxidized low-density lipoprotein (ox-LDL) is considered to be a key factor in the occurrence and development of AS, which can promote the formation of foam cells and the destruction of the homeostasis of cholesterol uptake and efflux in macrophages [2]. Macrophages play crucial roles in all stages of AS [3]. In the progressive stage of AS, the accumulation of ox-LDL, the promotion of inflammation responses and the activation of oxidative stress can facilitate the formation of vulnerable plaques through promoting macrophage apoptosis [4]. Vulnerable plaque rupture contributes to the formation of thrombus, leading to cardiovascular diseases [4]. Therefore, it is of great significance to explore effective treatment strategies to inhibit the abnormal characteristics of macrophages to inhibit the progression of AS.
Increasing studies have reported that circular RNAs (circRNAs) are implicated in the development of AS through multiple regulatory modes, including the modulation of vascular endothelial cell injuries and the mediation of vascular smooth muscle cell proliferation [5, 6]. However, the implication of circRNAs in macrophage activities and functions is still lacking, even though the great advances of other non-coding RNA (ncRNA) molecules, such as long ncRNA (lncRNA) and microRNA (miRNA), have been made in the functions of macrophage in AS [7,8,9]. Certain circRNAs are involved in specific physiological and pathological processes through transcriptional or post-transcriptional regulatory mechanisms [10], including the development of heart and blood vessels, cardiac abnormalities such as coronary heart disease and other cardiovascular diseases [11]. Ox-LDL is widely used to treat vascular endothelial cells, vascular smooth muscle cells, or macrophages to mimic AS cell conditions in vitro [12]. A previous study utilized ox-LDL to treat monocytic THP-1 cells pretreated with phorbol 12-myristate 13-acetate (PMA), and circRNA expression profiling (GSE107522) showed that several circRNAs were differently expression after treatment [13], including increased circ_0003645 (circ-C16orf62). The detailed role of circ-C16orf62 needs to be further explored in ox-LDL-treated THP-1 cells.
It is well canonical that certain circRNAs compete with mRNAs for target miRNA response elements (MREs) via a competing endogenous RNA (ceRNA) mechanism [14], and the circRNA-miRNA-mRNA networks are widely identified to expound the functional modes of circRNAs in human diseases [14]. The development of bioinformatics analysis makes it easy to predict potential targets of circRNA or miRNA [15]. Based on this, miR-377 is predicted to be one of the targets of circ-C16orf62, and miR-377 was documented to be involved in AS progression [16]. Thus, miR-377 was screened in our study for further exploration. Moreover, bioinformatics analysis shows that Ras-related protein (RAB22A) is a putative target of miR-377. It is worth exploring the interaction among them to construct a potential regulatory network for circ-C16orf62 in AS progression.
In the present study, we determined the expression of circ-C16orf62 in clinical AS serum samples and ox-LDL-treated THP-1 macrophages. Loss-function experiments were conducted to explore the function of circ-C16orf62 on apoptosis, inflammatory responses, oxidative stress and cholesterol accumulation in macrophages. Besides, the circ-C16orf62-miR-377-RAB22A network was proposed to elucidate the regulatory mechanism of circ-C16orf62, aiming to provide an additional therapeutic target for AS.
Materials and Methods
Serum Samples
Blood samples were collected from the Second Affiliated Hospital of Hainan Medical University, including blood samples from AS patients (n = 29) and blood samples from normal participants (n = 18, without AS). The clinical parameters of these subjects were listed in Table 1. The use of these samples was approved by each subject with written informed consent. AS patients were enrolled following the exclusion criteria: patients with severe infection or malignant disease, with severe liver or kidney damage, receiving any surgical intervention on the heart in history, body mass index over 35. Serum samples were obtained from blood samples by centrifugalization and then frozen in the − 80℃ conditions until use. Importantly, this study was approved by the Ethics Committee of the Second Affiliated Hospital of Hainan Medical University (LW2022241).
Cells and ox-LDL Treatment
Human monocytic THP-1 cells were purchased from EK-Bioscience (Shanghai, China) and cultured in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% fetal bovine serum (FBS; EK-Bioscience) at a 37℃ incubator with 5% CO2.
THP-1 cells plated into a 6-well plate were induced into macrophages using the phorbol-12-myristate acetate (PMA, 100 nM; Sigma-Aldrich, St. Louis, MO, USA) for 48 h at 37℃ prior to subsequent experimentation.
For dose effect analysis, THP-1 macrophages were treated with ox-LDL (Solarbio, Beijing, China) for 48 h, at the dose of 0, 25, 50 and 100 µg/mL. For time effect analysis, THP-1 macrophages were treated with 50 µg/mL ox-LDL for different time (0, 12, 24 and 48 h). The subsequent functional experiments were performed using THP-1 macrophages treated with 50 µg/mL ox-LDL for 48 h.
Real-time Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA isolated using a Trizol reagent (Invitrogen, Carlsbad, CA, USA) was checked using NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). For circ-C16orf62, C16orf62 and RAB22A, cDNA synthesis and RT-qPCR were performed using SuperScript™ III One-Step RT-PCR System (Invitrogen) and SYBR GreenER™ qPCR SuperMix (Invitrogen). For miR-377, cDNA synthesis and RT-qPCR were performed using miRcute Plus miRNA First-Strand cDNA Kit (TianGen, Beijing, China) and miRcute Plus miRNA qPCR Kit (SYBR Green) (TianGen). The relative expression was calculated using the 2−ΔΔCt method, with Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH, for circ-C16orf62 and RAB22A) or U6 (for miR-377) as the internal reference. The following PCR cycling conditions were used: initial denaturation at 95 °C for 5 min, 1 cycle; denaturation at 95 °C for 30 s, annealing at 61 °C for 30 s, and extension at 72 °C for 30 s, 35 cycles. The primers used were listed follows:
circ-C16orf62, F: 5’-CCTCTCCCAAGTGCACTCA-3’ and R: 5’-AGAACAGGCTCCAGATTGCA-3’; C16orf62, F: 5’-ATCAGAGGGATCGGAGACCC-3’ and R: 5’-CTTCCATTCCCACCCGGC-3’; miR-377, F: 5’-CGCGATCACACAAAGGCAAC-3’ and R: 5’-AGTGCAGGGTCCGAGGTATT-3’; RAB22A, F: 5’-TTTTGGCCGCTTTGTAGCAC-3’ and R: 5’-CAGAAGCTTGGCAAACCACC-3’; GAPDH, F: 5’-GAATGGGCAGCCGTTAGGAA-3’ and R: 5’-AAAAGCATCACCCGGAGGAG-3’; U6, F: 5’-CTCGCTTCGGCAGCACA-3’ and R: 5’-AACGCTTCACGAATTTGCGT-3’;
Subcellular Distribution
The RNA from cytoplasmic fraction or nuclear fraction of THP-1 cells using the Cytoplasmic & Nuclear RNA Purification Kit (Norgen Biotek, Ontario, Canada). The expression of circ-C16orf62 in cytoplasmic fraction or nuclear fraction was detected using RT-qPCR, with U6 as an interference in the nucleus and GAPDH as an interference in the cytoplasm.
RNase R Treatment
Total RNA was isolated and then exposed to RNase R (3 U/µg; Epicentre, Madison, WI, USA) for 15 min at 37℃. Subsequently, the expression of circ-C16orf62 and linear C16orf62 was detected by RT-qPCR using these treated RNA samples.
Cell Transfection
Small interference RNA (siRNA)-mediated circ-C16orf62 knockdown (si-circ-C16orf62) and its negative control (si-NC) were obtained from Integrated Biotech Solutions (IBSBIO; Shanghai, China). The mimic and inhibitor of miR-377 (miR-377 or anti-miR-377) were purchased from Ribobio (Guangzhou, China), using mimic control (miR-NC) or inhibitor control (anti-miR-NC) as corresponding control. pcDNA3.1(+) CircRNA Mini Vector was used for circ-C16orf62 overexpression (circ-C16orf62), which was constructed by EK-Bioscience, with empty pcDNA3.1(+) (pcDNA) as a control. Besides, RAB22A sequence was cloned into pcDNA vector for RAB22A overexpression (RAB22A) by EK-Bioscience. These sequences or plasmids were transfected into THP-1 cells for functional analyses using Lipofectamine 3000 transfection kit (Invitrogen).
Cell Counting kit-8 (CCK-8) Assay
To detect cell viability, THP-1 macrophages with treatment or transfection were planted into a 96-well plate (2000 cells/well) to incubate cells. At 48 h post-incubation, cells in each well were treated with 10 µL CCK-8 reagent (Sigma-Aldrich) for another 2 h. Subsequently, the absorbance at 450 nm was detected using a microplate reader (Thermo Fisher Scientific) to monitor cell viability.
EdU Assay
EdU proliferation was performed using Cell-Light EdU Apollo567 Kit (Ribobio). Simply put, cells with transfections were planted in 96-well plates (2000 cells/well). At 48 h post-incubation, cells were cultured with EdU for 4 h, fixed by 4% paraformaldehyde, and stained by Apollo567 and 4, 6-diamidino-2-phenylindole (DAPI). Images were taken by a fluorescence microscope (Leica, Wetzlar, Germany), and the number of EdU-positive cells was counted.
Flow Cytometry Assay
Cell apoptosis was assessed using the Annexin V-FITC Apoptosis Detection Kit (Beyotime, Shanghai, China). Briefly, the experimental cells were collected at 48 h post-transfection and suspended in phosphate-buffered saline (PBS). Then, cells (5 × 104) were used to incubate with 195 µL Annexin V-FITC binding buffer, followed by 5 µL Annexin V-FITC and 10 µL propidium iodide (PI). The apoptotic cells were sorted and analyzed using a FACScan (Beckman Coulter, Fullerton, CA, USA), and then processed using the FlowJo software.
Enzyme-linked Immunosorbent Assay (ELISA)
Human IL-8 ELISA Kit (ab214030; Abcam, Cambridge, MA, USA), Human IL-1 beta (Interleukin 1 beta) ELISA kit (ab214025; Abcam), and Human TNF alpha ELISA kit (ab181421; Abcam) were utilized to examine the release of proinflammatory factors in cell culture medium, including IL-8, IL-1β and TNF-α, according to the protocols. Cells with transfections were cultured for 48 h, and the supernatant cell culture medium was collected for detection.
Detection of Malondialdehyde (MDA) and Superoxide Dismutase (SOD)
The production of MDA and SOD was examined using MDA assay kit (Sigma-Aldrich) and SOD assay kit (Sigma-Aldrich) to assess oxidative stress. The procedures were implemented following the protocols.
Total Cholesterol (T-CHO) Level Detection and Cholesterol Efflux Level Analysis
T-CHO level in THP-1 macrophages was determined using a Total cholesterol quantitation kit (Sigma-Aldrich) by a colorimetric or fluorometric test according to the manufacturer’s instructions.
THP-1 cells were incubated in RPMI 1640 containing 0.5 µCi/mL [3 H] cholesterol (radiolabeled cholesterol; PerkinElmer, Waltham, MA, USA) for 24 h. Then, cells were washed with PBS and cultured in RPMI 1640 containing 0.1% BSA to equilibrate [3 H] cholesterol in all cell pools. Subsequently, cells were washed with PBS again and incubated in RPMI 1640 containing 0.1% BSA and 25 µg/mL apolipoprotein A-I (apoA-I; Sigma-Aldrich) for 6 h. The efflux medium was next collected to remove cell debris by centrifuging. Cells were treated with scintillation lysis, and the liquid scintillation counter was applied to assess medium and cell-associated [3 H] cholesterol.
Bioinformatics Analysis
Bioinformatics tools, such as circular RNA interactome (https://circinteractome.nia.nih.gov/) and starbase (http://starbase.sysu.edu.cn/), were used to analyze the putative targets of circ-C16orf62 and miR-377.
Dual-luciferase Reporter Assay
According to the binding sites between miR-377 and circ-C16orf62 or RAB22A, luciferase reporter plasmids were constructed using pmirGLO vector (Promega, Madison, WI, USA), including WT-circ-C16orf62 (circ-C16orf62 fragment harboring miR-377 binding sites), MUT-circ-C16orf62 (circ-C16orf62 fragment harboring mutated miR-377 binding sites), RAB22A 3’UTR-WT (RAB22A 3’UTR fragment harboring miR-377 binding site)and RAB22A 3’UTR-MUT (RAB22A 3’UTR fragment harboring mutated miR-377 binding site). Any one of these reporter plasmids was transfected with miR-377 or miR-NC into THP-1 cells. At 48 h post-transfection, luciferase activity in cells was checked using the dual-luciferase reporter assay system (Promega).
RNA Immunoprecipitation (RIP) Assay
RIP assay was implemented using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA). THP-1 cells were lysed using lysis buffer, and cell lysates were incubated with the beads coated with human Argonaute 2 antibody (anti-Ago2; Millipore) or mouse Immunoglobulin G antibody (anti-IgG; Millipore). The RNA compounds bound to conjugated beads were eluted for RT-qPCR analysis.
Western Blot Assay
The protein was extracted using RIPA reagent (Solarbio) and then separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Polyvinylidene fluoride (PVDF) membranes (Bio-Rad, Hercules, CA, USA) stained with protein blots were blocked by 5% skim milk and then incubated with the primary antibodies (all purchased from Abcam) against RAB22A (anti-RAB22A; ab138505; dilution: 1/3000), total caspase-3 (anti-caspase-3; ab32150; dilution: 1/1000), cleaved-caspase-3 (anti-cleaved-caspase-3; ab2302; dilution: 1/200), Bcl-2 (anti-Bcl-2; ab32124; dilution: 1/1000) or Bax (anti-Bax; ab182733; dilution: 1/2000), followed by the incubation with the secondary antibody (ab205718; Abcam; dilution: 1/5000). The protein visualized by enhanced chemiluminescent (ECL) Detection Reagents (Beyotime) were quantified using Image J software (NIH, Bethesda, MA, USA).
Statistical Analysis
Three independent experiments were included in this study to obtain the data in each figure. All data analyzed by GraphPad Prism 5.0 (GraphPad Software, San Diego, CA, USA) were expressed as the mean ± standard deviation (SD). Student’s t-test was used to analyze the significance between two groups, and multiple comparisons among multiple groups were performed using analysis of variance (ANOVA). The correlation between miR-377 expression and circ-C16orf62 expression or RAB22A expression in serum samples was determined using Pearson correlation analysis. It was considered statistically significant when p-value < 0.05.
Results
Circ-C16orf62 was Overexpressed in AS Serum Samples and ox-LDL-treated THP-1 Cells
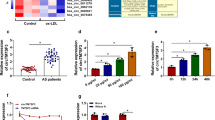
At first, we explored the expression levels of circ-C16orf62 in serum samples and ox-LDL-treated THP-1 cells. Noticeably, the expression of circ-C16orf62 in serum samples from AS patients (n = 29) was higher than that in serum samples from normal participants (n = 18) (Fig. 1A). As well, the expression of circ-C16orf62 was significantly increased in ox-LDL-administered THP-1 cells compared to Control in a dose-dependent manner, also in a time-dependent manner (Fig. 1B C). Further expression analysis showed that circ-C16orf62 was chiefly located in the cytoplasm but not in the nucleus of THP-1 cells (Fig. 1D). RNase R test manifested that circ-C16orf62, compared to linear C16orf62, was more resistant to RNase R digestion (Fig. 1E). We investigated the expression and characteristics of circ-C16orf62, showing that circ-C16orf62 might be involved in AS.
circ-C16orf62 was highly expressed in AS serum samples and ox-LDL-treated THP-1 cells. (A) RT-qPCR was performed to detect the expression of circ-C16orf62 in serum samples from AS patients (n = 29) and normal participants (n = 18). (B) THP-1 cells were treated with different concentrations of ox-LDL (Control, 25, 50 and 100 µg/mL) for 48 h, and the expression of circ-C16orf62 was detected using RT-qPCR. (C) THP-1 cells were treated with 50 µg/mL ox-LDL for different time (0, 12, 24 and 48 h), and the expression of circ-C16orf62 was detected using RT-qPCR. (D) Cytoplasmic RNA and nuclear RNA were isolated to detect the distribution of circ-C16orf62 using RT-qPCR. (E) The effect of RNase R on the expression of circ-C16orf62 and linear C16orf62 was determined by RT-qPCR. **P < 0.01, ***P < 0.001, and ****P < 0.0001
Ox-LDL Induced Apoptosis, Inflammation, Oxidative Stress and Cholesterol Accumulation in THP-1 Cells
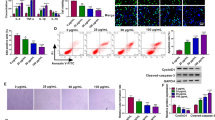
THP-1 cells were treated with different concentrations of ox-LDL. CCK-8 assay showed that cell viability was notably impaired in these treated cells in a dose-dependent manner (Fig. 2A). Flow cytometry assay introduced that the apoptotic rate of these treated cells was rapidly increased in a dose-dependent manner (Fig. 2B). EdU assay showed that ox-LDL treatment significantly reduced the number of EdU-positive cells in a dose-dependent manner, suggesting that ox-LDL inhibited cell proliferation (Fig. 2C). Additionally, the protein levels of cleaved-caspase-3 and Bax were strikingly enhanced, while the protein level of Bcl-2 was strikingly reduced in THP-1 cells treated with ox-LDL (Fig. 2D and E). ELISA led us to realize that ox-LDL enhanced the releases of proinflammatory factors, including IL-8, IL-1β and TNF-α, in a dose-dependent manner (Fig. 2F). In addition, the production of MDA and SOD was detected to assess oxidative stress. The level of MDA was markedly increased, while the level of SOD was lessened in ox-LDL-treated THP-1 cells in a dose-dependent manner (Fig. 2G H). Moreover, we monitored the levels of T-CHO and cholesterol efflux, and the data showed that T-CHO levels were markedly reinforced in ox-LDL-treated THP-1 cells in a dose-dependent manner, while cholesterol efflux levels were declined (Fig. 2I J). ABCA1, also known as the cholesterol efflux regulatory protein (CERP), is a major regulator of cellular cholesterol and phospholipid homeostasis [17]. Herein, we monitored that ABCA1 protein expression was gradually weakened in ox-LDL-treated THP-1 cells in a dose-dependent manner (Fig. 2K). These data suggested that ox-LDL induced a range of abnormal cellular functions and injuries, including apoptosis, inflammation, oxidative stress and cholesterol accumulation.
Ox-LDL induced THP-1 cell apoptosis, inflammation, oxidative stress and cholesterol accumulation. THP-1 cells were treated with different concentrations of ox-LDL (Control, 25, 50 and 100 µg/mL), and (A) cell viability was detected by CCK-8 assay. (B) Cell apoptosis was monitored by flow cytometry assay. (C) Cell proliferative capacity was determined by EdU assay. (D and E) The expression of cleaved-caspase-3, Bax and Bcl-2 was detected by western blot. (F) The levels of IL-8, IL-1β and TNF-α were examined using ELISA kits. (G and H) The levels of MDA and SOD production were checked using detection kits. (I) The levels of T-CHO were examined using a total cholesterol test kit. (J) The levels of cholesterol efflux were determined using a liquid scintillation counter. (K) The protein level of ABCA1 was examined by western blot. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001
Circ-C16orf62 Knockdown Relieved ox-LDL-induced Apoptosis, Inflammation, Oxidative Stress and Cholesterol Accumulation
Next, we addressed the function of circ-C16orf62 in ox-LDL-treated THP-1 cells. As shown in Fig. 3A, circ-C16orf62 expression was largely repressed in ox-LDL-treated THP-1 cells transfected with si-circ-C16orf62 compared to si-NC, suggesting the available efficiency of si-circ-C16orf62. In function, ox-LDL-blocked cell viability was rescued by combined circ-C16orf62 knockdown (Fig. 3B), while ox-LDL-induced cell apoptotic rate was largely inhibited by circ-C16orf62 knockdown (Fig. 3C). EdU assay indicated that ox-LDL-inhibited cell proliferation was recovered by circ-C16orf62 knockdown (Fig. 3D). The protein levels of cleaved-caspase-3 and Bax were largely repressed, while the protein level of Bcl-2 was largely restored in ox-LDL-treated THP-1 cells after circ-C16orf62 knockdown (Fig. 3E F). Besides, the transfection of si-circ-C16orf62 largely weakened the releases of IL-8, IL-1β and TNF-α that were promoted by ox-LDL treatment (Fig. 3G). In addition, circ-C16orf62 knockdown alleviated ox-LDL-induced oxidative stress because si-circ-C16orf62 transfection weakened the level of MDA and reinforced the level of SOD (Fig. 3H and 3I). Moreover, the level of T-CHO accumulated by ox-LDL was partly reduced by circ-C16orf62 knockdown, and the level of cholesterol efflux blocked by ox-LDL was recovered by circ-C16orf62 knockdown (Fig. 3J and 3K), suggesting that circ-C16orf62 knockdown inhibited cholesterol accumulation. ABCA1 protein expression impaired by ox-LDL was largely restored by circ-C16orf62 knockdown in THP-1 cells (Fig. 3L). In short, we characterized that circ-C16orf62 deficiency could alleviate ox-LDL-induced THP-1 cell dysfunctions and injuries.
circ-C16orf62 knockdown alleviated ox-LDL induced THP-1 cell apoptosis, inflammation, oxidative stress and cholesterol accumulation. THP-1 cells treated with ox-LDL were transfected with si-circ-C16orf62 or si-NC. (A) The expression of circ-C16orf62 in these cells was measured by RT-qPCR. (B) Cell viability was assessed by CCK-8 assay. (C) Cell apoptosis was assessed by flow cytometry assay. (D) Cell proliferation was detected by EdU assay. (E and F) The protein levels of cleaved-caspase-3, Bax and Bcl-2 were quantified by western blot. (G) The releases of IL-8, IL-1β and TNF-α were checked using ELISA kits. (H and I) The levels of MDA and SOD in these experimental cells were checked using detection kits. (J and K) The levels of T-CHO were examined using a total cholesterol test kit, and the levels of cholesterol efflux were determined using a liquid scintillation counter. (L) The protein level of ABCA1 was examined by western blot. **P < 0.01, ***P < 0.001, and ****P < 0.0001
Circ-C16orf62 Bound to miR-377, and miR-377 was Downregulated in AS Serum and ox-LDL-treated THP-1 Cells
Our previous data suggested that circ-C16orf62 was mainly distributed in the cytoplasm, hinting that circ-C16orf62 might regulate miRNA expression through ceRNA mechanism. From the prediction of circular RNA interactome, miR-377 was a putative target of circ-C16orf62, with special binding sites between their sequences (Fig. 4A). The putative relationship between miR-377 and circ-C16orf62 was then validated by dual-luciferase reporter assay and RIP assay. The data presented that only the cotransfection of miR-377 and WT-circ-C16orf62 could significantly reduce luciferase activity compared to other groups (Fig. 4B), and both circ-C16orf62 and miR-377 bound to Ago2 antibody but not lgG antibody could be enriched in the RIP assay (Fig. 4C). The expression of miR-377 was remarkably downregulated in serum samples from AS patients (n = 29) compared with that from normal participants (n = 18) (Fig. 4D), and miR-377 expression was also decreased in ox-LDL-administered THP-1 cells in a dose-dependent manner, also in a time-dependent manner (Fig. 4E F). In AS serum samples (n = 29), miR-377 expression was negatively correlated with circ-C16orf62 expression (Fig. 4G). The expression of circ-C16orf62 was elevated in ox-LDL-treated THP-1 cells, and circ-C16orf62 expression was further increased by the transfection of circ-C16orf62 compared to pcDNA (Fig. 4H). In ox-LDL-treated THP-1 cells, si-circ-C16orf62 transfection significantly enhanced the level of miR-377, and circ-C16orf62 transfection significantly impaired the level of miR-377 (Fig. 4I). All data verified that miR-377 was a target of circ-C16orf62.
MiR-377 was a target of circ-C16orf62. (A) MiR-377 was predicted as a target of circ-C16orf62 by circular RNA interactome. (B and C) The putative relationship between miR-377 and circ-C16orf62 was verified by dual-luciferase reporter assay and RIP assay. (D) RT-qPCR was performed to detect the expression of miR-377 in serum samples from AS patients (n = 29) and normal participants (n = 18). (E) The expression of circ-C16orf62 was detected using RT-qPCR in THP-1 cells treated with different concentrations of ox-LDL (Control, 25, 50 and 100 µg/mL) for 48 h. (F) The expression of circ-C16orf62 was detected using RT-qPCR in THP-1 cells treated with 50 µg/mL ox-LDL for different time (0, 12, 24 and 48 h). (G) The correlation between miR-377 expression and circ-C16orf62 expression in AS serum samples was analyzed by Pearson correlation analysis. (H) The efficiency of circ-C16orf62 overexpression in ox-LDL-treated THP-1 cells was checked by RT-qPCR. (I) The expression of miR-377 in ox-LDL-treated THP-1 cells with circ-C16orf62 knockdown or overexpression was measured by RT-qPCR. **P < 0.01, ***P < 0.001, and ****P < 0.0001
Circ-C16orf62 Knockdown Relieved ox-LDL-induced Apoptosis, Inflammation, Oxidative Stress and Cholesterol Accumulation by Increasing miR-377 Level
The expression of miR-377 was strikingly elevated in ox-LDL-treated THP-1 cells transfected with si-circ-C16orf62, while miR-377 expression was decreased in ox-LDL-treated THP-1 cells cotransfected with si-circ-C16orf62 + anti-miR-377 (Fig. 5A). In function, circ-C16orf62 knockdown promoted cell viability impaired by ox-LDL, while combined miR-377 inhibition largely reduced cell viability (Fig. 5B). Besides, in ox-LDL-treated THP-1 cells, the reintroduction of anti-miR-377 strengthened the apoptotic rate that was blocked by si-circ-C16orf62 alone (Fig. 5C). EdU assay showed that circ-C16orf62 knockdown-rescued cell proliferative capacity was partially repressed by miR-377 inhibition (Fig. 5D). The protein levels of cleaved-caspase-3 and Bax were inhibited by circ-C16orf62 knockdown but restored by miR-377 downregulation in ox-LDL-treated THP-1 cells, while the expression pattern of Bcl-2 was opposite to cleaved-caspase-3 and Bax (Fig. 5E). In addition, ox-LDL-induced the releases of IL-8, IL-1β and TNF-α were alleviated by circ-C16orf62 knockdown, while further miR-377 inhibition restored the releases of these factors (Fig. 5F). Moreover, the reintroduction of anti-miR-377 partly enhanced the level of MDA and lessened the level of SOD compared to single si-circ-C16orf62 transfection in ox-LDL-treated THP-1 cells (Fig. 5G H). The level of T-CHO repressed in ox-LDL-treated cells with si-circ-C16orf62 transfection alone was largely recovered in ox-LDL-treated cells with si-circ-C16orf62 + anti-miR-377 cotransfection, while the level of cholesterol efflux was opposite to the level of T-CHO (Fig. 5I J). Circ-C16orf62 deficiency recovered ABCA1 level that was repressed by ox-LDL, while further miR-377 inhibition largely reduced ABCA1 level (Fig. 5K). We concluded from these data that circ-C16orf62 knockdown alleviated ox-LDL-induced THP-1 cell dysfunctions and injuries by enriching the level of miR-377.
circ-C16orf62 knockdown alleviated ox-LDL induced THP-1 cell apoptosis, inflammation, oxidative stress and cholesterol accumulation by upregulating miR-377. THP-1 cells treated with ox-LDL were transfected with si-circ-C16orf62, si-NC, si-circ-C16orf62 + anti-miR-377 or si-circ-C16orf62 + anti-miR-NC. (A) The expression of miR-377 in these experimental cells was detected by RT-qPCR. (B) Cell viability was assessed by CCK-8 assay. (C) Cell apoptosis was monitored by flow cytometry assay. (D) Cell proliferation was assessed by EdU assay. (E) The expression of cleaved-caspase-3, Bax and Bcl-2 was detected by western blot. (F) The releases of IL-8, IL-1β and TNF-α in these experimental cells were checked using ELISA kits. (G and H) The levels of MDA and SOD in these experimental cells were checked using detection kits. (I) The levels of T-CHO in these experimental cells were examined using a total cholesterol test kit. (J) The levels of cholesterol efflux in these experimental cells were determined using a liquid scintillation counter. (K) The protein level of ABCA1 was examined by western blot. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001
MiR-377 Combined to RAB22A 3’UTR
RAB22A was predicted to be a target of miR-377 by starbase, and miR-377 bound to RAB22A 3’UTR through a special binding site (Fig. 6A). Next, dual-luciferase reporter assay showed that the luciferase activity was strikingly reduced in THP-1 cells cotransfected with miR-377 and RAB22A 3’UTR-WT but not RAB22A 3’UTR-MUT (Fig. 6B). Besides, both miR-377 and RAB22A could be abundantly detected in the compounds from the Ago2-RIP group compared to IgG-RIP group (Fig. 6C). The data demonstrated that RAB22A was a target of miR-377. The mRNA level and protein level of RAB22A were both significantly elevated in the serum samples from AS patients (n = 29) compared with that from normal participants (n = 18) (Fig. 6D and E). The mRNA level of RAB22A was negatively correlated with miR-377 level in the serum samples from AS patients (n = 29) (Fig. 6F). Likewise, the protein level of RAB22A was also remarkably promoted in ox-LDL-treated THP-1 cells in a dose-dependent manner (Fig. 6G), also in a time-dependent manner (Fig. 6H). Furthermore, miR-377 expression was enhanced in ox-LDL-treated THP-1 cells transfected with miR-377 but largely reduced in ox-LDL-treated THP-1 cells transfected with anti-miR-377 (Fig. 6I), and on the contrary, RAB22A expression was strikingly weakened in ox-LDL-treated THP-1 cells transfected with miR-377 but largely reinforced in ox-LDL-treated THP-1 cells transfected with anti-miR-377 (Fig. 6J), suggesting that miR-377 negatively regulated RAB22A.
RAB22A was a target of miR-377. (A) RAB22A was predicted as a target of miR-377 by starbase. (B and C) The putative interaction between RAB22A and miR-377 was verified by dual-luciferase reporter assay and RIP assay. (D and E) RT-qPCR and western blot were performed to detect the expression of RAB22A in serum samples from AS patients (n = 29) and normal participants (n = 18). (F) The correlation between RAB22A mRNA level and miR-377 level in serum samples from AS patients (n = 29) was analyzed by Pearson correlation analysis. (G) The expression of RAB22A was detected using western blot in THP-1 cells treated with different concentrations of ox-LDL (Control, 25, 50 and 100 µg/mL) for 48 h. (H) The expression of RAB22A was detected using western blot in THP-1 cells treated with 50 µg/mL ox-LDL for different time (0, 12, 24 and 48 h). (I) The efficiency of miR-377 mimic and inhibitor in ox-LDL-treated THP-1 cells was checked by RT-qPCR. (J) The expression of RAB22A in ox-LDL-treated THP-1 cells with miR-377 overexpression or inhibition was measured by western blot. **P < 0.01, ***P < 0.001, and ****P < 0.0001
MiR-377 Overexpression Alleviated ox-LDL-induced Apoptosis, Inflammation, Oxidative Stress and Cholesterol Accumulation by Suppressing RAB22A
THP-1 cells treated with ox-LDL were transfected with miR-377, miR-NC, miR-377 + RAB22A or miR-377 + pcDNA to monitor whether miR-377 could suppress the function of RAB22A. The protein level of RAB22A was notably declined in ox-LDL-treated cells transfected with miR-377 compared to miR-NC, while the protein level of RAB22A was largely recovered in ox-LDL-treated cells transfected with miR-377 + RAB22A compared to miR-377 + pcDNA (Fig. 7A). In function, ox-LDL-suppressed cell viability was recovered by single miR-377 transfection, while miR-377 + RAB22A cotransfection largely impaired cell viability (Fig. 7B). MiR-377 overexpression suppressed ox-LDL-induced cell apoptosis, while RAB22A reintroduction recovered the apoptotic rate of THP-1 cells (Fig. 7C). EdU assay displayed that ox-LDL-inhibited cell proliferation was recovered by miR-377 restoration but repressed by the reintroduction of RAB22A (Fig. 7D). The protein levels of cleaved-caspase-3 and Bax were reduced in ox-LDL-treated THP-1 cells transfected with miR-377 alone but largely increased in ox-LDL-treated THP-1 cells transfected with miR-377 + RAB22A, while the level of Bcl-2 was recovered by alone miR-377 transfection but largely repressed by miR-377 + RAB22A transfection in ox-LDL-treated THP-1 cells (Fig. 7E). The releases of IL-8, IL-1β and TNF-α were markedly suppressed in ox-LDL-treated THP-1 cells transfected with miR-377 but partly restored in ox-LDL-treated THP-1 cells cotransfected with miR-377 + RAB22A (Fig. 7F). MiR-377 restoration decreased the level of MDA and increased the level of SOD in ox-LDL-treated THP-1 cells, while RAB22A reintroduction recovered the level of MDA and diminished the level of SOD (Fig. 7G H), suggesting RAB22A overexpression recovered oxidative stress inhibited by miR-377. Additionally, the level of T-CHO was impaired, while the level of cholesterol efflux was elevated in ox-LDL-treated cells transfected with miR-377. However, RAB22A reintroduction reversed their levels (Fig. 7I J). MiR-377 enrichment increased ABCA1 protein level that was repressed by ox-LDL, while further RAB22A overexpression largely impaired ABCA1 expression (Fig. 7K). In short, we concluded from these results that miR-377 overexpression alleviated ox-LDL-induced THP-1 cell dysfunctions and injuries by sequestering RAB22A.
MiR-377 overexpression alleviated ox-LDL-induced THP-1 cell apoptosis, inflammation, oxidative stress and cholesterol accumulation by mediating RAB22A. THP-1 cells treated with ox-LDL were transfected with miR-377, miR-NC, miR-377 + RAB22A or miR-377 + pcDNA. (A) The expression of RAB22A in these transfected cells was detected by western blot. (B) Cell viability in these indicated cells was assessed by CCK-8 assay. (C) Cell apoptosis was monitored by flow cytometry assay. (D) Cell proliferation was assessed by EdU assay. (E) The expression of cleaved-caspase-3, Bax and Bcl-2 was detected by western blot. (F) The releases of IL-8, IL-1β and TNF-α in these indicated cells were evaluated using ELISA kits. (G and H) The levels of MDA and SOD in these indicated cells were checked using detection kits. (I) The levels of T-CHO in these indicated cells were examined using a total cholesterol test kit. (J) The levels of cholesterol efflux in these indicated cells were determined using a liquid scintillation counter. (K) The protein level of ABCA1 was examined by western blot. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001
Circ-C16orf62 Increased the Level of RAB22A by Targeting miR-377, thus Promoting AS Progression
In ox-LDL-treated THP-1 cells transfected with si-circ-C16orf62, we found the expression of RAB22A was largely declined compared to si-NC, while the expression of RAB22A was largely recovered in ox-LDL-treated THP-1 cells cotransfected with si-circ-C16orf62 + anti-miR-377 compared to si-circ-C16orf62 + anti-miR-NC at both mRNA and protein levels (Fig. 8A and 8B), suggesting that circ-C16orf62 positively regulated RAB22A expression by competitively targeting miR-377.
circ-C16orf62 was involved in AS progression by upregulating RAB22A via targeting miR-377. (A and B) The mRNA level and protein level of RAB22A in ox-LDL-treated THP-1 cells transfected with si-circ-C16orf62, si-NC, si-circ-C16orf62 + anti-miR-377 or si-circ-C16orf62 + anti-miR-NC were measured by RT-qPCR and western blot. **P < 0.01, ***P < 0.001, and ****P < 0.0001
Circ-C16orf62 Regulated the miR-377/RAB22A axis to Mediate NF-κB Signaling
Regarding the key regulatory effect of NF-κB on inflammatory cytokine expression, we investigated the activity of NF-κB signaling affected by circ-C16orf62 network. As depicted in Fig. 9A, the ratio of p-P65/P65 and p-IκBα/IκBα was considerably enhanced in ox-LDL-administered THP-1 cells. However, circ-C16orf62 downregulation effectively recovered the ratio of p-P65/P65 and p-IκBα/IκBα in ox-LDL-administered THP-1 cells. Further miR-377 inhibition or RAB22A overexpression partly reversed the effect of circ-C16orf62 downregulation, thus restoring the ratio of p-P65/P65 and p-IκBα/IκBα (Fig. 9B). Overall, circ-C16orf62 regulated the miR-377/RAB22A axis to mediate NF-κB signaling.
Circ-C16orf62 regulated the miR-377/RAB22A axis to mediate NF-κB signaling. (A) The levels of p-P65/P65 and p-IκBα/IκBα were quantified by western blot in ox-LDL-treated THP-1 cells. (B) The levels of p-P65/P65 and p-IκBα/IκBα were quantified by western blot in ox-LDL-treated THP-1 cells after si-circ-C16orf62, si-circ-C16orf62 + anti-miR-377 or si-circ-C16orf62 + RAB22A transfection. ***P < 0.001, and ****P < 0.0001
Discussion
Macrophages play a vital role in the development of AS and the formation and rupture of arterial plaques [18]. Restricted cholesterol efflux accelerates cholesterol accumulation and thus triggers the formation of foam cells, which is an early event in the development of AS [19]. Previous studies reported that ox-LDL treatment resulted in the promotion of macrophage apoptosis, inflammation and cholesterol accumulation [8, 9]. Consistently, in our study, we found that ox-LDL induced THP-1 cell apoptosis, stimulated the releases of inflammatory factors and promoted oxidative stress and cholesterol accumulation, suggesting that ox-LDL induced THP-1 cell injury and dysfunction.
Our data showed that circ-C16orf62 was highly expressed in serum samples from AS patients, as well as in ox-LDL-treated THP-1 macrophages, which was consistent with the data from a previous study [13], hinting that circ-C16orf62 might play crucial effects in AS. It was previously recorded that high expression of circ-C16orf62 was closed with unfavorable prognosis of non-small cell lung cancer (NSCLC), and circ-C16orf62 functioned as an oncogene to promote NSCLC progression through the miRNA/mRNA network [20]. Recently, silencing circ-C16orf62 was reported to inhibit ox-LDL-induced endothelial cell apoptosis and inflammation via the inactivation of the NF-κB pathway [21], suggesting that circ-C16orf62 also participated in AS progression by regulating endothelial cell phenotypes. LncRNA MEG3 was upregulated in ox-LDL-treated Raw264.7 macrophages, and MEG3 knockdown alleviated ox-LDL-triggered cell apoptosis and inflammatory responses [8]. Concerning our results, similarly, circ-C16orf62 knockdown ameliorated ox-LDL-induced apoptosis, the releases of proinflammatory factors, oxidative stress and T-CHO level in THP-1 macrophages, which contributed to the inhibition of AS progression, representing that circ-C16orf62 knockdown might be a strategy against AS.
MiR-377 was one of the targets of circ-C16orf62, which was verified by dual-luciferase reporter assay and RIP assay. Previous studies harbored the view that miR-377 inhibited the progression of AS because miR-377 restoration impaired plasma triglyceride levels and suppressed ox-LDL-induced vascular smooth muscle cell proliferation and migration [16, 22, 23], which contributed to the inhibition of plaque rupture. Our study illustrated that miR-377 deficiency reversed the effects of circ-C16orf62 knockdown and thereby recovered ox-LDL-induced apoptosis, inflammation, oxidative stress and cholesterol accumulation in THP-1 cells, while miR-377 overexpression remarkably blocked these injuries. Combined with our data and previous findings, miR-377 was a core mediator against the processes of AS.
MiRNAs are well-known to regulate gene expression and function by binding to 3’UTR of protein-coding transcripts [24]. The analysis from starbase showed that miR-377 could bind to RAB22A 3’UTR, and miR-377 restoration significantly inhibited the expression of RAB22A. Functional analysis revealed that RAB22A overexpression recovered miR-377-inhibited apoptosis, inflammation, oxidative stress and cholesterol accumulation in ox-LDL-treated THP-1 cells. RAB22A involving in ox-LDL-treated THP-1 cells was previously discussed [25]. RAB22A inhibition promoted cholesterol efflux and blocked inflammation in ox-LDL-treated THP-1 cells [25]. Besides, RAB22A was a target of miR-654-3p, involving in the lncRNA ZFAS1-miR-654-3p-RAB22A network in AS [25]. All of these findings supported that RAB22A was a promoter in AS. Importantly, in our study, we established the circ-C16orf62-miR-377-RAB22A network to reveal the new mechanism of circ-C16orf62, miR-377 and RAB22A function in ox-LDL-induced macrophages.
However, there still are limitations in our current study. For example, we mainly checked the role of circ-C16orf62 in ox-LDL-treated THP-1 cells, and the data in corresponding animal models were lacking. Animal model of AS should be constructed in future work to further verify the effects of circ-C16orf62 in vivo.
Overall, circ-C16orf62 was highly expressed in AS serum samples and ox-LDL-treated THP-1 cells. Ox-LDL induced macrophage apoptosis, inflammation, oxidative stress and cholesterol accumulation, which was partly attributed to increased expression of circ-C16orf62. In mechanism, circ-C16orf62 acted as a ceRNA to increase the expression of RAB22A by functioning as a miR-377 sponge, which was implicated in ox-LDL-mediated macrophage phenotypes. This new finding provides additional options for targeted therapy in patients with AS. We speculate that circ-C16orf62 targeted inhibition may be a promising therapeutic strategy for AS through improving macrophage functions.
Data Availability
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
References
Frohlich, J., & Al-Sarraf, A. (2013). Cardiovascular risk and atherosclerosis prevention. Cardiovascular Pathology : The Official Journal Of The Society For Cardiovascular Pathology, 22, 16–18.
Chistiakov, D. A., Bobryshev, Y. V., & Orekhov, A. N. (2016). Macrophage-mediated cholesterol handling in atherosclerosis. Journal Of Cellular And Molecular Medicine, 20, 17–28.
Shah, P., Bajaj, S., Virk, H., Bikkina, M., & Shamoon, F. (2015). Rapid Progression of Coronary Atherosclerosis: A Review. Thrombosis 2015, 634983
Linton, M. F., Babaev, V. R., Huang, J., Linton, E. F., Tao, H., & Yancey, P. G. (2016). Macrophage apoptosis and efferocytosis in the pathogenesis of atherosclerosis. Circulation Journal, 80, 2259–2268.
Wang, G., Li, Y., Liu, Z., Ma, X., Li, M., Lu, Q., et al. (2020). Circular RNA circ_0124644 exacerbates the ox-LDL-induced endothelial injury in human vascular endothelial cells through regulating PAPP-A by acting as a sponge of miR-149-5p. Molecular And Cellular Biochemistry, 471, 51–61.
Yang, L., Yang, F., Zhao, H., Wang, M., & Zhang, Y. (2019). Circular RNA circCHFR facilitates the Proliferation and Migration of Vascular smooth muscle via miR-370/FOXO1/Cyclin D1 pathway. Mol Ther Nucleic Acids, 16, 434–441.
Sun, C., Fu, Y., Gu, X., Xi, X., Peng, X., Wang, C., et al. (2020). Macrophage-enriched lncRNA RAPIA: a novel therapeutic target for atherosclerosis. Arteriosclerosis, Thrombosis, And Vascular Biology, 40, 1464–1478.
Yan, L., Liu, Z., Yin, H., Guo, Z., & Luo, Q. (2019). Silencing of MEG3 inhibited ox-LDL-induced inflammation and apoptosis in macrophages via modulation of the MEG3/miR-204/CDKN2A regulatory axis. Cell Biology International, 43, 409–420.
Zhen, Z., Ren, S., Ji, H., Ding, X., Zou, P., & Lu, J. (2019). The lncRNA DAPK-IT1 regulates cholesterol metabolism and inflammatory response in macrophages and promotes atherogenesis. Biochemical And Biophysical Research Communications, 516, 1234–1241.
Qu, S., Zhong, Y., Shang, R., Zhang, X., Song, W., Kjems, J., et al. (2017). The emerging landscape of circular RNA in life processes. Rna Biology, 14, 992–999.
Li, M., Ding, W., Sun, T., Tariq, M. A., Xu, T., Li, P., et al. (2018). Biogenesis of circular RNAs and their roles in cardiovascular development and pathology. Febs Journal, 285, 220–232.
Kattoor, A. J., Kanuri, S. H., & Mehta, J. L. (2019). Role of Ox-LDL and LOX-1 in atherogenesis. Current Medicinal Chemistry, 26, 1693–1700.
Wang, L., Zheng, Z., Feng, X., Zang, X., Ding, W., Wu, F., et al. (2019). circRNA/lncRNA-miRNA-mRNA network in oxidized, Low-Density, Lipoprotein-Induced Foam cells. Dna And Cell Biology, 38, 1499–1511.
Xiong, D. D., Dang, Y. W., Lin, P., Wen, D. Y., He, R. Q., Luo, D. Z., et al. (2018). A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med, 16, 220.
Cai, X., Lin, L., Zhang, Q., Wu, W., & Su, A. (2020). Bioinformatics analysis of the circRNA-miRNA-mRNA network for non-small cell lung cancer. Journal Of International Medical Research, 48, 300060520929167.
Chen, L. Y., Xia, X. D., Zhao, Z. W., Gong, D., Ma, X. F., Yu, X. H., et al. (2018). MicroRNA-377 inhibits atherosclerosis by regulating triglyceride metabolism through the DNA methyltransferase 1 in apolipoprotein E-Knockout mice. Circulation Journal, 82, 2861–2871.
Yin, K., Liao, D. F., & Tang, C. K. (2010). ATP-binding membrane cassette transporter A1 (ABCA1): a possible link between inflammation and reverse cholesterol transport. Molecular Medicine, 16, 438–449.
Cao, Q., Guo, Z., Du, S., Ling, H., & Song, C. (2020). Circular RNAs in the pathogenesis of atherosclerosis. Life Sciences, 255, 117837.
Yu, X. H., Fu, Y. C., Zhang, D. W., Yin, K., & Tang, C. K. (2013). Foam cells in atherosclerosis. Clinica Chimica Acta, 424, 245–252.
An, J., Shi, H., Zhang, N., & Song, S. (2019). Elevation of circular RNA circ_0003645 forecasts unfavorable prognosis and facilitates cell progression via miR-1179/TMEM14A pathway in non-small cell lung cancer. Biochemical And Biophysical Research Communications, 511, 921–925.
Qin, M., Wang, W., Zhou, H., Wang, X., Wang, F., & Wang, H. (2020). Circular RNA circ_0003645 silencing alleviates inflammation and apoptosis via the NF-kappaB pathway in endothelial cells induced by oxLDL. Gene, 755, 144900.
Guo, Y., Huang, S., Ma, Y., Zhang, J., Wen, Y., Zhou, L., et al. (2019). MiR-377 mediates the expression of syk to attenuate atherosclerosis lesion development in ApoE(-/-) mice. Biomedicine & Pharmacotherapy, 118, 109332.
Wang, H., Wei, Z., Li, H., Guan, Y., Han, Z., Wang, H., et al. (2020). MiR-377-3p inhibits atherosclerosis-associated vascular smooth muscle cell proliferation and migration via targeting neuropilin2. Bioscience Reports, 40, BSR20193425.
O’Brien, J., Hayder, H., Zayed, Y., & Peng, C. (2018). Overview of MicroRNA Biogenesis, Mechanisms of actions, and circulation. Front Endocrinol (Lausanne), 9, 402.
Tang, X., Yin, R., Shi, H., Wang, X., Shen, D., Wang, X. (2020). LncRNA ZFAS1 confers inflammatory responses and reduces cholesterol efflux in atherosclerosis through regulating miR-654-3p-ADAM10/RAB22A axis. Int. J. Cardiol.
Acknowledgements
None.
Funding
There is no funding to report.
Author information
Authors and Affiliations
Contributions
G.S. designed research, X.Y. performed experiments, H.C. analyzed data, and wrote the manuscript. Y.X. contributed the methodology, L.W. edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
This study was approved by the Ethics Committee of the Second Affiliated Hospital of Hainan Medical University (Number: LW2022241).
Consent for Publication
Not applicable.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xuejiao Yin and Hongdan Chen contributed equally to this work
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yin, X., Chen, H., Sun, G. et al. Circ-C16orf62 Regulates Oxidized low-density Lipoprotein-induced Apoptosis, Inflammation, Oxidative Stress and Cholesterol Accumulation of Macrophages via Mediating RAB22A Expression by Targeting miR-377. Appl Biochem Biotechnol 195, 6586–6606 (2023). https://doi.org/10.1007/s12010-023-04320-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-023-04320-4