Abstract
MicroRNAs are reported to be involved in tumor development. This study aims to investigate the biological functions and molecular mechanisms of microRNA-19a-3p in gastric cancer cells. TCGA-based expression analysis and qRT-PCR assay illustrated that microRNA-19a-3p was overexpressed in gastric cancer. MTT and Transwell assays indicated that microRNA-19a-3p could strengthen the proliferation, migration, and invasion of gastric cancer cells. SMOC2 was bioinformatically predicted as the target of microRNA-19a-3p, followed by identified using a dual-luciferase assay. The effects of microRNA-19a-3p/SMOC2 regulatory axis on gastric cancer cells were examined by MTT and Transwell assays as well. Concludingly, this study demonstrated that microRNA-19a-3p could promote the aggressive cell phenotypes of gastric cancer cells by targeting SMOC2.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Previous studies reported that dysregulation of miRNAs was involved in GC occurrence and development. Hu et al. [1] reported that microRNA-532 fostered GC cell migration and invasion via repressing NKD1 and activating the Wnt/β-catenin pathway, providing a novel idea for GC targeted therapy. Wang and his colleagues indicated that the exosomes delivering anti-microRNA-214 to tumor cells could reverse chemotherapy resistance of GC to cisplatin, providing the potential therapeutic options for GC patients with acquired resistance to cisplatin [2]. Some researchers found that exosomes secreted by tumor fibroblasts with microRNA-522 being internalized by GC cells, thus hampering lipid ROS accumulation and iron death in GC cells [3]. Collectively, modulating microRNA expression may contribute to developing novel GC therapeutic strategies. The biofunctions and molecular mechanisms of microRNA-19a-3p have not been fully understood, though some studies pointed out its role in several tumors [4,5,6]. In this study, microRNA-19a-3p, a noticeably increased miRNA in GC cells, was selected for examination.
SMOC2 is a SPARC family member and contains two thyroglobulin-like domains [7, 8]. In papillary carcinoma, the expression of SMOC2 is declined and SMOC2 positivity is closely related to better clinical outcomes [9]. Also, in Chang Lim Hyun’s study, bioinformatics analysis results illustrated that SMOC2 could be applied as an independent biomarker for risk prediction in GC [10]. As above, it could be suspected that SMOC2 may play an important role in GC progress.
Combining the bioinformatics predictions and in vitro cellular/molecular experiments, we believe that microRNA-19a-3p hastens GC malignant progression via targeting SMOC2. Our findings may provide promising evidence for novel GC therapy development.
Materials and Methods
Obtaining Differential Genes in GC
From The Cancer Genome Atlas (TCGA) database, we downloaded expression data of mature microRNA (444 normal samples, 45 cancer samples) and mRNA (373 normal samples, 32 cancer samples). microRNA-19a-3p expression was analyzed based on the downloaded data, using the R package edgeR mRNA to identify differential mRNAs (|logFC|> 2, padj < 0.05).
The microRNA Target Prediction
After microRNA was identified, TargetScan (http://www.targetscan.org/vert_72/), mirDIP (http://ophid.utoronto.ca/mirDIP), miRWalk (http:// miRWalk. Umm. Uni—Heidelberg. DE/), miRDB (http://mirdb.org/), and starBase (http://starbase.sysu.edu.cn/) databases were utilized for the miRNA target prediction. The correlation between microRNA and mRNA was calculated. The mRNA with the highest inverse correlation was deemed as the gene of interest.
Cell Cultivation and Transfection
The normal gastric epithelial cell line GES-1 and 4 GC cell lines (SGC-7901, HGC-27, MKN45 and BGC-823) were purchased from the Chinese Academy of Sciences (Shanghai) Cell Bank. The cells were maintained in RPMI-1640 medium (Invitrogen, USA) containing 10% FBS under routine conditions. The sequence fragments involved in cell transfection were microRNA-19a-3p-mimic (microRNA-mimic) and the control NC-mimic. The plasmids used were oe-SMOC2 and the control plasmid oe-NC. Lipofectamine 2000 (Invitrogen, USA) was utilized for cell transfection referring to instructions.
qRT-PCR
Total RNA extraction was done with QIAzol (Qiagen, Germany) and reverse transcription was done with Expand™ Reverse Transcriptase (Sigma-Aldrich, USA). qPCR amplification was completed using microRNA-specific forward primers, universal reverse primers and TaqMan probes. GAPDH and U6 genes were applied as internal reference genes for normalization of mRNA and microRNA, respectively. The relative expression software tool (REST®) was utilized to measure the fold change of gene expression. The primers used in this study are depicted in Table 1.
Western Blot
Cells were lysed with Pierce IP Lysis Buffer (ThermoFisher), and the protein concentration was determined by Micro BCA Protein Detection Kit (ThermoFisher). The proteins were electrophoresed by SDS-PAGE and transferred to a PVDF membrane which was blocked by 5% BSA for 2 h. The primary antibodies, anti-SMOC2 (1 μg/mL, ab56088) and anti-GAPDH (1:10000, ab181602), purchased from Abcam, UK, were maintained with the membrane at 4 °C overnight. Subsequently, the membrane was kept with an enzyme-labeled secondary antibody for 2 h. Next, an ECL kit was employed to detect the signals. ImageJ was utilized to quantify protein expression.
MTT
MTT was applied to assess the impact of microRNA-19a-3p-mimic on proliferative ability of GC cells. Ten thousand cells per well were seeded in 96-well plates, and then, microRNA-19a-3p mimics were transfected into the seeded cells and cultured to about 80% confluency. After 48 h, the cell proliferation was assayed through MTT assay kit (Sigma, USA). The experiment was repeated 3 times.
Transwell
For invasion assay, cells were seeded in the coated substrate of the upper chamber and cultured with serum-free medium. Ten percent fetal calf serum (FCS) and medium were dropped into the lower chamber. After 24 h, the invaded cells were fixed and then stained with crystal violet. Five fields were randomly selected to count cells under the microscope. The migration assay was conducted under the same procedures with exception that the upper chamber was not coated with Matrigel. Each experiment was done 3 times.
Dual-Luciferase Assay
Cells were cultured in 96-well plates and transfected with Lipofectamine 2000. At 48 h after transfection, GC cells were collected, and the luciferase activity was assayed with dual-luciferase reporter kit (Promega, USA). The experiment was repeated 3 times.
Statistical Analysis
The SPSS16.0 statistical software (Scarborough, Canada) was utilized for data analysis. All data were subjected to the F test before analysis and were considered to meet Gaussian distribution, and thus, the MEAN ± SD form of statistics was utilized. For comparisons between multiple groups, we used multiple comparison tests (Bonferroni) to control type I errors. For the comparison between the two groups, we utilized the t test. A two-sided p < 0.05 was considered statistically significant for all analyses.
Results
MicroRNA-19a-3p Expression is Increased in GC
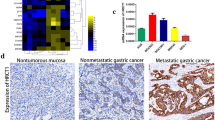
Previous studies showed that microRNA-19a-3p participates in the progression of colorectal cancer and myeloma [6, 11]. Expression analysis of microRNAs based on TCGA-STAD presented that microRNA-19a-3p expression was markedly enhanced in GC tissues compared to the normal tissues (Fig. 1A). The above results were confirmed in the cell lines by qRT-PCR, in which microRNA-19a-3p expression level was prominently upregulated in the GC cell lines (SGC-7901, HGC-27, MKN-45, BGC-823) (Fig. 1B). We selected HGC-27 cell line for the subsequent experiments. To sum up, miR-19a-3p was upregulated at post-transcriptional level in GC.
MicroRNA-19a-3p expression is increased in GC. A Violin plot of microRNA-19a-3p expression level. Red represents tumor group, and green represents normal group. B qRT-PCR detected the expression of microRNA-19a-3p in normal gastric epithelial cell line GES-1 and 4 GC cell lines (SGC-7901, HGC-27, MKN45, and BGC-823); The experiments above were triplicated; *P < 0.05
Overexpression of microRNA-19a-3p Boosts Malignant Phenotypes of GC Cells
To understand the effects of microRNA-19a-3p on the biological functions of the GC cells, microRNA-19a-3p mimic was transfected into HGC-27 cells. qRT-PCR assay demonstrated that microRNA-19a-3p expression was remarkably elevated by microRNA-19a-3p mimic transfection (Fig. 2A). MTT assay was introduced to detect the impact of microRNA-19a-3p on proliferation of the GC cells. As shown in the results, overexpression of microRNA-19a-3p significantly promoted proliferative ability of GC cells (Fig. 2B). The impact of microRNA-19a-3p on migration and invasion of GC cells was assayed by Transwell method. As manifested in the results, in comparison with the control group, HGC-27 cells with microRNA-19a-3p mimic exhibited a significant increase in the migration and invasion (Fig. 2C, D). Collectively, overexpression of microRNA-19a-3p could promote the malignant phenotypes of GC cells.
The influence of microRNA-19a-3p on the proliferation, migration and invasion of GC cells. A Transfection efficiency of miR-mimic in HGC-27 cells. B Detection of HGC-27 cell proliferation by MTT. C, D Transwell detected the number of migration and invasion of HGC-27 cells with miR-mimic or NC-mimic (100 ×); the experiments above were triplicated; *P < 0.05
SMOC2 is Identified as a Direct Target of microRNA-19a-3p
To identify the underlying mechanism of microRNA-19a-3 involved in tumor-promoting effects in GC cells, we employed TargetScan, mirDIP, miRDB, miRWalk, and starBase to predict the targets of microRNA-19a-3p. The predicted results were overlapped with the significantly downregulated genes in GC, determining 3 target genes finally (Fig. 3A). Among them, SMOC2 showed the strongest negative correlation with microRNA-19a-3p (Fig. 3B) at expression level, as well as the expression analysis displayed an evidently low expression of SMOC2 in GC tissues (Fig. 3C). The bioinformatics database predicted that SMOC2 contained the binding site in the 3′-untranslated regions (UTR) of microRNA-19a-3p (Fig. 3D). To validate the targeting relationship between miR-19a-3p and SMOC2, we transfected the luciferase reporter vector containing SMOC2 wild-type (WT) or mutant (MUT) 3′-UTR and microRNA-19a-3p mimic or microRNA-negative control into HGC-27 cells concurrently. As revealed by the results, forced expression of microRNA-19a-3p evidently reduced WT luciferase activity, while MUT luciferase activity was not suppressed (Fig. 3E). To verify whether microRNA-19a-3p could regulate SMOC2 expression, we transfected microRNA-19a-3p mimic or microRNA-negative control into HGC-27 cells, followed by measuring SMOC2 expression levels using qRT-PCR and western blot. As presented, forced expression of microRNA-19a-3p conspicuously constrained mRNA and protein expression of SMOC2 in GC cells (Fig. 3F, G). The above results indicated that SMOC2 was a target of microRNA-19a-3p, and microRNA-19a-3p negatively modulated SMOC2 level in GC cells.
MicroRNA-19a-3p represses SMOC2 level in GC cells. A Venn plot of predicted target gene of microRNA-19a-3p and differential mRNAs. B Correlation analysis between microRNA-19a-3p and target mRNA. C Violin plot of SMOC2 level in the normal and the tumor groups. Red represents the tumor group, and green represents the normal group. D SMOC2 was identified as the target of microRNA-19a-3p through the miRDB database. E Forced expression of microRNA-19a-3p reduced luciferase activity of SMOC2 WT 3′-UTR but had no impact on the MUT 3′-UTR. F, G Transfection of microRNA-19a-3p in HGC-27 hampered SMOC2 mRNA and protein expression; the experiments above were triplicated; *P < 0.05
Overexpression of SMOC2 Reverses the Influence of microRNA-19a-3p on GC Cell Growth
To clarify the mechanism of microRNA-19a-3p/SMOC2 regulatory axis in GC onset and progress, HGC-27 cells were used to construct 4 differently treated cell groups (NC-mimic + oe-NC, miR-mimic + oe-NC, NC-mimic + oe-SMOC2, miR-mimic + oe-SMOC2). Based on the constructed cell lines, qRT-PCR and western blot assays were introduced to examine SMOC2 mRNA and protein expressions in the 4 groups, in which miR-mimic treatment alone could reduce SMOC2 expression; oe-SMOC2 alone could increase SMOC2 expression; the co-treatment of miR-mimic and oe-SMOC2 could attenuate the role of miR-mimic (Fig. 4A). MTT assay was applied to investigate the cell proliferative abilities in the 4 groups, indicating miR-mimic could significantly strengthen the cell proliferation, while overexpression of SMOC2 simultaneously could hamper the role of miR-mimic in cell proliferation (Fig. 4B). A similar trend was observed in the Transwell assays for cell migratory and invasive examination. Both the cell migration and invasion conditions were strengthened in the miR-mimic + oe-NC group, while simultaneous treatment of miR-mimic and oe-SMOC2 could weaken the above phenotypes (Fig. 4C, D). Concludingly, microRNA-19a-3p could boost the malignant phenotypes of GC cells by modulating SMOC2 expression.
MicroRNA-19a-3p regulates tumor-aggressive associated cell phenotypes by targeting SMOC2. A qRT-PCR and western blot assays were introduced to measure the SMOC2 expression at mRNA and protein levels. B Cell proliferation was tested via MTT. C, D Cell migration and cell invasion conditions were examined via Transwell assay; the experiments above were triplicated; *P < 0.05
Discussion
Despite the increasing understanding of genetic and molecular basis of GC development in recent years, the established molecular diagnostic markers are still limited. Thus, studies focused on developing new molecular diagnostic markers for GC are urgently required [12]. In many cancer cases, microRNA expression is dysregulated to serve as a cancer suppressor or oncogene [13, 14]. MicroRNA-19a-3p level is markedly boosted in ovarian cancer [15], hepatocellular carcinoma [16], and colorectal cancer [6] to accelerate their malignant progression. In this paper, we unveiled that microRNA-19a-3p was prominently fostered in GC through bioinformatics analysis. The qRT-PCR results denoted that microRNA-19a-3p was increased in GC cells, and it was speculated that it may participate in GC progression. MicroRNA-19a-3p affects the malignant phenotypes of a variety of cancer cells, so it has attracted more attention in recent years. Sun et al. [17] uncovered that forced expression of microRNA-19a-3p prominently promoted proliferation of hepatocellular carcinoma cells. Upregulation of microRNA-19a-3p inhibited the invasion and migration of prostate cancer cells in vitro [18]. It can be concluded that microRNA-19a-3p has different roles in different cancer types. A previous study showed that forced expression of microRNA-19a-3p hastens GC cell proliferation while suppressing cell apoptosis [19]. Our study also confirmed that forced expression of microRNA-19a-3p significantly hastened proliferation and metastasis of GC cells, which was accompanied by malignant progression of GC cells.
microRNA-19a-3p modulates the malignant phenotypes of tumors through different mechanisms. It regulates the chemosensitivity of osteosarcoma cells through modulating level of PTEN, a tumor suppressor [20]. In rectal cancer, it induces tumor cell apoptosis by targeting FAS [21]. We unraveled that microRNA-19a-3p level in GC cell lines was evidently accelerated while SMOC2 level was repressed. SMOC2 was a direct target of microRNA-19a-3p. This finding was verified via dual-luciferase assay, and it could be negatively modulated by microRNA-19a-3p. A study uncovered that SMOC2 overexpression can constrain malignant progression of hepatocellular carcinoma [22]; hence, SMOC2 may participate in GC onset and progression. Next, we treated HGC-27 cells with NC-mimic and oe-NC, or co-transfected with miR-mimic and oe-NC or miR-mimic and oe-SMOC2 to perform a series of cell function experiments. As displayed in the results, overexpression of SMOC2 evidently reduced promoting impact of overexpression of microRNA-19a-3p on cell malignant phenotypes.
In summary, we proved the oncogenic role of microRNA-19a-3p in GC by targeting SMOC2. We disclosed that microRNA-19a-3p was a promising diagnostic marker and therapeutic target. This is the first study to discuss the function of microRNA-19a-3p/SMOC2 axis in GC progression, which may be a potential therapy for GC management.
Nonetheless, microRNA-19a-3p is not a classic oncogenic factor. In this study, microRNA-19a-3p plays a pivotal role in fostering cell malignant behaviors by mediating SMOC2. Therefore, we believe that microRNA-19a-3p is an oncogenic factor. We have confirmed the above conclusions to a certain extent through in vitro experiments, but considering complex expression and modification of these genes in the human body, these results still need verification for clinical treatment.
Conclusion
This study unveiled the novel role of microRNA-19a-3p in modulating GC cell progression via targeting SOMC2 to transmit signals. It also sheds new light on repressing tumor progression via blocking microRNA-19a-3p level.
Data Availability
The datasets generated and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
References
Hu, S., et al. (2017). miR-532 promoted gastric cancer migration and invasion by targeting NKD1. Life Sciences, 177, 15–19. https://doi.org/10.1016/j.lfs.2017.03.019
Wang, X., et al. (2018). Exosomes serve as nanoparticles to deliver Anti-miR-214 to reverse chemoresistance to cisplatin in gastric cancer. Molecular Therapy, 26, 774–783. https://doi.org/10.1016/j.ymthe.2018.01.001
Zhang, H., et al. (2020). CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Molecular Cancer, 19, 43. https://doi.org/10.1186/s12943-020-01168-8
Dai, W., Zeng, W., & Lee, D. (2021). lncRNA MCM3AP-AS1 inhibits the progression of colorectal cancer via the miR-19a-3p/FOXF2 axis. The Journal of Gene Medicine, 23, e3306. https://doi.org/10.1002/jgm.3306
Cheng, J., et al. (2020). Circulating miR-19a-3p and miR-483-5p as novel diagnostic biomarkers for the early diagnosis of gastric cancer. Medical Science Monitor, 26, e923444. https://doi.org/10.12659/MSM.923444
Yu, F. B., et al. (2020). MiR-19a-3p regulates the Forkhead box F2-mediated Wnt/beta-catenin signaling pathway and affects the biological functions of colorectal cancer cells. World Journal of Gastroenterology, 26, 627–644. https://doi.org/10.3748/wjg.v26.i6.627
Vannahme, C., Gosling, S., Paulsson, M., Maurer, P., & Hartmann, U. (2003). Characterization of SMOC-2, a modular extracellular calcium-binding protein. The Biochemical Journal, 373, 805–814. https://doi.org/10.1042/BJ20030532
Pazin, D. E., & Albrecht, K. H. (2009). Developmental expression of Smoc1 and Smoc2 suggests potential roles in fetal gonad and reproductive tract differentiation. Developmental Dynamics, 238, 2877–2890. https://doi.org/10.1002/dvdy.22124
Kim, H. S., et al. (2020). Downregulation of SMOC2 expression in papillary thyroid carcinoma and its prognostic significance. Science and Reports, 10, 4853. https://doi.org/10.1038/s41598-020-61828-z
Hyun, C. L., et al. (2021). The intestinal stem cell marker SMOC2 is an independent prognostic marker associated with better survival in gastric cancer. Anticancer Research, 41, 3689–3698. https://doi.org/10.21873/anticanres.15160
Zhang, X., et al. (2017). MicroRNA-19a functions as an oncogene by regulating PTEN/AKT/pAKT pathway in myeloma. Leukaemia & Lymphoma, 58, 932–940. https://doi.org/10.1080/10428194.2016.1213827
Tao, J., et al. (2015). miR-27b-3p suppresses cell proliferation through targeting receptor tyrosine kinase like orphan receptor 1 in gastric cancer. Journal of Experimental & Clinical Cancer Research, 34, 139. https://doi.org/10.1186/s13046-015-0253-3
Mou, T., et al. (2019). MiR-345-5p functions as a tumor suppressor in pancreatic cancer by directly targeting CCL8. Biomedicine & Pharmacotherapy, 111, 891–900. https://doi.org/10.1016/j.biopha.2018.12.121
Sun, X., et al. (2018). miR-652 Promotes tumor proliferation and metastasis by targeting RORA in endometrial cancer. Molecular Cancer Research, 16, 1927–1939. https://doi.org/10.1158/1541-7786.MCR-18-0267
Bai, R., et al. (2019). The NF-kappaB-modulated miR-19a-3p enhances malignancy of human ovarian cancer cells through inhibition of IGFBP-3 expression. Molecular Carcinogenesis, 58, 2254–2265. https://doi.org/10.1002/mc.23113
Jiang, X. M., et al. (2018). microRNA-19a-3p promotes tumor metastasis and chemoresistance through the PTEN/Akt pathway in hepatocellular carcinoma. Biomedicine & Pharmacotherapy, 105, 1147–1154. https://doi.org/10.1016/j.biopha.2018.06.097
Sun, H. X., et al. (2020). MicroRNA-19a-3p regulates cell growth through modulation of the PIK3IP1-AKT pathway in hepatocellular carcinoma. Journal of Cancer, 11, 2476–2484. https://doi.org/10.7150/jca.37748
Wa, Q., et al. (2018). Downregulation of miR19a3p promotes invasion, migration and bone metastasis via activating TGFbeta signaling in prostate cancer. Oncology Reports, 39, 81–90. https://doi.org/10.3892/or.2017.6096
Li, X., et al. (2019). Down-regulated lncRNA SLC25A5-AS1 facilitates cell growth and inhibits apoptosis via miR-19a-3p/PTEN/PI3K/AKT signalling pathway in gastric cancer. Journal of Cellular and Molecular Medicine, 23, 2920–2932. https://doi.org/10.1111/jcmm.14200
Zhang, B., Liu, Y., & Zhang, J. (2019). Silencing of miR-19a-3p enhances osteosarcoma cells chemosensitivity by elevating the expression of tumor suppressor PTEN. Oncology Letters, 17, 414–421. https://doi.org/10.3892/ol.2018.9592
Su, Y. F., Zang, Y. F., Wang, Y. H., & Ding, Y. L. (2020). MiR-19-3p Induces tumor cell apoptosis via targeting FAS in rectal cancer cells. Technology in Cancer Research & Treatment, 19, 1533033820917978. https://doi.org/10.1177/1533033820917978
Huang, X. Q., et al. (2017). Overexpression of SMOC2 attenuates the tumorigenicity of hepatocellular carcinoma cells and is associated with a positive postoperative prognosis in human hepatocellular carcinoma. Journal of Cancer, 8, 3812–3827. https://doi.org/10.7150/jca.20775
Author information
Authors and Affiliations
Contributions
HX participated in the design and interpretation of the data and revising the manuscript. GCL and SKZ conceived of the study and participated in its design and interpretation and helped to draft the manuscript. FF performed the statistical analysis and revised the manuscript critically. HX, GCL, SKZ, and FF participated in its design and interpretation and helped to revise the manuscript critically.
All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
No animal/human cell used.
Competing Interests
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Xu, H., Lu, G., Zhou, S. et al. MicroRNA-19a-3p Acts as an Oncogene in Gastric Cancer and Exerts the Effect by Targeting SMOC2. Appl Biochem Biotechnol 194, 3833–3842 (2022). https://doi.org/10.1007/s12010-022-03944-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-022-03944-2