Abstract
Lipases have a characteristic folding pattern of α/β-hydrolase with mostly parallel β-sheets, flanked on both sides by α-helixes in the structure. The active site is formed by a catalytic triad (serine, aspartic/glutamic acid, and histidine), which is highly conserved. In this study, we have used an integrated experimental and computational approach to identify the extremophilic microbial lipases from the saline habitats of the Thar Desert of Rajasthan. Lipase-producing bacteria were screened and a few samples showed significant lipase activity in both quantitative and qualitative experiments. 16S rRNA sequence analysis of the isolate F1 showed that its sequence is quite similar to that of Bacillus licheniformis and Bacillus haynesii, indicating that this isolate belongs to a new subspecies of Bacillus. The isolate F7 showed maximum sequence identity with Bacillus tequilensis strain 10b. The isolate F7 sequence analysis provided a clear testimony that it can be a new strain of Bacillus tequilensis. The F7 lipase exhibited optimal activity at 60 °C and pH 9. Structural modeling of the F7 lipase revealed that it has a highly conserved alpha/beta hydrolase fold at the sequence and structural level except for the N-terminal region. Interestingly, residue Glu128 was different from the template structure and showed the hydrogen bonding between the side chain of Glu128 and side chains of Asn35 and Gln152 amino acids. Besides, this amino acid also showed salt bridge interaction between Glu128--Lys101. These interactions may be assisting in preserving the stability and activity of lipase at high temperatures and in alkaline pH conditions. The information gathered from this investigation will guide in the rational designing of new more potential extremophilic lipase.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lipases (triacylglycerol acyl hydrolase, EC 3.1.1.3) are ubiquitous enzymes that catalyze the cleavage of ester bonds of fats (oils, animal fats, etc.) as well as other synthetic esters in the presence of water, and subsequent release of diacylglycerols, monoacylglycerols, free fatty acids, and glycerol in case of fats and other products in case of synthetic esters [1]. Lipases belong to serine hydrolases with versatility in catalyzing a variety of reactions that include fat hydrolysis, interesterification, transesterification, alcoholysis, acidolysis, and aminolysis [2]. In addition, these enzymes also carry out their catalytic function with greater selectivity viz. chemo-, regio-, and enantioselectivity [3]. Lipases are produced by several microbes, namely bacteria, fungi, and archaea, as well as animals and plants. Lipases are widely used in different industries, e.g., oleochemical, food, dairy, detergent, cosmetic, leather processing, pulp and paper, biodiesel, biopolymer, and pharmaceutical [3, 4]. As a consequence of the high industrial demand of microbial lipases, a number of researchers are interested to discover new potential lipase enzymes [5,6,7]. Many lipase-mediated industrial processes are performed under harsh conditions and most of them are carried out at higher temperatures and alkaline pH conditions. Thus, the requirement of thermostable and alkaliphilic lipases are rapidly increasing in a number of biotechnological industries/processes. Generally, high temperature decreases the risk of microbial contamination in industrial production processes and also enhances the solubility of organic compounds and the reaction rates [8].
Compared with other lipases, the bacterial lipases are more economical and stable and several bacterial species such as Bacillus thermoleovorans, B. stearothermophilus, B. acidocaldarius, and Bacillus sp. have been reported for lipase production and their lipases are commonly used for different applications. There are some reports available that prove the bacterial lipase has optima in both lower and higher ranges of pH (4 to 11) and temperature (30–60 °C) [9,10,11,12]. Bacillus species shows remarkable properties which make them a more potential candidate for industrial purposes. The most common Bacillus species reported for lipase production are Bacillus subtilis, Bacillus pumilus, Bacillus licheniformis, Bacillus coagulans, Bacillus stearothermophilus, and Bacillus alcalophilus. Some bacterial species have been reported as extracellular lipolytic enzyme producers, such as Pseudomonas sp., Pseudomonas aeruginosa, Burkholderia multivorans, Burkholderia cepacia, and Staphylococcus caseolyticus. Some bacterial strains can grow in high saline conditions, they are considered as a halophilic; for example, Staphylococci are one of them which produce an enzyme with remarkable tolerance to high salt concentrations [13].
The objective of this research work was to discover and characterize new extremophilic lipolytic enzymes from unexplored saline habitats of arid regions of Rajasthan (India), specifically the Thar Desert and their biochemical and structural characterization for further use for lipase improvement and applications.
Methods
Sample Collection and Enrichment of Halophilic Cultures
Water and soil samples were collected from different locations of the Great Indian Thar Desert in the summer months (May and June). The five different salt lakes, (i) Lunkaransar (Bikaner district), (ii) Badopal (Hanumangarh district), (iii) Didwana (Nagaur district), (iv) Sambher (Jaipur districts), and (v) Pachpadra (Barmer district), were selected for sample collection. The temperature range of sampling sites was 43–50 °C, the salinity range was 70–95‰ (i.e., part per thousand), and the pH range was 8–10. Samples were collected aseptically, for which hands, trowels, etc. were treated with 70% ethanol before use and autoclaved plastic bottles and plastic bags were used for sample collection. All collected water/soil samples were subjected to enrichment for halophilic cultures using different enrichment media. The inoculated media viz. M-X1, ATCC medium, and nutrient broth (NB) were incubated at 37 °C, 45 °C, and 50 °C for 96 h for the enrichment of the halophilic bacteria [14, 15].
Screening for Lipase Producing Cultures
Qualitative Screening
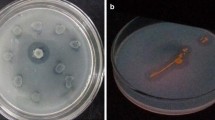
For qualitative screening of halophilic bacteria for lipase production, culture samples from the enriched media were streaked onto the medium MX2 and modified ATCC medium each containing tributyrin (1% v/v) as a substrate for lipase and NaCl in the range of 3–15%. The inoculated plates were incubated at 45 °C for 24 h. The plates were then observed for the hydrolytic zones (in the form of clear halos) around the bacterial colonies. Bacterial cultures which gave hydrolytic zones on TBA plates with ≥ 5% salinities were selected for further investigation.
Quantitative Screening
For quantitative screening, the lipase-positive cultures were grown in modified ATCC medium having 1% (v/v) olive oil. The inoculated medium flasks were incubated at 45 °C and 150 rpm for 24 h. The cell-free broth was evaluated for lipase activity using p-nitrophenyl palmitate lipase assay [16]. The samples showing high lipase activity with p-nitrophenyl palmitate lipase assay were selected for further investigation.
16S rRNA Sequencing
The selected lipase-positive bacterial isolates were identified based on 16S rRNA sequencing. 16S rRNA sequencing was followed by phylogenetic analyses for the selected bacterial cultures to determine their closely related members. The NCBI BLAST program (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to get their closely related ribosomal RNA sequences from the 16S ribosomal RNA sequences database (Bacteria and Archaea). After multiple sequence alignment, a phylogenetic tree was constructed by the neighbor-joining method.
Amplification, Cloning, and Gene Expression
For the development of the lipase expression vector, first, the primers were designed for the selected bacterial lipase according to the corresponding gene sequence available on NCBI. Around 639 bp lipase gene was amplified by polymerase chain reaction (PCR) using the forward primer (lipase: BamH1-F: CTAGGATCCATGAAATTTGTAAAAAGAAGAATC) and the reverse primer (lipase: Xho1-R: GATCTCGAGTTAATTCGTATTCTGGCCCCC). The expression vector used was pET29b–6xHis–TEV having compatible restriction sites for BamH1 and Xho1-R so that the gene could be cloned in the frame. The final lipase gene construct was designated as pET29b–6xHis–TEV-LIP. The expression vector was transformed into BL21 (DE3) expression hosts. The transformed host cells were streaked onto TBA plates having the antibiotics kanamycin (50 μg/mL) and ampicillin (100 μg/mL) for maintenance of the transformed plasmids into the host cells. The streaked plates were incubated at 37 °C overnight. A single colony from the overnight incubated plate was used for preculture raising in LB medium containing antibiotics kanamycin (50 μg/mL) and ampicillin (100 μg/mL). The inoculated LB medium was incubated at 37 °C and 200 rpm until its OD660nm reached 0.6–0.8. For lipase production, 1% of the preculture was inoculated into 50 mL of fresh LB medium (contained in 250-mL capacity Erlenmeyer flask) containing antibiotics kanamycin (50 μg/mL) and ampicillin (100 μg/mL). The inoculated media were incubated at 37 °C and 200 rpm until the biomass reached the early log-phase growth (OD 600nm ~ 0.5–0.6). The lipase production was then induced with IPTG which was added to a final concentration of 0.5 mM (to induce the T7 RNA polymerase-mediated expression of the protein (i.e., the lipase production). After 6 h of post-induction incubation, the culture was harvested by centrifugation at 10000 rpm for 10 min. The harvested cells were lysed by ultrasonication (3 × 30 s with a 2-min interval between each sonication). The lysate was subjected to centrifugation at 13,000 rpm for 30 min at 4 °C and the pellet and supernatant fractions obtained were analyzed by SDS–PAGE.
Lipase Activity Assay
Lipase activity assay was performed according to the colorimetric method of Winkler and Stuckmann (1979). To 2.4 mL of freshly prepared p-NPP solution, 0.1 mL of culture supernatant/filtrate was added. The reaction mixture was incubated at 37 °C (or optimal temperature of enzyme activity) in a water bath shaker for 5 min. The reaction was terminated by the addition of 0.2 mL of 100 mM CaCl2 (fused) solution. The reaction mixture was then centrifuged to clarify the solution. The absorbance of the yellow color was read at 410 nm. The process was repeated with the heat-inactivated culture filtrate (supernatant) samples, which served as control. Lipase activity was calculated from a standard curve of p-nitrophenol prepared in the range of 10-100 μmol/mL. The Michaelis–Menten constant (Km) and maximum velocity for the reaction (Vmax) with pNP palmitate as substrate was calculated by Lineweaver–Burk plot and further kcat and kcat/Km values were calculated.
Biochemical Characterization
Effect of Temperature on Enzyme Activity and Stability
The optimum temperature for lipase activity was determined by performing the enzyme assay at different temperatures viz. 20–80 °C. The heat-deactivated enzyme was used for performing the control experiment. For determination of the thermostability of the lipase, the enzyme was incubated at different temperatures viz. 20–80 °C, for 30 min. The residual lipase activity was determined by performing the lipase assay at the optimal temperature as per the method mentioned above. The enzyme activity determined under standard assay conditions without prior incubation at any temperature was taken as control (100%).
Effect of pH on Enzyme Activity and Stability
The optimum pH for lipase activity was determined by assaying the purified lipase in buffers of different pH (sodium acetate, pH 5.0; sodium phosphate, pH 6.0–8.0; Tris HCl, pH 9.0; glycine NaOH, pH 10.0–11.0) at the optimum temperature of enzyme activity. The pH stability of the lipase was determined by incubating the enzyme with 0.05 M buffer of different pHs (5.0–11.0) for 1 h at room temperature. The enzyme was assayed as per standard protocol at the optimum activity temperature.
Thermal Inactivation of Enzyme
Thermal denaturation profile of the selected enzyme was carried out by pre-incubating the lipase separately at different temperatures such as 45 °C, 50 °C, 55 °C, 60 °C, 65 °C, and 70 °C. The aliquots were taken out at different time intervals followed by cooling at 4 °C for 15 min. The enzyme assay was carried out according to standard assay conditions. The enzyme activity determined under standard assay conditions without incubation at any pH was taken as control (i.e., its activity was considered as 100%). The enzyme activity at the start of the experiment was taken as 100% and the residual lipase activity after incubation was determined. The reaction mixture without enzyme served as blank.
Evolutionary Conservation Analysis
The evolutionary sequence conservation analyses were performed to identify the functionally and structurally important amino acid residues of the lipase under investigation using the ConSurf Program [17]. For lipase query sequences, the CSI-BLAST (Context-Specific Iterated-Basic Local Alignment Search Tool) homolog search algorithm was used with E value cutoff of 0.00001, maximal identity of 95%, and minimal identity of 35% between the sequences was used for homology searches. The protein database UNIREF-90 (http://www.uniprot.org/help/uniref) was used for homologous search; other parameters were kept at default values for calculation of the conservation scores. The primary goal was to reveal the highly conserved regions and functionally and structurally important residues in lipase.
Structural Modeling of Target Lipase
The identified lipase sequence showed significant sequence similarity with already reported enzyme crystal structures, so its 3D structure was deduced using the ModellerV9.22 program [18] and SWISS-MODEL (https://swissmodel.expasy.org) computational comparative modeling server. The SWISS-MODEL is a fully automated and user-friendly protein structure homology-modeling server [19]. The initial modeled structure was further energetically minimized and MD (molecular dynamics) simulation was performed with CABS-flex 2.0 server. The CABS-flex server (http://212.87.3.12/CABSflex2) is an efficient simulation engine that allows for modeling of the large-scale conformational change of proteins [20]. It is based on the CABS coarse-grained model representation. In CABS coarse-grained model representation, the single amino acid residue is represented by 4 pseudo-atoms: C-alpha (CA), C-beta (CB), side-chain group (SC), and peptide bond (cp). The modeled structure was MD simulated with a default mode of CABS-flex. The output trajectory file was further analyzed for protein flexibility. Additionally, modeled structures especially N-terminal (modeled region: 1–34 amino acids long) part that did not show any significant similarity with the template, was validated using the MD simulation. The UCSF Chimera program was used for protein visualization and trajectory analysis (https://www.rbvi.ucsf.edu/chimera/).
Results and Discussion
Enrichment of Halophilic Cultures
As mentioned in the methodology section, soil and water samples were collected from five different sites having similar environmental conditions such as high temperatures, high pH, and salinity. The sample sites were lakes of Rajasthan: Lunkaransar, Badopal, Didwana, Sambhar, and Pachpadra. The edible salt NaCl concentrations were higher in Sambhar and Pachpadra lakes. The high concentration of fluoride salt has been reported in the Didwana Lake (Govt. of Rajasthan, India). Sampling site temperature ranged from 43 to 50 °C. A total of 30 water and soil samples were obtained. The pH and salinity were examined for each sample. Soil and water samples from Luankaransar and Badopal showed a pH of ~ 7.95 (± 0.15) while Didwana and Pachpadra showed a pH of ~ 9.10 (± 0.1). The highest pH of ~ 10.00 was observed for the Sambhar samples. Furthermore, the salinity of each sample was found in the range of 70–95‰. More details about samples’ pH and salinity are given in Table 1.
Screening of Lipase Producing Culture and Their Characterization
Qualitative Screening
The bacterial cultures showed lipase production from 37 °C onwards, but higher lipase activity was observed in the temperature range of 40–45°. Subsequently, lipase screening was performed at different salinities and pHs. Lipase production was also determined at different salinities. This was performed by preparing media with different concentrations of NaCl. Some of the pure colonies were selected at higher salinity and pH parameters. The results of the experiment showed that some cultures were able to grow at pH higher than 10.0 but did not produce lipase, as they did not produce any hydrolytic zones on TBA plates with pH above 10.0.
On the basis of qualitative screening at different salinities, pHs, and temperatures, only a few colonies were selected for further analysis. Only 7 bacterial isolates, which grew at temperatures > 45 °C, salinity ≥ 5% with the hydrolytic zone on TBA and ATCC plates, were selected for further investigation. Out of 7 bacterial isolates, 5 isolates (F3–F7) were from water samples and two (F1 and F2) were from soil samples. The results of the study showed that both water and soil samples from Lunkaransar showed lipase production under all the conditions discussed above. Out of the remaining four sites, only water sample gave lipase-positive bacterial isolates (Table 2).
From the above Table 2, it becomes evident that the isolates F1 and F7 showed hydrolytic zones at high salinity. Growth and lipase production at the highest salinity was observed for isolate F7 while a larger hydrolytic zone was observed for F1 and F5 on TBA plates. It was also observed that the hydrolytic zones produced by the eight isolates were bigger on TBA plates as compared with those produced on the ATCC agar plates. This may be attributed to the fact that the ATCC medium was designed for highly halophilic bacterial cultures and was less preferred by the moderately halophilic bacteria. Morphological and biochemical characterization showed all those seven bacterial isolates were Gram positive, except F4. Microscopic examination of the bacterial cells showed that all were rod shaped, except F5 which was coccus/round in shape. Details about morphological, physiological, and biochemical characterization are given in Table 2.
Various studies have been reported the use of tributyrin agar for primary/qualitative screening. For example, lipolytic Bacillus spp. and Pseudomonas spp. have been screened and isolated using the TBA medium [21, 22]. Meghwanshi et al. (2006) have also reported the use of rhodamine B agar medium for the screening of lipase producers. Other screening approaches have also been used for the determination of lipolytic activity, but agar plate is an easier method and economical. Another reason to use agar plates assay is that the lipase activity determination in liquid substrates is difficult as these enzymes are water soluble but the substrates are insoluble [23].
Quantitative Screening
In order to select the best lipase producer from lipase producing bacterial cultures (as determined by agar plate assay), the lipase-positive isolates were cultivated in the fermentation medium (ATCC medium containing olive oil instead of tributyrin). After 24 h of fermentation, the broths were centrifuged at 10000 rpm for 10 min and the supernatants were collected and evaluated of the lipase titres by p-nitrophenyl palmitate assay [16]. All the isolates showed good lipase activity (lipase activity in the range of 4–15 IU, given in Table 2). The highest lipase activity was shown by the isolate F7 (15.12 IU) while the isolate F3 showed the least activity (3.97 IU). Remarkably, isolate F1 and F7 showed bigger hydrolytic zones on TBA medium at high salinity; similarly, they also showed higher enzymes activities as compared with other isolates. Interestingly, the isolate F7 showed higher enzyme activity (15.117 IU) as compared with hydrolytic zones on the TBA medium (Table 2). This is maybe due to the fact that the tributyrin present in TBA medium is a short-chain fatty acid, while palmitic acid present in the p-NPP is a long-chain fatty acid and the lipase from isolate F7 may have preference for long-chain fatty acids but not for the short-chain fatty acids.
Rahman et al. (2005) studied and characterized lipase from Pseudomonas sp. S5 and performed a colorimetric assay for the measurement of lipase activity [24]. Zhou et al. (2011) performed copper soap colorimetric assay using n-hexane–olive oil microemulsions with slight modifications to investigate the lipase activity from Aspergillus oryzae CJLU-31 [25]. Among the isolates analyzed, samples F1 and F7 were higher lipase producers. Therefore, these highly lipid-degrading bacterial isolates were selected for further analysis.
Identification of the Isolates Using 16S rRNA Gene Sequencing
The 16S rRNA gene sequence of isolate F1 (Lunkaransar) showed maximum sequence identity (99.79%) with Bacillus licheniformis, Bacillus paralicheniformis, and Bacillus haynesii species and their strains. The 16S rRNA-based phylogenetic analysis of isolate F1 is shown in Supplementary Fig. 1. This isolate is closely related to members of the Bacillus licheniformis clade. This strain is phylogenetically positioned between the Bacillus licheniformis and Bacillus haynesii species. The 16S analysis clearly indicates that this is a new strain or species of the Bacillus genus. Bacillus licheniformis is well adapted to grow in alkaline conditions and high pH levels. Optimal growth temperature is around 50 °C and it can survive at much higher temperatures. Improvement in the thermostability of lipase produced by various strains of Bacillus licheniformis by directed evolution has been reported by different researchers [26]. From these findings, it can be deduced that the F1 isolate may be an excellent source for thermoalkaliphilic lipase. The other closely related Bacillus haynesii species was isolated from the soil habitat of Evolution Canyon III (Israel) in a survey of ecological diversification [27]. This phylogenetic analysis study confirmed that our isolate similarly belongs to the extremophilic environmental condition of the Thar Desert.
The 16S rRNA gene sequence of isolate F7 showed maximum (99.63%) sequence identity with 16S ribosomal RNA gene sequence of Bacillus tequilensis strain 10b and other various Bacillus subtilis strains. Phylogenetic analysis of the 16S rRNA showed that this isolate is closely related to members of the Bacillus clade (see in the supplementary Fig. 2). The 16S rRNA analysis clearly indicates that this isolate F7 might be a new species or a new strain of Bacillus tequilensis or Bacillus subtilis. Several thermophlic and halophilic lipases were reported from the genus Bacillus [28]. The isolate F7 also showed close homology to halotolerant species of Bacillus. Further the analysis of the 16S rRNA sequence of the bacterial isolate, F7 confirms that it belongs to the extremophilic Bacillus species. Based on its consistent lipase production and its extremophilic characteristics, this isolated was selected for further investigations viz. lipase gene cloning, expression, purification, and characterization.
Gene Cloning, Expression, and Sequencing
The primer was designed using the lipase gene as a reference sequence from NCBI. The 639 bp lipase gene was amplified by PCR using both the forward primer (lipase: Bamh1-F: CTAGGATCCATGAAATTTGTAAAAAGAAGAATC) and the reverse primer (lipase: Xho1-R: GATCTCGAGTTAATTCGTATTCTGGCCCCC). The selected gene was successfully cloned into the pET29b vector, with 6× His-tag at the N-terminal. SDS-PAGE showed that the lipase had a molecular weight of ~ 21 KDa. Cloning and sequencing of the lipase gene of isolate F7 revealed an open reading frame corresponding to 241 amino acids (726 bp) long including the His-tag and TEV sequences. The lipase coding gene mined using the NCBI Smart BLAST program using the UniprotKB/Swiss-Port database indicated starting sequence from 1 to 29 included His-tag and TEV, etc. The true lipase gene sequence demonstrated that it consists of coding 212 amino acids long showing high identity with lipases of multispecies Bacillales [Bacillales lipase LipA]. This highly homologous clone sequence has three amino acids which were different from the Bacillus multispecies lipase. The extracellular crude lipase was used for characterization and kinetic studies.
Biochemical Characterization of Lipase
Effect of Temperature on Enzyme Activity and Stability
The enzyme activity was determined in the temperature range of 20–80 °C. The lipase activity was observed to increase with increasing temperature up to 60 °C; however, above 60 °C, there was a decline in the activity. The maximum enzyme activity of 66.013 IU was observed at the optimal temperature of 60 °C (Supplementary Fig. 3). Even at higher temperatures of 70 and 80 °C, 56.28 IU and 38.65 IU, respectively, were observed which were 85.26% and 58.54%, respectively of the activity obtained at the optimal temperature of 60 °C (66.013 IU). Similar thermophilic lipase has been reported from Bacillus licheniformis that showed optimum activity at 60 °C [29]. Another reported extracellular lipase from Bacillus subtilis 168 is stable for at least 30 min at 40 °C [30]. For the determination of the stability of the lipase at different temperatures, it was incubated in the temperature range of 20–80 °C for 30 min. The residual activity of the enzyme was then determined at the optimal temperature of 60 °C. Results showed that the enzyme was stable in the temperature range of 20–60 °C, with no decline in activity observed up to 60 °C. However, there was a drastic decline in the lipase stability above 60 °C, and almost 50% activity of the enzyme was lost at temperatures above 70 °C (Supplementary Fig. 3).
Effect of pH on Enzyme Activity and Stability
The cloned lipase enzyme did not show any activity in the pH range of 5–6 but at pH 7, it showed the least activity. The enzyme exhibited good activity in the pH range of 8–11 (Supplementary Fig. 4). Lipase stability at different pHs was determined by incubating it in buffers of different pHs viz. 5–11 for 60 min. The residual lipase activity was then determined at the optimal pH of 9.0. The enzyme seemed almost stable at pH 5–11 (Supplementary Fig. 4). But the maximum stability was observed at pH 9.0. Microbial lipases show activity in a wide range of pH and temperature [31]. Generally, bacterial lipases are preferred over other microbial lipases as they have neutral or alkaline pH optima and are often thermostable [32, 33]. Extracellular lipase from Bacillus subtilis 168 exhibited optimum activity at pH 10. Drastic losses in lipase activity were reported above pH 10.5 and below pH 6.5. Our cloned lipase showed high similarity to the sequence of lipase from Bacillus subtilis 168, so it also showed quite similar biochemical properties. Extracellular lipase from Bacillus subtilis is remarkably stable at alkaline pH, showing maximum stability at pH 12 and maintaining > 65% of its activity after 24 h at pH 13 [30].
Thermal Inactivation of Enzyme
Thermal inactivation of the lipase was examined by incubating it for 30–240 min at various temperatures (45–70 °C). It can be observed from Fig 1 that with increasing temperature there was a decline in lipase activity. A decline in lipase activity of ~ 25% and 15% were observed after 240-min treatment at 45 °C and 50 °C, respectively. At 55 °C and 60 °C, enzyme activity decreases by 50% after 180 min of treatment (Supplementary Fig. 5). At 65 °C and 70 °C, ~ 34% residual activity was observed after 30 min. However, after 120 min of treatment, no residual activity was observed at 65 °C and 70 °C. The inactivation of the enzyme was rapid at temperatures > 70 °C. Similarly, lipase from B. subtilis showed stability at higher temperatures as well as maximum activity at 50 °C. Another extracellular lipase reported from B. megaterium AKG-1 showed optimum activity at 55 °C [34].
The p-Nitrophenol Palmitate Kinetic Analysis
The kinetic parameters Km and Vmax (maximum velocity) values of p-nitrophenyl palmitate were determined from the Lineweaver–Burk double-reciprocal plot. It was observed that the Km and Vmax values for extracellular lipase were 4.76 μmol and 400 μmol/mL/min, respectively (Fig. 1). Brabcova´ et al. (2010) stated that the low Km value of an enzyme for its substrate indicates that the enzyme needs a small quantity of the substrate to reach saturation point. Henceforth, the Vmax is achieved at comparatively low substrate concentrations. Brabcova´ et al. (2010) compared the kinetic parameters of cell-bound lipase and extracellular lipase and concluded that the extracellular lipase had a higher affinity for the substrate as compared with cell-bound lipase [35].
Analysis of Lipase Gene for Evolutionarily Conserved Regions
The gene sequence was analyzed for the highly conserved regions, functionally and structurally important residues. Analysis of the sequence clearly indicated that it has conserved regions except for the N-terminal region (1–34 amino acids). It can be observed from Fig. 2 that both functionally and structurally important residues are highly conserved during the evolution. The highly conserved pentapeptide [G-X-S-X-G] motif is a key feature of the lipase family [36]. This key motif was also found in the lipase sequence of F7 with a highly conserved [A-H-S-M-G] pattern. The active site catalytic triad (Ser108, Asp164, and His187) is also conserved. Intestinally, the one residue Glu128 which is different from the other Bacillus species lipase is moderately conserved (Fig. 2). At this position, the amino acid Ala was found in another bacterial species which is highly conserved. An overall sequence conservation score, and functionally and structurally important residues are given in Fig. 2.
Structural Modeling
Computational structural modeling of target sequence (cloned sequence) was performed for secondary structure topology and structural level characterization. The target lipase sequence showed a high sequence identity with the 3D crystal structure (PDB ID 1I6W) of Bacillus subtilis lipase [37]. Only few amino acid variations were observed in the minimal alpha/beta-hydrolase fold region at the sequence level. The N-terminal 34 amino acids long region (from 1 to 34 amino acids) did not show any sequence identity with the crystal structure. Initial sequence analysis indicated; this region belongs to signal peptide. Furthermore, this was also confirmed by structural modeling. The N-terminal (1–34 amino acids) region was separately modeled. Initially, the secondary structure was predicted. It was confirmed by various servers that the N-terminal part from the amino acids ranging from 3rd to 24th formed helical secondary structure while the rest 10 amino acids formed the random coil secondary structure. The predicted secondary structure viz. N-terminal helix and coil were converted into 3D coordinate via applying their corresponding Phi and Psi angle values. Finally, the complete modeled structure was obtained by building a peptide bond between two modeled structures with minimal energy. The optimum conformation of modeled structure especially the N-terminal part was obtained after molecular simulations. An overall modeled structure is highly constraint except the N-terminal modeled structure. The minimal alpha/beta hydrolase fold showed to be highly conserved at the structural and sequence level. Interestingly, residue Glu128 that was different from the template structure showed the hydrogen bonding interaction between the side chain of Glu128 and side chains of Asn35 and Gln152 amino acids. Remarkably, this amino acid also showed salt bridge interaction between Glu128--Lys101 during the MD simulation. Those interactions may be assisting in preserving the stability and activity of lipase at high temperatures and alkaline pHs. The detailed information about modeling is given in Fig. 3a–e.
This is an overall structural modeling of identified lipase (a) highly homologous 3D structure (PDB) BLAST hits. (b) Initial modeled structure of target sequence. (c) Predicted secondary structure of N-terminal region (predicted as a signal peptide). (d) Integrated predicted secondary structure with core alpha/beta-hydrolyses fold. (e) Modeled structure after energy minimization and MD simulation
It was observed the N-terminal helix structure showed flexibility due to a long-loop region (from 24 to 34 amino acids long). Notably, the predicted N-terminal helix structure (3–23 amino acids long) was not distorted after the MD simulation. This helix structure did not impinge on the remaining minimum alpha/beta hydrolase fold structure and no significant interactions were observed between the helix and the rest of the protein. It seems that this helix structure independently maintained the secondary structure topology. At the sequence level, this helix was predicted as a signal peptide that may be helpful in the secretion of extracellular lipase. Another functional possibility of this stable secondary structure may be that it plays an important role in providing extra stability to the minimum α/β-hydrolase fold of lipase structure in the realistic model structure. The actual conformation and position of this helix are hard to predict by the computational approach with limited sources.
Computational structural modeling and molecular dynamics might help to predict the accurate 3D structure of macro-biomolecules. Computational modeling is also used to predict the impact of mutations in structural stability, lid opening, and closing behavior in order to understand the importance of an individual residue [38]. Recently, Haque and Prabhu (2016) performed computational structural modeling and MD simulation studies to understand the dynamics behavior of lipase active site lid at different temperatures and in organic solvents [39]. The computational structural modeling studies also help in exploring the mechanism of thermostability and thermoactivity and the role of particularly conserved amino acid tryptophan in bacterial thermoalkaliphilic lipases [40]. Similarly, Rehm et al. (2010) also performed computational modeling studies on different lipases from Candida rugosa, Thermomyces lanuginosa, and Rhizomucor miehei [41]. The results indicated that in all those lipases, lid opening and closing were driven by the organic solvents but was independent of a bound substrate.
Concluding Remarks
This is a first report (to the best of our knowledge) on an extremophilic lipase isolated and characterized from the Indian Thar Desert. The lipase from Bacillus tequilensis (F7) is an alkaliphilic and thermophilic one with substantial activity and stability in a wide range of temperature and pH. Furthermore, molecular-level characterization of this lipase showed that it can be used for various industrial applications such as food processing, dairy, oleochemical, leather processing, polymer synthesis, biodiesel synthesis, and many more. The enzyme can further be engineered to make it a potential candidate for many novel applications based on the chemo-, regio-, and enantioselective properties of the enzyme.
References
Ghosh, P. K., Saxena, R. K., Gupta, R., Yadav, R. P., & Davidson, S. (1996). Microbial lipases: Production and applications. Science Progress, 79, 119–157.
Haki, G. D., & Rakshit, S. K. (2003). Developments in industrially important thermostable enzymes: A review. Bioresource Technology, 89(1), 17–34.
Saxena RK, Agarwal L, Meghwanshi GK (2005) Diversity of fungal and yeast lipases: Present and future scenario for the 21st century. In: Microbial diversity: Current perspectives and potential applications, 791-814.
Meghwanshi GK and Vashishtha A (2018) Biotechnology of fungal lipases. In Fungi and their role in sustainable development: Current perspectives. Springer Nature Singapore, 383–411.
Shu, Z. Y., Jiang, H., Lin, R. F., Jiang, Y. M., Lin, L., & Huang, J. Z. (2010). Technical methods to improve yield, activity and stability in the development of microbial lipases. Journal of Molecular Catalysis B: Enzymatic, 62(1), 1–8.
Treichel, H., de Oliveira, D., Mazutti, M. A., Di Luccio, M., & Oliveira, J. V. (2009). A review on microbial lipases production. Food Bioprocess Tech, 3, 182–196.
Verma, S., Kumar, R., & Meghwanshi, G. K. (2019). Identification of new members of alkaliphilic lipases in archaea and metagenome database using reconstruction of ancestral sequences. 3Biotech, 9(5), 165.
Demirjian, D., Morís-Varas, F., & Cassidy, C. (2001). Enzymes from extremophiles. Current Opinion in Chemical Biology, 5(2), 144–151.
Khyami-Horani, H. (1996). Thermotolerant strain of Bacillus licheniformis producing lipase. World Journal of Microbiology and Biotechnology, 12(4), 399–401.
Kaur, R., Kumar, R., Verma, S., Kumar, A., Rajesh, C., & Sharma, P. K. (2020). Structural and functional insights about unique extremophilic bacterial lipolytic enzyme from metagenome source. International Journal of Biological Macromolecules, 152, 593–604. https://doi.org/10.1016/j.ijbiomac.2020.02.210.
Sharma, P. K., Kumar, R., Garg, P., & Kaur, J. (2015). Insights into controlling role of substitution mutation, E315G on thermostability of a lipase cloned from metagenome of hot spring soil. 3Biotech, 4, 189–196.
Sharma, P. K., Kumar, R., Kumar, R., Mohammad, O., Singh, R., & Kaur, J. (2011). Engineering of a metagenome derived lipase towards thermal tolerance: Effect of aspargine to lysine mutation on the protein surface. Gene, 491, 264–271.
Daoud, L., Kamoun, J., Ali, M. B., Jallouli, R., Bradai, R., Mechichi, T., Gargouri, Y., Ali, Y. B., & Aloulou, A. (2013). Purification and biochemical characterization of a halotolerant Staphylococcus sp. extracellular lipase. In J Biol Macromol, 57, 232–237.
Schneegurt, M. A. (2012). Media and conditions for the growth of halophilic and halotolerant bacteria and archaea. In R. H. Vreeland (Ed.), Advances in understanding the biology of halophilic microorganisms. Dordrecht: Springer.
Ventosa, A., Nieto, J. J., & Oren, A. (1998). Biology of moderately halophilic aerobic bacteria. Microbiology and Molecular Biology Reviews, 62(2), 504–544.
Winkler, U. K., & Stuckmann, M. (1979). Glycogen, hyaluronate and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. Journal of Bacteriology, 138(3), 663–670.
Ashkenazy, H., Abadi, S., Martz, E., Chay, O., Mayrose, I., Pupko, T., & Ben-Tal, N. (2016). ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Research, 44, 344–350.
Webb, B., & Sali, A. (2016). Comparative protein structure modeling using MODELLER. Current Protocols in Bioinformatics, 54, 5.6.1–5.6.37.
Bertoni, M., Kiefer, F., Biasini, M., Bordoli, L., & Schwede, T. (2017). Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Scientific Reports, 7(1), 10480.
Kuriata, A., Gierut, A. M., Oleniecki, T., Ciemny, M. P., Kolinski, A., Kurcinski, M., & Kmiecik, S. (2018). CABS-flex 2.0: A web server for fast simulations of flexibility of protein structures. Nucleic Acids Research, 46, 338–343.
Meghwanshi, G., Agarwal, L., Dutt, K., & Saxena, R. (2006). Characterization of 1-3 regiospecific lipases from new Pseudomonas and Bacillus isolates. J Mol Catal B Enzyme, 40(3-4), 127–131.
Takaç, S., & Marul, B. (2008). Effects of lipidic carbon sources on the extracellular lipolytic activity of a newly isolated strain of Bacillus subtilis. Journal of Industrial Microbiology & Biotechnology, 35(9), 1019–1025.
Kanchana, R., Muraleedharan Dr, U., & Raghukumar, S. (2011). Alkaline lipase activity from the marine protists, thraustochytrids. World Journal of Microbiology and Biotechnology, 27(9), 2125–2131.
Rnzra, R., Baharum, S. N., Basri, M., & Salleh, A. B. (2005). High-yield purification of an organic solvent-tolerant lipase from Pseudomonas sp. strain S5. Analytical Biochemistry, 341, 267–274.
Zhou, J., Chen, W. W., Jia, Z. B., Huang, G. R., Hong, Y., Tao, J. J., & Luo, X. B. (2012). Purification and characterization of lipase produced by Aspergillus oryzae CJLU-31 isolated from waste cooking oily soil. American Journal of Food Technology, 7(10), 596–608.
Madan, B., & Mishra, P. (2014). Directed evolution of Bacillus licheniformis lipase for improvement of thermostability. Biochemical Engineering Journal, 91, 276–282.
Dunlap, C., Schisler, D., Perry, E., Connor, N., Cohan, F., & Rooney, A. (2017). Bacillus swezeyi sp. nov. and Bacillus haynesii sp. nov., isolated from desert soil. International Journal of Systematic and Evolutionary Microbiology, 67(8), 2720–2725.
Chakravorty D, Patra S (2012) Attaining extremophiles and extremolytes: Methodologies and limitations. Extremophiles: Sustainable resources and biotechnological implications, 29–74.
Kaur, G., Singh, A., Sharma, R., Sharma, V., Verma, S., & Sharma, P. K. (2016). Cloning, expression, purification and characterization of lipase from Bacillus licheniformis, isolated from hot spring of Himachal Pradesh, India. 3 Biotech, 6(1), 49.
Lesuisse, E., Schanck, K., & Colson, C. (1993). Purification and preliminary characterization of the extracellular lipase of Bacillus subtilis 168, an extremely basic pH-tolerant enzyme. European Journal of Biochemistry, 216(1), 155–160.
Gupta, R., Gupta, N., & Rathi, P. (2004). Bacterial lipases: An overview of production, purification and biochemical properties. Applied Microbiology and Biotechnology, 64(6), 763–781.
Cadirci, B. H., & Yasa, I. (2010). An organic solvent tolerant and thermotolerant lipase from Pseudomonas fluorescens P21. Journal of Molecular Catalysis B: Enzymatic, 64(3-4), 155–161.
Aysun, A. G., & Alper, A. (2013). Purification and biochemical characterization of an extracellular lipase from psychrotolerant Pseudomonas fluorescens KE38. Turkish Journal of Biology, 37, 538–546.
Sekhon, A., Dahiya, N., Tiwari, R. P., & Hoondal, G. S. (2005). Properties of a thermostable extracellular lipase from Bacillus megaterium AKG-1. Journal of Basic Microbiology, 45(2), 147–154.
Brabcova, J., Zarevucka, M., & Mackova, M. (2010). Difference in hydrolytic activities of two crude lipases from Geotrichum candidum 4013. Yeast, 27(12), 1029–1038.
Verma, S., Meghwanshi, G. K., & Kumar, R. (2018). Structural homogeneity in microbial lipases. Microbiol Curr Res, 2, 12–13.
Pouderoyen, G. V., Eggert, T., Jaeger, K. E., & Dijkstra, B. (2001). The crystal structure of Bacillus subtilis lipase: A minimal alpha/beta hydrolase fold enzyme. J Mol Bio, 309(1), 215–226.
Khan, F. I., Lan, D., Durrani, R., Huan, W., Zhao, Z., & Wang, Y. (2017). The lid domain in lipases: Structural and functional determinant of enzymatic properties. Frontiers in Bioengineering and Biotechnology, 5, 16.
Haque, N., & Prabhu, N. P. (2016). Lid dynamics of porcine pancreatic lipase in non-aqueous solvents. Biochimica et Biophysica Acta, 1860(10), 2326–2334.
Timucin, E., & Sezerman, O. U. (2013). The conserved lid tryptophan, W211, potentiates thermostability and thermoactivity in bacterial thermoalkalophilic lipases. PLoS One, 8(12), 85186.
Rehm, S., Trodler, P., & Pleiss, J. (2010). Solvent-induced lid opening in lipases: A molecular dynamics study. Protein Science, 19(11), 2122–2130.
Funding
The author Gautam Kumar Meghwanshi sincerely acknowledge the financial support provided by SERB, New Delhi (Sanction order NO. SB/YS/LS-146/2014, dated 25 May 2015), for carrying out this research work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 723 kb)
Rights and permissions
About this article
Cite this article
Verma, S., Kumar, R., Kumar, P. et al. Cloning, Characterization, and Structural Modeling of an Extremophilic Bacterial Lipase Isolated from Saline Habitats of the Thar Desert. Appl Biochem Biotechnol 192, 557–572 (2020). https://doi.org/10.1007/s12010-020-03329-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-020-03329-3