Abstract
Yeast surface display has emerged as a viable approach for self-immobilization enzyme as whole-cell catalysts. Herein, we displayed Candida rugosa lipase 1 (CRL LIP1) on the cell wall of Pichia pastoris for docosahexaenoic acid (DHA) enrichment in algae oil. After a 96-h culture, the displayed CRL LIP1 achieved the highest activity (380 ± 2.8 U/g) for hydrolyzing olive oil under optimal pH (7.5) and temperature (45 °C) conditions. Additionally, we improved the thermal stability of displayed LIP1, enabling retention of 50% of its initial bioactivity following 6 h of incubation at 45 °C. Furthermore, the content of DHA enhanced from 40.61% in original algae oil to 50.44% in glyceride, resulting in a 1.24-fold increase in yield. The displayed CRL LIP1 exhibited an improved thermal stability and a high degree of bioactivity toward its native macromolecule substrates algae oil and olive oil, thereby expanding its potential for industrial applications in fields of food and pharmaceutical. These results suggested that surface display provides an effective strategy for simultaneous convenient expression and target protein immobilization.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
Long-chain polyunsaturated omega 3 fatty acid (PUFA) demonstrated to be vital for human growth, nervous system development, and reproduction [1, 2]. As important essential fatty acids, PUFAs also play important biological roles in human health and disease resistance. Therefore, dietary supplementation is beneficial to avoiding cardiovascular disease, dyslipidemia, diabetes mellitus, inflammatory diseases, and metabolic syndromes [2,3,4]. Docosahexaenoic acid (DHA) is the primary active ingredient in functional oils and has drawn increasing attention in recent years, with DHA enrichment from PUFA-containing oils becoming a significant area of study. Numerous chemical process has been investigated for PUFA enrichment in fish oil, marine algae, or fishery byproducts, including high-performance liquid chromatography, liquid–liquid extraction fraction, low-temperature fractional crystallization, molecular distillation, salt-solubility methods, and urea-inclusion complexation [5,6,7]; however, most of these methods require high-energy consumption and are non-selective for fatty acids [8]. The selective hydrolysis catalyzed by triacylglycerol acyl hydrolases (lipases) represents a straightforward and energy-efficient method for production of PUFA concentrates [9,10,11].
Candida rugosa lipase (CRL) is one kind of the most important and versatile commercial catalysts, exhibiting broad substrate specificity, acyl chain length selectivity, fatty acid specificity, triglyceride specificity, and stereo-selectivity. It has been widely utilized in industrial processes, involving food, pharmaceuticals, cosmetics, flavor enhancement, and fragrances. CRL is regarded as safe and has been employed to enrich DHA in oil following hydrolysis by free CRL due to its substrate specificity for the C18–C22 acyl chain lengthened PUFAs [12,13,14,15,16]. However, applications of the free biocatalyst have not yet reached the industrial level due to its high production costs, low operational stability, and tedious downstream-purification procedure [14, 17, 18]. According to previous reports, the immobilized enzyme is capable of providing enzyme reusability and reducing operational costs as compared with use of the free enzyme. Therefore, many strategies have been applied to immobilize the CRL lipase 1 (LIP1) to overcome bottleneck problems [19, 20]. Unfortunately, processes involving traditional immobilization and subsequent purification remain too complex for industrial production.
Recently, yeast surface-display technology, a novel immobilization strategy, was investigated to produce whole-cell catalysts based on the simplicity of the process and mechanism of post-translational modification [21, 22]. Candida antarctica lipase B [23, 24], Geotrichum sp. lipase [25], Rhizomucor miehei lipase [26], Rhizopus oryzae lipase [27, 28], Thermomyces lanuginosus lipase [29], and Yarrowia lipolytica lipase [30] have been self-immobilized on the cell surface of Pichia pastoris and employed for different utilization. To our best knowledge, CRL whole-cell catalyst obtained by yeast displaying strategy was rarely reported in the existing literature.
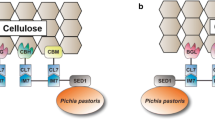
Based on our previous studies [25], we immobilized CRL LIP1 on the P. pastoris cell wall using a-agglutinin derived from Saccharomyces cerevisiae to produce active whole-cell biocatalyst. Herein we first report its preparation and application for concentration DHA from algae oil.
Materials and Methods
Materials
Escherichia coli DH5α cells were stored in our laboratory for plasmid amplification. P. pastoris strain X-33 was from Invitrogen (Carlsbad, CA, USA) and used to express target protein as the host cells. The plasmid pMD18-T from TaKaRa Biotechnology was utilized for cloning. The plasmid pPICZXFa was constructed as a vector for lipase expression in our laboratory [25]. In brief, two expression cassettes were constructed based on the vector pPICZα: one is AOX1 promoter-Aga1-AOX1 terminator, which is used to expressed the Aga1 anchor protein on the surface of yeast. The other is FLD1 promoter-Aga2-Xa-HA-MCS-AOX1 terminator. The target gene can be inserted into the multiple cloning site (MCS), and expressed as the “Aga2-Xa-HA-target gene” fusion protein, which can attach to the Aga1 through disulfide bond. E. coli was cultured in Luria–Bertani (LB) including ampicillin at a final concentration of 100 μg/mL at 37 °C. P. pastoris strain X-33 and recombinant X-33 strains were cultivated in YPD medium (1% yeast extract, 2% peptone, 2% dextrose) including 100 μg mL−1 zeocin, BMGY medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate, pH 7.0, 1.34% yeast nitrogen base [YNB], 4 × 10−5% biotin, and 1% glycerol) or BMMY medium (2% peptone, 1% yeast extract, 100 mM potassium phosphate, pH 7.0, 1.34% YNB, 4 × 10−5% biotin, and 1% methanol). BMMY plates containing 2 × 10−4% (w/v) Rhodamine B and 1% (v/v) olive oil were utilized to detect the bioactivity of the displayed CRL LIP1.
Refined algae oil was obtained from Yuwang Pharmaceutical (Shandong, China). Endonucleases, PrimeSTAR HS DNA polymerase, DNA purification kits, and DNA ligation kit version 2.0 were products from Takara Bio (Shiga, Japan). Factor-Xa protease was product from New England BioLabs (Ipswich, MA, USA). All other reagents and chemicals were analytical grade products from Promega (Durham, NC, USA) or Sigma (Sigma-Aldrich, USA).
Gene Design and Assembly of the Codon-Optimized C. rugosa lip1
According to a polymerase chain reaction (PCR)–based accurate synthesis (PAS) protocol [31, 32], the optimized full-length CRL lip1 gene was amplified using two outer pair primers (P1: 5′-ATAGCTAGCGCCCCCACCGCCACGC-3′; Nhe I site is underlined; and P2: 5′-TATCTCGAGTTACACAAAGAAAGACGGCGG-3′; Xho I site is underlined) based on the obtained F1, F2, and F3 from the first round of PCR [33]. Products from the second of PCR were then inserted into Nhe I-Xho I sites of the vector pMD18-T from TaKaRa (Dalian, China), and sequenced (Sangon Biotech, Shanghai, China).
Plasmid Construction
The optimized mature CRL lip1 gene without the signal peptide was introduced into Nhe I/Xho I-digested pPICZXFa, resulting in plasmid pPICZXFa-lip1. All plasmids were transformed into competent cells of E. coli DH5α and amplified, which were further sequenced after coarse screening on LB ampicillin plates (50 μg mL−1).
Yeast Transformation and Inducible Expression of Displayed CRL LIP1
Plasmids pPICZXFa-lip1 and pPICZXFa were linearized with BstX I and electro-transformed into P. pastoris X-33 competent host cells by electroporation following Invitrogen’s instructions. For 0.2-cm cuvette, the electroporation pulse parameters included 1500 V, 5 ms, 25 μF, and 200 Ω [25]. The resulting transformants were selected on YPD plates with sorbitol containing zeocin at 100 μg mL−1 at 30 °C for 3 days.
A single colony was picked and pre-cultured into YPD medium for 15 h at 30 °C and shaking at 150 rpm. This culture was then inoculated into BMGY medium and cultivated until the OD600 reached 2.0–6.0. The cells were harvested and cultured in BMMY medium for inducible expression, supplemented with a final concentration of 1% (v/v) methanol at 24-h intervals. The P. pastoris X-33 cells displaying CRL LIP1 were collected by centrifugation (2000g, 8 min), washed, re-suspended, and divided into two equal parts. One equivalent was utilized to measure lipase bioactivity. Meanwhile, another equivalent was dried at 105 °C to a constant weight as control.
Immunofluorescence Assay
Localization of displayed CRL LIP1 was analyzed by indirect immunofluorescence assay. The harvested P. pastoris X-33 whole cells displaying CRL LIP1 were re-suspended in phosphate-buffered saline (PBS, pH 7.4) supplemented with 1 mg/mL bovine serum albumin (BSA) and treated with the primary antibody anti-HA-tag mouse monoclonal antisera (1:150) on ice for 3 h. Subsequently, these P. pastoris X-33 whole cells were exposed to the fluorescein isothiocyanate (FITC)–conjugated goat anti-mouse second antibody (1:300) for half of an hour in darkness followed by washing 3 times with PBS. One drop of cell suspension loaded on glass slide was detected using a fluorescence microscope (Nikon, Tokyo, Japan). At the same time, most of cell suspension was utilized for a Cell Lab flow cytometry analysis (Quanta SC, Beckman Coulter, Fullerton, CA, USA). P. pastoris X-33 whole cells of treated samples were counted and analyzed fluorescence value with the CXP software (Beckman Coulter). In the same manner, the immunolabeling of transformants harboring pPICZXFa was carried out as the background.
Western Blot Analysis
The harvested P. pastoris X-33 whole cells displaying CRL LIP1 were digested by 1-μg Factor-Xa protease overnight as described previously [34, 35]. The targeted fusion protein CRL LIP1 with the HA-tag was collected, boiled, separated by 12% sodium dodecyl sulfate polyacrylamide (SDS-PAGE), and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA) [36]. The membranes were blocked with Tris-buffered saline and Tween 20 (TBST) containing 5% (w/v) non-fat dried milk and incubated with the primary antibody anti-HA mouse monoclonal IgG (1:800). The PVDF membranes were washed twice, and subsequently exposed to the horseradish peroxidase–conjugated goat anti-mouse secondary antibody (1:2000). The target protein bands were visualized by electrogenerated chemiluminescence (Bio-Rad) after the PVDF membranes were incubated with diaminobenzidine (Pierce Biotechnology, Rockford, IL, USA) and 0.01% H2O2.
Activity Assay for the Displayed CRL LIP1
The resulting transformants were cultivated on olive oil-BMMY plates with Rhodamine B, as a screening indicator [37], for 3 days. The expression of fusion protein AGA2-lipase was induced by supplemented methanol in the interval of every 24 h. The activity of the lipase was estimated by the halo around the yeast colony. The hydrolysis activity of the displayed CRL LIP1 was measured by the acid-base titration method according to previous literatures [25, 38].
Characterization of the Displayed CRL LIP1
To determine the optimal temperature for the displayed CRL LIP1, reactions were performed in the same substrate solution described previously and at various temperatures range from 30 to 60 °C. Based on the results of these experiments, the displayed CRL LIP1 was used to catalyze reactions to determine its optimal pH at 45 °C in 50 mM potassium phosphate buffer (pH 6.0–7.5) and 50 mM Tris–HCl buffer (pH 8.0–9.0). To determine thermostability, re-suspended cells were incubated at 45 °C for up to 6 h in 50 mM potassium phosphate buffer (pH 7.5), and the residual activity was measured every 1 h by titrimetry using an olive oil emulsion as the substrate. Additionally, substrate spectra was determined by the relative hydrolysis activities toward a serials acyl chain length p-nitrophenyl esters (C2, acetate; C4, butyrate; C8, caprylate; C10, decanoate; C14, myristate; and C16, palmitate) as substrates.
DHA Enrichment by Displayed CRL LIP1
First, 1 mL algae oil and 3 mL polyvinyl alcohol solution (2% (w/v)) was homogenized completely and placed in a conical flask with a stopper [25]. Then, the 5 mL buffer of K2HPO4/KH2PO4 (pH 7.5) was added to 4 mL emulsified fish oil and pre-incubated at 45 °C for 15 min. Subsequently, 90 mg dry weight whole-cell catalyst with the displayed CRL LIP1 per milliliter cell suspension was utilized to hydrolyze algae oil in the emulsified system. The reaction was performed at 150 rpm and 45 °C on a rocking incubator. Aliquots were extracted from the reaction mixture at designated times every 2 h to analyze the hydrolysis degree and fatty acid composition in glycerides. The initial rate of reaction was analyzed at the first 2 h. The glyceride fraction in the reaction system was treated with n-hexane (100 mL) following additional 500 mM ethanoic KOH (50 mL). DHA in glycerides were quantified and analyzed by Agilent 7890A-5975C gas chromatography–mass spectrometry (GC-MS) (Agilent, Santa Clara, CA, USA) equipped with a DB-WAX capillary column (30 m × 250 μm × 0.25 μm) after methylation according to the official method Ce-1b 89 of American Oil Chemists’ Society (2007).
The degree of hydrolysis was calculated according to the following equation:
where SV0 represents the value of saponification in crude algae oil, and AV0 and AVt represent the acid values of algae oil before and after hydrolysis [8], respectively.
Results
Preparation of the Whole-Cell Catalyst
As shown in Fig. 1, the displaying plasmid pPICZXFa-lip1 for self-immobilization of CRL LIP1 was generated. The self-immobilized active lipase fused to the AGA1 via disulfide linkages, and the AGA1 protein, was bonded covalently on the cell surface via the glycosyl-phosphatidylinositol anchor-attachment signal peptide sequence. When induced by methanol, the resulting pPICZXFa-lip1 transfectants efficiently expressed AGA1 and the fusion protein AGA2-LIP1.
To determine the bioactivity and its relative molecular weight of recombinant CRL LIP1, we performed a halo assay and western blotting of CRL LIP1 containing an HA-tag following its removal from the cell surface of pPICZXFa-lip1 transfectants using the Factor-Xa protease. After 4 days of cultivated on Rhodamine B-BMMY agar plates supplemented with olive oil, the lipase activities were preliminary estimated according to the surrounding florescence halos. As shown in Fig. 2a, whole cells P. pastoris X-33/pPICZXFa-lip1 hydrolyzed olive oil, yielded fluorescent complexes, and produced clear halos, while no halos were observed in control cells (pPICZXFa transfectants). P. pastoris X-33/pPICZXFa-lip1 produced a target protein of ~ 63 kDa representing CRL LIP1 with the HA-tag (Fig. 2b). These results suggested that active CRL LIP1 with the HA peptide tag were co-expressed on the pPICZXFa-lip1 transfectants’ surface.
a Halos of transfectants P. pastoris X-33 on Rhodamine B-BMMY agar plates supplemented with olive oil. Halo circled the colony of P. pastoris X-33/pPICZXFa-lip1 cells; in contrast, no halos were observed around P. pastoris X-33/pPICZXFa colonies. b Western blotting confirmed the expression of CRL LIP1 containing an HA peptide tag. The fusion proteins of CRL LIP1-HA-tag cleaved from P. pastoris X-33/pPICZXFa-lip1 by the protease Factor-Xa were separated by 12% SDS-PAGE and blotted onto a polyvinylidene fluoride (PVDF) membrane. Blots were detected by mouse monoclonal antibodies anti-HA and secondary antibody horseradish peroxidase–linked goat anti-mouse IgG
CRL LIP1 Localization in P. pastoris
CRL LIP1 localization in P. pastoris X-33/pPICZXFa-lip1 cells was determined by indirect immunofluorescence microscopy and flow cytometry. The localization of CRL LIP1 containing an HA peptide tag in P. pastoris X-33/pPICZXFa-lip1 cells was detected by using mouse monoclonal antisera to HA-tag and FITC-conjugated secondary antibody goat anti-mouse IgG. As shown in Fig. 3, P. pastoris X-33/pPICZXFa-lip1 transfectants exhibited bright green fluorescence at their cell surface, whereas the cell surface of P. pastoris X-33/pPICZXFa as the control emitted no fluorescence.
Immunofluorescence microscopy demonstrated the surface expression of CRL LIP1 in P. pastoris X-33/ pPICZXFa-lip1. The surface displaying of CRL LIP1-HA in pPICZXFa-lip1 transfected P. pastoris X-33 cells was detected by immunologically labeling with the anti-HA antibody and FITC-linked secondary antibody. No green fluorescence was detected in control pPICZXFa transfected P. pastoris X-33 cells
Moreover, flow cytometry analysis (FCA) indicated that > 98.89% of pPICZXFa-lip1 transfected P. pastoris X-33 cells efficiently expressed the fusion proteins of CRL LIP1-HA on the cell surface, but the emitted green fluorescence signals associated with P. pastoris X-33/pPICZXFa cells were indistinguishable from background (Fig. 4). From the abovementioned results, we could conclude that the fusion protein of CRL LIP1-HA-AGA2 was successfully surface immobilized via the AGA1 through disulfide bonds.
Flow cytometry analysis of surface expression CRL LIP1-HA. The expression of target protein CRL LIP1 was quantitatively analyzed after these plasmids pPICZXFa-lip1 and pPICZXFa transfected P. pastoris X-33 cells labeled with the anti-HA antibody and FITC-linked anti-IgG. About 98.89% cells expressed the fusion protein CRL LIP1-HA on the surface of P. pastoris X-33/pPICZXFa-lip1 transfectants (black-filled histograms). Negative controls X-33/pPICZXFa transfectants are marked with gray-filled histograms
Functional Bioactivity of Surface-Displayed CRL LIP1
We then performed quantitative functional tests to evaluate its practical application of surface-displayed CRL LIP1 as a whole-cell enzyme catalyst. The hydrolytic bioactivity of surface-displayed CRL LIP1 on the P. pastoris X-33/pPICZXFa-lip1 transfectants reached at 380 ± 2.8 U/g dry cells toward olive oil. The maximal hydrolytic activity of 380 U/g of dry cells for surface-displayed CRL LIP1 was higher than 273 U/g of dry cells’ activity for surface-displayed Geotrichum sp. lipase [25], and 257.8 U/g dry cells for surface-displayed T. lanuginosus lipase toward the same substrate olive oil [29]. The functional result showed that the surface-displayed CRL LIP1 on the cell wall of P. pastoris achieved an active structural conformation and exhibited a high bioactivity toward its native substrate.
Characteristics of Surface-Displayed CRL LIP1
To evaluate enzymatic characteristics of the displayed CRL LIP1 including its optimum pH, optimum temperature, and thermostability, these catalytic parameters were measured using substrate olive oil. As presented in Fig. 5a, the optimum pH of the self-immobilized CRL LIP1 was pH 7.5 as compared to that for the P. pastoris–secreted CRL LIP1 which was 7.0 [39, 40]. Moreover, the self-immobilized CRL LIP1 showed relative high stability over a wide range of pH values and retained > 70% of its maximal activity at pH 7.0–8.0. In Fig. 5b, the optimum temperature for surface-displayed lipase activity was 45 °C, as compared to that for the P. pastoris–secreted CRL LIP1 which was 40 °C. Importantly, the surface-displayed CRL LIP1 exhibited excellent thermostability, retaining 50% of its native activity toward olive oil after 45 °C treatment for 6 h at pH 7.5 (Fig. 5c), whereas P. pastoris–secreted CRL LIP1 retained only 26.4% activity after a 6-h incubation at its optimal temperature and pH. This enhancement of thermostability agreed with previous results reporting the protective effect from its proximity to the cell wall [25, 26]. The enhancement of lipase stability can improve its interactions with substrates and likely resulting in higher degrees of enzyme activity, which could potentially broaden its application. As shown in Fig. 5d, the self-immobilized CRL LIP1 demonstrated similar substrate specificity as those exhibited by native CRL LIP1 or commercial CRL LIP1 preparations [16, 40, 41]. Additionally, surface-displayed CRL LIP1 exhibited optimum substrate hydrolysis on p-nitrophenyl (pNP) caprate (C10), as well as relatively high levels of lipolytic activity toward pNP-caprylate (C8) and short-chain pNP-butyrate (C4), which constituted 89% and 90% of the activity observed on pNP-caprate (C10), respectively. These results indicated that surface-displayed CRL LIP1 constitutes a promising catalyst for practical applications for industrial-scale production.
Enzymatic characteristics of the self-immobilized CRL LIP1 catalyst. a The pH effect on the bioactivity of self-immobilized CRL LIP1 was assessed by evaluating hydrolysis activity toward olive oil at 45 °C at pH 6.0–10.0 after a 10-min incubation. Relative bioactivity was assessed by setting to 100% the bioactivity detected at pH 7.5. Activities are presented as the means ± SD of three independent assays. b Temperature effect on the activity of self-immobilized CRL LIP1 was determined by setting to 100% the activity obtained at 45 °C and pH 7.5. c Thermostability of the self-immobilized CRL LIP1 was evaluated by the residual lipase activity every 1 h after treatment at 45 °C in 50 mM potassium phosphate buffer (pH 7.5) for up to 6 h, which was assayed by titrimetry using olive oil. Relative lipase activity was determined by defining the activity of the displayed CRL LIP1 without heat incubation as 100%. Activities are presented as the means ± SD of three independent assays. d Substrate specificity of self-immobilized CRL LIP1 was determined by monitoring the hydrolysis bioactivity toward pNP-acetate (C2), pNP-butyrate (C4), pNP-octanoate (C8), pNP-decanoate (C10), pNP-myristate (C14), and pNP-palmitate (C16) substrates. Relative activity was evaluated by assuming the hydrolysis activity toward pNP-decanoate (C10) substrate was 100%
Enzymatic DHA Enrichment via Surface-Displayed CRL LIP1
After a 4-day culture, the whole cells X-33/pPICZXFa-lip1 were collected to selective hydrolyze algae oil for enrichment DHA as whole-cell biocatalysts. Similar to free CRL LIP1 [16], self-immobilized CRL LIP1 exhibited fatty acid specificity toward algae oil, resulting in increased DHA content in glycerides based on the high degree of hydrolysis (Fig. 6). After a 14-h incubation, the maximum degree of hydrolysis reached 30.6%, and the DHA content increased from 40.61% in crude algae oil to 50.44% in the glyceride fraction using the self-immobilized CRL LIP1 as lipase catalyst, resulting in a 1.24-fold increase.
Relationship between degree of hydrolysis and the relative contents of DHA and EPA compared with their own contents in original algae oil before hydrolysis, respectively. DHA and EPA relative contents were measured by setting that the original content in algae oil was 100% before hydrolysis. All values are the mean values ± standard deviation (SD) from triplicate experiments
Discussion
C. rugosa lipases have been widely applied due to their acyl chain length specificity, fatty acid specificity, triglyceride specificity, and stereo-selectivity toward different substrates. Moreover, the chemical immobilization of CRLs aimed at different catalytic reactions has been investigated to improve catalytic performance [42,43,44]. Given its fatty acid specificity, free CRLs have been employed to enrich DHA by selective hydrolysis or ethanolysis [14, 15]. However, the strategy of surface-displaying has not been adopted to produce self-immobilized CRLs as whole-cell catalysts until now. In the context, we constructed a eukaryotic expression vector for displaying CRL LIP1 on the yeast cell wall and evaluated the potential application of self-immobilized CRL LIP1 for DHA enrichment in algae oil.
It is interesting to observe that the maximal hydrolytic activity of 380 U/g of dry cells for surface-displayed CRL LIP1 toward natural substrate olive oil was higher than 273 U/g of dry cells’ activity for surface-displayed Geotrichum sp. lipase toward olive oil [25], 4.1 U/g of dry cells toward triolein [26], and 60 U/g dry cell for surface-displayed R. oryzae lipase toward 4-nitrophenyl dodecanoate (pNPD) [27], 220 U/g of dry cells’ activity reported for surface-displayed CALB toward p-nitrophenyl butyrate (pNPB) [28], 257.8 U/g dry cells for surface-displayed T. lanuginosus lipase toward olive oil [29], but lower than 106 U/mg DCW toward tributyrin reported for surface-displayed CALB [24] and 495.8 U/g dry cell toward 4-nitrophenyl caprylate (pNPC) [45]. We could conclude that the hydrolytic bioactivity in this work is within the higher rank coupled with better catalytic properties [46].
Moreover, the surface-displayed CRL LIP1 exhibited relatively high stability under mildly acidic pH conditions, with an optimal pH of 7.5 versus 8.0 for that of P. pastoris–secreted CRL LIP1 [33]. As shown in Fig. 5, the optimal temperature for immobilized CRL LIP1 was 45 °C, while that of P. pastoris–secreted CRL LIP1 was 40 °C. Additionally, surface-displayed CRL LIP1 exhibited improved thermostability as compared with that of free CRL LIP1. This finding agreed with previous reports indicating that protection by the cell wall enhanced immobilized LIP1 thermostability [22, 23, 25]. The enhancement of lipase stability can be improved through surface-display technology, thereby improving its interactions with substrates and likely resulting in higher degrees of enzyme activity, which could potentially broaden its application.
In conclusion, this study represents the first report of surface-displayed CRL LIP1 used as a thermostable biocatalyst for enrichment DHA from algae oil. Its utilization increased DHA content from 40.61% before hydrolysis to 50.44% after hydrolysis, with maximal hydrolysis of algae oil reaching 30.6% and suggesting CRL LIP1 as a promising self-immobilized enzyme for more desired applications in processes of biotransformation and catalysis.
Conclusion
This study represents the first report of surface-displayed CRL LIP1 used as a thermostable biocatalyst for enrichment DHA from algae oil. Its utilization increased DHA content from 40.61% before hydrolysis to 50.44% after hydrolysis, with maximal hydrolysis of algae oil reaching 30.6% and suggesting CRL LIP1 as a promising self-immobilized enzyme for more desired applications in processes of biotransformation and catalysis.
References
Bazinet, R. P., & Layé, S. (2014). Polyunsaturated fatty acids and their metabolites in brain function and disease. Nature Reviews Neuroscience, 15(12), 771–785.
Wison, D. W., Nash, P., Buttar, H. S., Griffiths, K., Singh, R., De Meester, F., Horiuchi, R., & Takahashi, T. (2017). The role of food antioxidants, benefits of functional foods, and influence of feeding habits on the health of the older person: an overview. Antioxidants (Basel), 6(4), 81.
Riediger, N. D., Othman, R. A., Suh, M., & Moghadasian, M. H. (2009). A systemic review of the roles of n-3 fatty acids in health and disease. Journal of the American Dietetic Association, 109(4), 668–679.
von Schacky, C. (2015). Omega-3 fatty acids in cardiovascular disease-an uphill battle. Prostaglandins, Leukotrienes, and Essential Fatty Acids, 92, 41–47.
Chakraborty, K., & Paulraj, R. (2007). Eicosapentaenoic acid enrichment from sardine oil by argentation chromatography. Journal of Agricultural and Food Chemistry, 55(18), 7586–7595.
Guil-Guerrero, J. L., López-Martínez, J. C., Rincón-Cervera, M. A., & Campra-Madrid, P. (2007). One-step extraction and concentration of polyunsaturated fatty acids from fish liver. Journal of the American Oil Chemists' Society, 84(4), 357–361.
Rubio-Rodríguez, N., Beltrán, S., Jaime, I., de Diego, S. M., Sanz, M. T., & Carballido, J. R. (2010). Production of omega-3 polyunsaturated fatty acid concentrates: a review. Innovative Food Science and Emerging Technologies, 11(1), 1–12.
Yan, J. Y., Liu, S. X., Hu, J., Gui, X. H., Wang, G. L., & Yan, Y. J. (2011). Enzymatic enrichment of polyunsaturated fatty acids using novel lipase preparations modified by combination of immobilization and fish oil treatment. Bioresource Technology, 102(14), 7154–7158.
Chakraborty, K., Vijayagopal, P., Chakraborty, R. D., & Vijayan, K. K. (2010). Preparation of eicosapentaenoic acid concentrates from sardine oil by Bacillus circulans lipase. Food Chemistry, 120(2), 433–442.
Hayes, D. (2004). Enzyme-catalyzed modification of oilseed materials to produce eco-friendly product. Journal of the American Oil Chemists' Society, 81(12), 1077–1103.
Mala, J. G., & Takeuchi, S. (2008). Understanding structural features of microbial lipases ---an overview. Analytical Chemistry Insights, 3, 9–19.
Huang, S., Li, X., Xu, L., Ke, C., Zhang, R., & Yan, Y. J. (2015). Protein-coated microcrystals from Candida rugosa lipase: its immobilization, characterization, and application in resolution of racemic ibuprofen. Applied Biochemistry and Biotechnology, 177(1), 36–47.
Wanasundara, U. N., & Shahidi, F. (1998). Concentration of omega-3 polyunsaturated fatty acids of marine oils using Candida cylindracea lipase: optimization of reaction conditions. Journal of the American Oil Chemists' Society, 75(12), 1767–1774.
Kahveci, D., & Xu, X. B. (2011). Repeated hydrolysis process is effective for enrichment of omega 3 polyunsaturated fatty acids in salmon oil by Candida rugosa lipase. Food Chemistry, 129(4), 1552–1558.
Tanaka, Y., Hirano, J., & Funada, T. (1992). Concentration of docosahexaenoic acid in glyceride by hydrolysis of fish oil with Candida cylindracea lipase. Journal of the American Oil Chemists' Society, 69(12), 1210–1214.
Domínguez de María, P., Sánchez-Montero, J. M., Sinisterra, J. V., & Alcántara, A. R. (2006). Understanding Candida rugosa lipases: an overview. Biotechnology Advances, 24(2), 180–196.
Sun, T., Pigott, G. M., & Herwig, R. P. (2002). Lipase-assisted concentration of n-3 polyunsaturated fatty acids from viscera of farmed Atlantic Salmon (Salmo salar L.). Journal of Food Science, 67, 130–136.
Okada, T., & Morrissey, M. T. (2008). Production of n-3 polyunsaturated fatty acid concentrate from sardine oil by immobilized Candida rugosa lipase. Journal of Food Science, 73(3), C146–C150.
Kahveci, D., & Xu, X. B. (2011). Enhancement of activity and selectivity of Candida rugosa lipase and Candida antarctica lipase A by bioimprinting and/or immobilization for application in the selective ethanolysis of fish oil. Biotechnology Letters, 33(10), 2065–2071.
Liu, Y., Zhou, H., Wang, L. Y., & Wang, S. H. (2016). Stability and catalytic properties of lipase immobilized on chitosan encapsulated magnetic nanoparticles cross-linked with genipin and glutaraldehyde. Journal of Chemical Technology and Biotechnology, 91(5), 1359–1367.
Liu, Z., Ho, S. H., Hasunuma, T., Chang, J. S., Ren, N. Q., & Kondo, A. (2016). Recent advances in yeast cell-surface display technologies for waste biorefineries. Bioresource Technology, 215, 324–333.
Andreu, C., & Del Olmo, M. L. (2018). Yeast arming systems: pros and cons of different protein anchors and other elements required for display. Applied Microbiology and Biotechnology, 102(6), 2543–2561.
Su, G., Zhang, X., & Lin, X. (2010). Surface display of active lipase in Pichia pastoris using Sed1 as an anchor protein. Biotechnology Letters, 32(8), 1131–1136.
Moura, M. V., da Silva, G. P., Machado, A. C., Torres, F. A., Freire, D. M., & Almeida, R. V. (2015). Displaying lipase B from Candida antarctica in Pichia pastoris using the yeast surface display approach: prospection of a new anchor and characterization of the whole cell biocatalyst. PLoS One, 28, e0141454.
Pan, X. X., Xu, L., Zhang, Y., Xiao, X., Wang, X. F., Liu, Y., Zhang, H. J., & Yan, Y. J. (2012). Efficient display of active Geotrichum sp. lipase on Pichia pastoris cell wall and its application as a whole-cell biocatalyst to enrich EPA and DHA in fish oil. Journal of Agriculture and Food Chemistry, 60(38), 9673–9679.
Han, Z. L., Han, S. Y., Zheng, S. P., & Lin, Y. (2009). Enhancing thermostability of a Rhizomucor miehei lipase by engineering a disulfide bond and displaying on the yeast cell surface. Applied Microbiology and Biotechnology, 85(1), 117–126.
Li, W., Shi, H., Ding, H., Wang, L., Zhang, Y., Li, X., & Wang, F. (2015). Cell surface display and characterization of Rhizopus oryzae lipase in Pichia pastoris using Sed1p as an anchor protein. Current Microbiology, 71(1), 150–155.
Washida, M., Takahashi, S., Ueda, M., & Tanaka, A. (2001). Spacer-mediated display of active lipase on the yeast cell surface. Applied Microbiology and Biotechnology, 56(5-6), 681–686.
Dai, M., Ji, C., Wang, X., Zhi, X., Shao, H., Xu, L., & Yan, Y. (2012). Cell surface display of Thermomyces lanuginosus lipase in Pichia pastoris and its characterization. Wei Sheng Wu Xue Bao (Chinese), 52, 857–865.
Jiang, Z. B., Song, H. T., Gupta, N., Ma, L. X., & Wu, Z. B. (2007). Cell surface display of functionally active lipases from Yarrowia lipolytica in Pichia Pastoris. Protein Expression and Purification, 56(1), 35–39.
Hoover, D. M., & Lubkowski, J. (2002). DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Research, 30(10), e43.
Xiong, A. S., Yao, Q. H., Peng, R. H., Duan, H., Li, X., Fan, H. Q., Cheng, Z. M., & Li, Y. (2006). PCR-based accurate synthesis of long DNA sequences. Nature Protocols, 1(2), 791–797.
Xiao, X., Liu, Z. Y., Chen, Y., Wang, G. L., Li, X., Fang, Z. G., Huang, S. S., Liu, Z. M., Yan, Y. J., & Xu, L. (2015). Over-expression of active Candida rugosa lip1 in Pichia pastoris via high cell-density fermentation and its application to resolve racemic ibuprofen. Biocatalysis and Biotransformation, 33(5-6), 260–269.
Eaton, D., Rodriguez, H., & Vehar, G. A. (1986). Proteolytic processing of human factor VIII. Correlation of specific cleavages by thrombin, factor Xa, and activated protein C with activation and inactivation of factor VIII coagulant activity. Biochemistry, 25(2), 505–512.
Laemmli, U. K., & Favre, M. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227(5259), 680–685.
Towbin, H., Staehlin, T., & Gordon, J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedures and some applications. Proceedings of the National Academy of Sciences of the United States of America, 76, 4350–4354.
Kouker, G., & Jaeger, K. E. (1987). Specific and sensitive plate assay for bacterial lipases. Applied and Environmental Microbiology, 53(1), 211–213.
Kojima, Y., & Shimizu, S. (2003). Purification and characterization of the lipase from Pseudomonas fluorescens HU380. Journal of Bioscience and Bioengieering, 96(3), 219–226.
Brocca, S., Schmidt-Dannert, C., Lotti, M., & Alberghina, L. (1998). Design, total synthesis, and functional overexpression of the Candida rugosa lip1 gene coding for a major industrial lipase. Protein Science, 7(6), 1415–1422.
Chang, S. W., Shieh, C. J., Lee, G. C., & Shaw, J. F. (2006). Codon optimization of Candida rugosa LIP1 gene for improving expression in Pichia pastoris and biochemical characterization of the recombinant LIP1 lipase. Journal of Agriculture and Food Chemistry, 54(3), 815–822.
López, N., Pernas, M. A., Pastrana, L. M., Sánchez, A., Valero, F., & Rúa, M. L. (2004). Reactivity of pure Candida rugosa lipase isoenzymes (Lip1, Lip2, and Lip3) in aqueous and organic media, Influence of the isoenzymatic profile on the lipase performance in organic media. Biotechnology Progress, 20, 65–73.
Fan, Y. L., Wu, G. Y., Su, F., Li, K., Xu, L., Han, X. T., & Yan, Y. J. (2016). Lipase oriented-immobilized on dendrimer-coated magnetic multi-walled carbon nanotubes toward catalyzing biodiesel production from waste vegetable oil. Fuel, 178, 172–178.
Yilmaz, E., Can, K., Sezgin, M., & Yilmaz, M. (2011). Immobilization of Candida rugosa lipase on glass beads for enantioselective hydrolysis of racemic naproxen methyl ester. Bioresource Technology, 102(2), 499–506.
Zheng, M. M., Dong, L., Lu, Y., Guo, P. M., Deng, Q. C., Li, W. L., Feng, Y. Q., & Huang, F. H. (2012). Immobilization of Candida rugosa lipase on magnetic poly (allyl glycidyl ether-co-ethylene glycol dimethacrylate) polymer microsphere for synthesis of phytosterol esters of unsaturated fatty acids. Journal of Molecular Catalysis B: Enzymatic, 74(1-2), 16–23.
Liang, X. X., Wang, B. B., Sun, Y. F., Lin, Y., Han, S. Y., Zheng, S. P., & Cui, T. B. (2013). Quantitative evaluation of Candia antarctica lipase B displayed on the cell surface of a Pichia pastoris based on an FS anchor system. Biotechnology Letters, 35(3), 367–374.
Liu, Y., Zhang, R., Lian, Z., Wang, S., & Wright, A. T. (2014). Yeast cell surface display for lipase whole cell catalyst and its applications. Journal of Molecular Catalysis B: Enzymatic, 106, 17–25.
Acknowledgments
All authors would like to thank the Analytical and Testing Center of Huazhong University of Science and Technology for their valuable assistances in the gas chromatography–mass spectrometry measurements.
Funding
This research was funded by the National Natural Science Foundation of China (grant number: NSFC31170078), the National High Technology Research and Development Program of China (grant numbers: 2013AA065805 and 2014AA093510), the National Natural Science Foundation of Hubei Province (grant number: 2015CFA085), and the Fundamental Research Funds for HUST (grant numbers: 2014QN119 and 2014NY007).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PNG 175 kb)
Rights and permissions
About this article
Cite this article
Xu, L., Xiao, X., Wang, F. et al. Surface-Displayed Thermostable Candida rugosa Lipase 1 for Docosahexaenoic Acid Enrichment. Appl Biochem Biotechnol 190, 218–231 (2020). https://doi.org/10.1007/s12010-019-03077-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-019-03077-z