Abstract
The intensive production of polycyclic aromatic hydrocarbons by anthropogenic activities is a serious environmental problem. Therefore, new bioremediation methods are required to avoid widespread contamination. In this work, Serratia sp. AC-11 strain isolated from a tropical peat was selected for immobilization into chitosan beads, which were employed in the biodegradation of fluoranthene. The sizes of the produced beads were relatively uniform with an average diameter of 3 mm. The material was characterized by SEM and FT-IR, confirming the cells immobilization and the protective barrier formed by the chitosan surrounding the biomass. The immobilized bacteria were able to degrade 56% of fluoranthene (the initial concentration was 100 mg L−1) in just 1 day at twice the degradation rate achieved by free-living cells. Furthermore, the immobilized bacteria showed excellent removal during five reuse cycles, from 76% to 59% of biodegradation. These results showed the potential of this approach for remediation of contaminated sites.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Polycyclic aromatic hydrocarbons (PAHs) are a class of compounds with aromatic fused rings that possess toxic properties, such as carcinogenicity and bioaccumulation. They are ubiquitous environmental compounds considered persistent organic pollutants (POPs) due to their low bioavailability and high hydrophobicity which makes them difficult to treat and eliminate. The importance of minimizing the release of PAHs and their dangerous effectsresulted in a list of 16 PAHs designated as priority pollutants by the Unites States Environmental Protection Agency (USEPA), including fluoranthene [1, 2].
A four-ring compound that is the major component of petroleum sludge [3], one of the commonest PAH in the environment and has been reported as a recalcitrant, genotoxic, cytotoxic, mutagenic, and potentially carcinogenic compound [4]. Therefore, the removal of fluoranthene by different techniques has been studied due to its spreading and toxicity, including biodegradation processes by bacteria [5], yeasts [6], and fungi [7].
Remediation of PAHs contaminated sites is conducted to minimize risks to the environment and human health. Compared to the conventional physical–chemical treatments, the use of microorganisms has been highly efficient, stable, economic, and eco-friendly [8, 9]. However, finding strains capable of degrading PAHs is quite difficult, since high concentrations of these compounds and environmental conditions can affect the microbial growth and survival [10]. Therefore, to overcome this problem and improve the bioremediation efficiency, the use of microorganisms immobilized in supports showed several advantages over the use of free cells [11, 12].
There are different types of cells immobilization, but it is important to note that bacterial entrapment provides greater protection against changes of environmental conditions, as well as improved viability of the employed microorganisms [13]. In addition, this technique is a low cost, fast, and easily applicable method for biodegradation processes [14].
A diversity of natural and synthetic polymers are available for immobilization of microorganisms, as nylon [15], silica gel, silica xerogel, chitosan [16], polyurethane foam, sodium alginate, polyacrylamide, and agar [17]. However, the preference for natural polysaccharides lies on their abundance in nature, economical viability and low toxicity in comparison with those of synthetic origin [18]. Among these supports, chitosan is known for its biodegradability, biological compatibility, and non-toxic properties [19].
Chitosan is one of the most abundant polysaccharide in nature, and a linear co-polymer industrially obtained from the deacetylation of chitin, which is found in the exoskeletons of crustaceans and insects [13]. The different functional groups present in the chemical structure of chitosan, and the possibility of modify them enables the production of biomaterials with different morphological configurations, including beads, films, membranes, gels, and powders [20]. Moreover, this material can be used in many fields as medicine [21], food [22], environment management [23], agriculture [24], and others.
Previous studies have reported the application of microorganisms immobilized into chitosan beads for the degradation of phenol [25], hexadecane [13], n-hexadecane [11], and oil [26]. However, there are no reports of the employment of this method for PAHs biodegradation.
Therefore, a new bioproduct containing a bacterial strain, Serratia sp. AC-11, isolated from a tropical peat and entrapped into chitosan beads was developed and evaluated for fluoranthene biodegradation. A comparison between the rates of fluoranthene consumption using entrapped and free bacteria was carried out, including the reuse of the obtained beads in high efficiency for future applications in environmental remediation.
Materials and Methods
Reagents, Solvents, and Culture Media
Fluoranthene (98%) was acquired from Sigma-Aldrich (Steinheim, Germany), anthracene (99%) was obtained from Acros Organics (New Jersey, USA) and chitosan (74% deacetylated) was obtained from Polymar (Fortaleza, Brazil). Salts, reagents, and solvents were obtained from Sigma-Aldrich (Steinheim, Germany) and Synth (São Paulo, Brazil). Whereas, agar and nutrient broth was purchased from Himedia Laboratories (Mumbai, India). The Taq DNA polymerase and others molecular supplies were obtained from Invitrogen (CA, USA) and Integrated DNA Technologies (IL, USA).
Isolation and Molecular Identification of Serratia sp. AC-11
The Serratia sp. AC-11 strain was isolated from a peatland in Santo Amaro das Brotas, Sergipe State, Brazil (N 07° 20.729′/ W 880° 35.113′) using fluoranthene as sole carbon source, as described by Madueno et al. [27]. The molecular identification was performed using the 16S rRNA gene sequence. The bacterial genomic DNA was extracted and purified using an alkaline lysis protocol, then the 16S rRNA was amplified by polymerase chain reaction (PCR) using 968F (5′-AACGCGAAGAACCTTAC-3′) as the forward primer and 1401R (5′-CGGTGTGTACAAGACCC-3′) as the reverse primer.
Amplification reactions were prepared in a final volume of 50 μL using 50 ng of DNA, 5 μL of buffer, 3 mM of MgCl2, 25 μM of each dNTP, 0.4 μM of each primer, 2 U of Taq DNA polymerase (Invitrogen), 5% of dimethylsulfoxide (DMSO), and sterile deionized water. The amplification was carried out as follows: one cycle at 95 °C for 3 min, followed by 35 cycles at 94 °C for 40 s, 56 °C for 40 s, 72 °C for 40 s, and a final extension step at 72 °C for 3 min. The PCR product was analyzed by electrophoresis, purified using a GE Healthcare kit and then sequenced by the sequencing service of São Paulo University (USP).
The forward nucleotide sequences were determined using a Big Dye Terminator v. 3.1 Cycle Sequencing Kit and an automated ABI 3730 DNA Analyzer (Applied Biosystems). The sequence was viewed, edited, and aligned using Geneious v. 6.1.8 software. Numts and stop codons were absent in the sequence. The determined sequence was compared with the GenBank using the BLAST/NCBI software. The neighbor-joining phylogenetic tree was constructed in MEGA v. 6.0 using p-distances, complete deletion, and bootstrapping with 1000 replications. In addition, the sequence was then deposited in GenBank.
Effect of Chitosan on the Bacterial Strain Serratia sp. AC-11
The effect of the chitosan solution on the bacterial strain was evaluated according to Angelim et al. [26] with modifications. A bacterial suspension (100 μL) with an optical density of 0.1 at 600 nm (108 CFU mL−1) was used to inoculate tubes with 1 mL of a solution containing 1% of acetic acid and 2, 3, or 4% of chitosan. After 24 h, aliquots of these cultures were transferred to nutrient agar medium and incubated at 32 °C for 24 h to determine the effect of the tested solution on the bacterial growth.
Chitosanase Production
Chitosanase production was evaluated following the protocol described by Choi et al. [28]. The bacterial strain was grown in nutrient medium for 24 h at 32 °C, and subsequently, 15 μL of the culture was transferred to the surface of the chitosanase detection agar. Chitosanase production was identified by the formation of a hydrolysis halo around the colony after incubation for 48 h at 32 °C. All experiments were conducted in triplicate.
Entrapment of Bacteria into Chitosan Beads
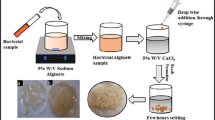
The entrapment procedure was performed following the protocol described by Costa et al. [13] with some modifications. The bacterial strain was grown for 24 h in 30 mL of nutrient liquid medium at 32 °C and 150 rpm. Subsequently, the cells were harvested by centrifugation for 10 min at 4 °C and 5000 rpm.
Then, the bacterial pellet was resuspended in 0.5 mL of sterile 0.9% NaCl aqueous solution and added to 30 mL of a 4% chitosan solution. This preparation was previously performed with 1% of acetic acid in distilled water, which was then sterilized for 10 min at 120 °C in an autoclave. The suspension was stirred for 10 min and kept standing for 10 min. After that, this mixture was dripped into a 1% sodium tripolyphosphate (TPP) solution (pH 9) for beads formation using a sterile burette. After stirring for 40 min, the beads were removed from the solution and washed three times for 10 min with 200 mL of sterile 0.9% NaCl aqueous solution.
The consistency of the beads was improved by treating them with 0.2% of glutaraldehyde (previously sterilized at 120 °C for 10 min) for 1 h and then washed three times with 0.9% (w/v) NaCl aqueous solution. The beads were then ready for use in the biodegradation assays. Moreover, to determine the number of viable cells inside the beads, 1 g of beads was macerated in 10 mL of 0.9% (w/v) NaCl aqueous solution, and serially diluted on nutrient agar plates for determination of colony-forming units (CFU/g). The entire procedure was carried out in a biological safety cabinet.
Characterization of the Beads with Immobilized Bacteria
FT-IR
FT-IR spectra of the chitosan samples and the chitosan beads with immobilized bacteria were obtained using KBr tablets (1:100, sample:KBr) in a Varian Model 640 IR spectrometer. The spectra were acquired from 4000 to 400 cm−1 with 32 scans per sample and a resolution of 4 cm−1.
SEM
Photomicrographs of the chitosan spheres with and without the immobilized bacteria were obtained. The spheres were previously dried in oven at 35 °C for 48 h and coated with gold (ion sputter, E-1010, Hitachi, Japan). The images were obtained using a Hitachi Model TM 3000 scanning electron microscope operated under high vacuum with an acceleration voltage of 15 kV and a filament current of 1850 mA.
Fluoranthene Biodegradation
Biodegradation assays were performed with free-living and immobilized cells. For the free-living cells, 200 μL of a culture with 108 CFU mL−1 was used as inoculum, whereas 2 g of chitosan beads was used in the immobilized assays. Both experiments were carried out in 50-mL Erlenmeyer flasks containing 20 mL of nutrient liquid medium with 100 mg L−1 of fluoranthene for 1 to 10 days of biodegradation. In addition, chitosan beads without immobilized bacteria were employed as a control of the fluoranthene loss by adsorption and/or sublimation. All experiments were performed in an orbital shaker at 150 rpm and 32 °C in triplicate.
The assay containing the immobilized chitosan beads was filtered in a Büchner apparatus to separate the beads and the reactional medium. The beads were washed with ethyl acetate (10 mL) and employed for viable cells determination. For the free-living bacterial assays, 10 mL of ethyl acetate was added to the Erlenmeyer flask containing the bacterial suspension and the resulting mixture was vigorously stirred for 30 min. The obtained liquid phases (both immobilized and free-living cells) were centrifuged at 5000 rpm for 10 min at 4 °C, followed by liquid–liquid extraction with 3 steps of 20 mL of ethyl acetate each in a separatory funnel. Anhydrous Na2SO4 for drying was added to the organic phase, which was then filtered and evaporated under vacuum. The residue was resuspended in ethyl acetate.
Reuse Cycle of the Immobilized Biocatalyst
The chitosan beads with immobilized bacteria were tested for five reuse cycles of 5 days each in fluoranthene biodegradation. The same procedure described in “Fluoranthene Biodegradation.” was followed. However, the beads filtered in the Büchner apparatus after the incubation period of 5 days were immediately collected with a sterile spatula and transferred to a new liquid medium containing 100 mg L−1 of fluoranthene.
Determination of Fluoranthene Degradation by GC-FID
The quantitative analyses of fluoranthene were carried out in a Shimadzu GC 2010 gas chromatograph equipped with an AOC 20i auto-injector, a flame ionization detector (FID), and a DB5 column (J&W Scientific, 30 m × 0.25 mm × 0.25 μm). Hydrogen was used as the carrier gas at a flow rate of 1.0 mL min−1. The oven program initiated at 100 °C and maintained in this condition for 1 min, then a temperature increase at a rate of 8 °C min−1 up to 280 °C was employed, and this final temperature was kept for 7.5 min. A split injection ratio of 5 was used and the injector temperature was 250 °C.
An analytical curve was obtained employing solutions of 50, 350, 650, 950, and 1250 mg L−1 of fluoranthene with 1000 mg L−1 of anthracene as internal standard. Moreover, all the solutions were injected in triplicate and a linear equation was obtained: Fluoranthene concentration = {[(Fluoranthene area/ Anthracene area) + 0.0267] / 0.001}, with a R2 of 0.998.
A control experiment was carried out employing chitosan beads without immobilized bacteria, and a method recovery of 96.3% was observed. Therefore, only 3.7 ± 0.8% % of the fluoranthene reduction can be attributed to the absorption/adsorption in the chitosan beads.
Results and Discussion
Molecular Identification
The Serratia sp. AC-11 was isolated from a tropical peat using fluoranthene as sole carbon source, showing the potential of this environment as source of important biocatalysts. The peat of Santo Amaro das Brotas in Sergipe State has been studied in terms of chemical composition [29] and adsorption of organic [30, 31] and inorganic compounds [32, 33], but few investigations about its microbiota were carried out. Although peatlands are known for containing large microbial populations with a wide metabolic diversity, which should be evaluated for potential biotechnological applications [34, 35].
The partial 16S rRNA gene sequence of the AC-11 strain was determined and deposited in GenBank under the accession number KP264565. A BLAST search revealed that the determined sequence had 100% identity with 16S sequences from different strains of Serratia sp. (KX215147.1, KU555937.1, KU535676.1, KT183566.1 and KX527573.1), thus this bacteria was identified as Serratia sp. AC-11. The neighbor-joining tree showed the clade of Serratia in which AC-11 is included (Fig. 1).
The Serratia genus is ubiquitous in nature and has been isolated from water, soil, animals (including man), and plants [36]. Previous studies have shown the ability of Serratia sp. to degrade compounds such as methyl parathion, p-nitrophenol [37], chitin [38], organic compounds present in sewage sludge (such as tributryin), phosphoric acid [39], pulp and paper mill effluent [40]. In the case of PAHs, Machin-Ramirez et al. [41] described the capacity of Serratia marcescens to consume benzo[a]pyrene by almost 40%.
Effect of Chitosan on Bacterial Growth and Chitosanase Production
The evaluation of bacterial resistance to the chitosan solution revealed that Serratia sp. AC-11 tolerated up to 4% of chitosan during 24 h. Therefore, this concentration was employed to produce stable beads suitable for orbital stirring during the biodegradation assays.
The next step was the determination of chitosanase activity by the employed strain, since this is an undesirable characteristic for the reuse of the chitosan beads. The evaluation of enzymatic activity was performed by inoculating the bacteria onto chitosan agar plates in order to observe any halo formation after 48 h. The absence of a halo confirmed that no chitosanase was produced, confirming that the strain fulfilled the requirements for immobilization into the chitosan.
Immobilization of Cells into Chitosan Beads
The beads containing immobilized bacteria were relatively uniform with an average diameter of 3.0 ± 0.3 mm and a content of 1 × 107 CFU/g of beads. However, disintegration of the beads occurred at the end of the fifth day of the biodegradation assays.
To understand the motive of the beads instability, pH measurements of the culture medium were performed at the end of the experiment, since chitosan is readily soluble at acidic pH. No substantial change in pH was observed between the initial and final days, with values of 7.0 ± 0.2 and 6.81 ± 0.4, respectively. Moreover, a new investigation of chitosanase production was conducted. At the end of the fifth day of the experiment, an aliquot of the bacterial strain was inoculated on chitosan agar and incubated for 48 h at 32 °C. After this period, the formation of a hydrolysis halo around the inoculum was observed, confirming the presence of chitosanase activity under these experimental conditions.
Probably, the prolonged assay with immobilized cells induced the expression of a gene responsible for chitosan consumption. According to Gupta et al. [42], the production of most hydrolytic enzymes (including chitosanases) is controlled by metabolic induction and repression. Therefore, inductive expression of the enzyme occurred under these experimental conditions. The digestibility of chitosan or any other immobilizing support is interesting only when the function of the support is to carry cells to a specific site of action [43], which unfortunately was not the case.
Barreto et al. [11] also observed disintegration of chitosan beads during hexadecane biodegradation due to chitosanase production by trapped bacterial cells. To overcome this problem, it was necessary to treat the beads with a cross-linker (glutaraldehyde) in order to increase their stability. Glutaraldehyde is a low cost compound and the most widely used cross-linker, which can increase the strength of the support by forming a spatial grid structure [44], which may increase the resistance of the matrix to chitosanase.
This compound quickly reacts with amine groups at near-neutral pH generating thermally and chemically stable cross-links and increasing stability and reusability of supported biocatalysts [44, 45]. The covalent linkages between the amine groups of chitosan and the aldehyde groups of glutaraldehyde are stable in wide ranges of pH, temperature and resistant to enzymatic hydrolysis [11].
In this study, the beads were treated with 0.2% of glutaraldehyde for 1 h immediately after the cells immobilization process, which allowed the microorganism cells to survive. The cross-linking process generated a color change from beige to orange in the beads, confirming the reaction of the aldehyde groups of glutaraldehyde with the amine groups of chitosan [46].
A CFU determination was performed after this procedure and 3.4 × 106 CFU/g of beads was determined. This result showed that cell losses occurred due to the cross-linking procedure, although these losses did not affect the use of the beads in the biodegradation tests. After 10 days of biodegradation experiment, the cross-linked beads remained intact and undamaged, differently from the beads without this treatment, confirming the resistance promoted by the employed procedure. The integrity of the beads is essential for their reuse under controlled conditions in large scale processes, where characteristics such as stability and rigidity are essential [13].
Characterization of Chitosan Beads
SEM
The use of SEM enabled differentiation between the beads with and without immobilized bacteria (Fig. 2). The micrographs showed that the bacteria were firmly immobilized into chitosan and that the presence of the bacteria generated a wrinkled bead surface, which promotes an increased superficial area. This wrinkle effect was enhanced by the employment of glutaraldehyde, facilitating the colonization and production of microbial biomass, and resulting in biofilm formation [25]. The porosity observed in the beads surface allowed better adsorption of the cells onto the chitosan and improves the rate of mass transfer of fluoranthene, oxygen, and water to the cells [12].
Images of chitosan beads: (A1) bead without bacteria at 100 x of magnification; (A2) bead without bacteria at 1000 x of magnification; (B1) bead with immobilized bacteria at 100 x of magnification; (B2) bead with immobilized bacteria at 1000 x of magnification; (C1) bead with immobilized bacteria employing glutaraldehyde at 100 x of magnification; and (C1) bead with immobilized bacteria employing glutaraldehyde at 1000 x of magnification
FT-IR
FT-IR spectra for the Serratia sp. AC-11 strain, chitosan, and chitosan beads with immobilized bacteria are shown in Fig. 3 . The Serratia sp. AC-11 spectrum showed a broad absorption band centered at 3417 cm−1 and a shoulder at 3280 cm−1, attributed to peaks overlapping associated with hydrogen bonds of O–H groups from lipids and carbohydrate structures, and N–H bonds of proteins. Bands at 2962–2855 cm−1 were attributed to the stretching of C–H bonds in aliphatic structures, as confirmed by the asymmetric folding bands at 1459 cm−1 and 1387 cm−1, both related to the presence of lipids. The presence of a shoulder at 1743 cm−1 was due to the C=O stretching of aliphatic esters associated with the present lipid structures. Bands at 1655 cm−1 and 1548 cm−1 were attributed to amide C=O stretching and N–H folding, respectively. It is important to note that these bands are present in almost all FT-IR spectra of bacteria [47]. An absorption band at 1238 cm−1 was attributed to the symmetric stretching of phosphate groups (P=O) characteristic of nucleic acids [48] and a broadband centered at 1075 cm−1 was associated with complex vibrational modes of C–O bonds of polysaccharides [47].
In the chitosan infrared absorption spectrum, an intense broadband at 3435 cm−1 was related to the peaks overlapping of O–H and N–H hydrogen bonds. Bands at 2968–2857 cm−1 were assigned to aliphatic C–H stretching and were confirmed by asymmetric and symmetric C–H bond folding bands at 1465 cm−1 and 1383 cm−1, respectively. Bands at 1654 cm−1 and 1315 cm−1 were due to the amide bonds C=O and C–N stretching, respectively, while a band at 1565 cm−1 was assigned to the folding of the amine N–H bond. An intense band at 1080 cm−1 and a shoulder at 1158 cm−1 were attributed to the symmetric and asymmetric stretching of C–O–C from the glucopyranoside ring, respectively [49].
The infrared spectrum of the chitosan beads with immobilized bacteria did not show any changes in bond peaks that could indicate any specific chemical interactions between the bacterial strain and the support. The spectrum showed similar features in terms of bands overlapping with increased intensity of the 1651 cm−1 and 1565 cm−1 bands related to the contribution of amides, and a broadening of the band at 3435 cm−1 attributed to O–H and N–H groups in both samples.
Therefore, it can be inferred that the chitosan effectively surrounded the bacterial biomass, which was captured by a physical process during immobilization. Probably, an electrostatic interaction that immobilizes the cells without a strong chemical effect occurred, since chitosan is positively charged and the bacterial strain presents a negative surface [50].
Fluoranthene Biodegradation
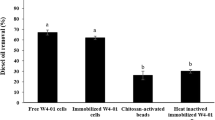
Firstly, the chitosan beads were tested for the adsorption of fluoranthene in the absence of immobilized bacteria (control) for 10 days. A removal of only 3.7 ± 0.8% of fluoranthene was observed and considered negligible for this study. The biodegradation assays were performed for 1, 5 and 10 days. In just 1 day, the immobilized cells were able to degrade 56 ± 1.9% of fluoranthene, followed by a progressive increase of the biodegradation percentage to 76 ± 2.1% and 84 ± 3.4% for 5 and 10 days, respectively.
A comparison of fluoranthene biodegradation using free-living cells of Serratia sp. AC-11 (108 CFU mL−1) under similar experimental conditions showed that the use of immobilized cells resulted in a 100% increase in the biodegradation of this PAH for the three tested periods (Fig. 4 ). These findings showed that the microbial system formed by Serratia sp. AC-11 entrapped into chitosan beads was more effective in the fluoranthene biodegradation than free cells of the same strain.
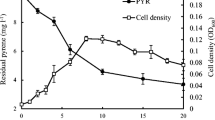
The growth curve obtained for the free cells presented a maximum value followed by a decline phase, as described in the literature [51]. Whereas immobilized cells were able to continue multiplying during longer incubation periods without reaching this decline phase and maintaining high bacterial mass (Fig. 5 ). Therefore, the beads provided an appropriated environment for cellular growth and metabolism, resulting in increased microbial activity and stability, which enables the high degradation activity for longer periods than by free cells [25, 52, 53].
The increase of bacterial biomass in the beads analyzed by the number of viable cells (CFU/g) explains the higher rate of biodegradation during the biodegradation period (Fig. 6 ). The fluoranthene biodegradation rate increased until the fifth day, but this rate gradually stabilized whereas the number of viable cells continued to raise. This phenomenon has also been reported by Hsieh et al. [25] and Banerjee et al. [54], who explained that the larger amounts of entrapped cells may lead to saturation within the beads, decreasing the mass transfer efficiency and consequently the biodegradation rates.
Fluoranthene may repress microbial metabolism by toxic effects, inhibiting the biodegradation by free cells due to lower efficiency and/or cell death. In the immobilization process employed in this study, the physical barrier created by chitosan avoided the direct contact of the cells with the pollutant, providing protection against environmental stress and increasing the microbial survival and biomass. According to Sarma and Pakshirajan [55], immobilized cell systems can be used efficiently in the biodegradation of highly hydrophobic compounds without any inhibitory effect of contaminants present in the outside of the beads.
Reuse of Chitosan Beads with Immobilized Bacteria
The use of Serratia sp. AC-11 immobilized into chitosan beads was further evaluated in continuous biodegradation cycles. Cycles of 5 days each were employed, since the biodegradation achieved after 10 days (84%) was similar to that obtained after 5 days (76%). This reaction period was chosen considering energy consumption, process time and others cost-effective factors, which would be important in future applications on large scale. The obtained results of biocatalyst reuse are shown in Fig. 7 .
The fluoranthene biodegradation continued throughout five continuous reuse cycles, in which was observed a 18% difference between the biodegradation rates of the first cycle (77%) and the fifth one (59%). The first three cycles presented non-significant differences (< 3%) in the rates of biodegradation, and most of the beads remained intact at the end of the fifth reuse cycle with 1.9 × 106 CFU/g of beads. The chitosan beads conferred great stability to the cells and enabled a high continuous degradation activity. It is important to note the good integrity of the beads at the end of the experiment, which is an essential characteristic for their reuse.
The reusability of immobilized cells is one of the most important aspects of this method, especially when the chosen support is inert and has high mechanical resistance [56], as reported for chitosan [57]. The produced beads with immobilized bacteria were suitable for continuous reuse cycles with satisfactory degradation of fluoranthene and stable viable cells number. The reuse of immobilized cells reduces operational costs and improves efficiency by the activity of the bacteria over longer reaction periods [13].
Several studies concerning the biodegradation of fluoranthene by free bacteria have been reported. Mishra et al. [3] studied the degradation of 200 mg L−1 of fluoranthene by two bacterial strains, solely and in consortium. The use of Rhodococcus sp. NJ2 resulted in 64% degradation, while 51% of degradation was achieved with Pseudomonas aeruginosa PSA5. The use of the two bacterial strains in consortium resulted in 87% degradation in 10 days [3]. Whereas Kafilzadeh et al. [51] achieved 73% of degradation in 10 days of incubation using a strain of Bacillus circulans. In another study, Kumar et al. [8] isolated four bacterial strains and, the best of them (Acinetobacter sp. PSM11 strain) achieved 61% degradation of this PAH (at 100 mg L−1) in 7 days. In addition, Xu et al. [58] reported a bacterial strain, Herbaspirillum chlorophenolicum FA1, which achieved 58.5% degradation of 20 mg L−1 fluoranthene in 30 days. These results can be compared with the fluoranthene degradation rates obtained in this study, in which the immobilized bacteria was capable of degrading 56% of fluoranthene in just 1 day and 84% in 10 days, representing a considerable improvement over the earlier findings.
According to Tao et al. [52], the use of free microorganisms for bioremediation of contaminated sites can fail because the inoculants must be able to overcome biotic and abiotic stresses in the environment that they were introduced. The advantages of immobilized microorganisms rather than free bacteria include better adaptation, greater control of bioprocesses, lower susceptibility to contamination by undesirable microorganisms, greater resistance to extreme environmental conditions, possibility of reutilization and improved degradation efficiency [10, 13]. In addition, immobilized microorganisms can be used in bioreactors, increasing these biocatalysts applications. Sarma and Pakshirajan [55] investigated the use of Mycobacterium frederiksbergense NRRL B-24126 immobilized in calcium alginate for complete degradation of pyrene with a high degradation rate of 250 mg L−1 day−1. These findings identified potential applications of immobilized cells, as developed and reported in our work.
Conclusion
The successful immobilization of the bacterial strain Serratia sp. AC-11 into chitosan beads was innovative in at least three aspects. The isolation of a strain from a peatland which had never been previously explored for biotechnological purposes. The high fluoranthene biodegradation rate that was achieved using the beads with immobilized bacteria, showing better results than earlier findings and, finally, the reusability of the modified beads during continuous cycles, reducing costs by the great stability of the viable cells number and integrity of the beads.
The support chitosan was a favorable environment for microbial activity and cellular growth. In addition, interesting characteristics such as great surface area, porosity, and a physical barrier that avoids the direct contact of the bacteria with the pollutant allowed an increased bacterial biomass during the biodegradation process. The high number of viable cells into the beads enabled a high rate of fluoranthene removal from the medium when compared to free cells.
This novel bioproduct developed in this study is a low-cost, efficient, eco-friendly, practical, and potentially suitable for an industrial scale, offering an innovative approach for bioremediation of areas contaminated with fluoranthene.
References
Pugazhendi, A., Qari, H., Basahi, J. M. A., Godon, J. J., & Dhavamani, J. (2017). Role of a halothermophilic bacterial consortium for the biodegradation of PAHs and the treatment of petroleum wastewater at extreme conditions. International Biodeterioration and Biodegradation, 121, 44–54.
Singh, P., & Tiwary, B. N. (2017). Optimization of conditions for polycyclic aromatic hydrocarbons (PAHs) degradation by Pseudomonas stutzeri P2 isolated from Chirimiri coal mines. Biocatalysis and Agricultural Biotechnology, 10, 20–29.
Mishra, S., Singh, S. N., & Pande, V. (2014). Bacteria induced degradation of fluoranthene in minimal salt medium mediated by catabolic enzymes in vitro condition. Bioresource Technology, 164, 299–308.
Ahmed, R. Z., & Ahmed, N. (2014). Effect of yeast extract on fluoranthene degradation and aromatic ring dioxygenase expressing bacterial community structure of a fluoranthene degrading bacterial consortium. International Biodeterioration and Biodegradation, 88, 56–61.
Ma, Y. L., Lu, W., Wan, L. L., & Luo, N. (2015). Elucidation of fluoranthene degradative characteristics in a newly isolated Achromobacter xylosoxidans DN002. Applied Biochemistry and Biotechnology, 175(3), 1294–1305.
Kamyabi, A., Nouri, H., & Moghimi, H. (2017). Synergistic effect of Sarocladium sp. and Cryptococcus sp. co-culture on crude oil biodegradation and biosurfactant production. Applied Biochemistry and Biotechnology, 182(1), 324–334.
Birolli, W. G., Santos, D. A., Alvarenga, N., Garcia, A. C. F. S., Romão, L. P. C., & Porto, A. L. M. (2017). Biodegradation of anthracene and several PAHs by the marine-derived fungus Cladosporium sp. CBMAI 1237. Marine Pollution Bulletin, 129, 525–533.
Kumar, S., Upadhayay, S. K., Kumari, B., Tiwari, S., Singh, S. N., & Singh, P. K. (2011). In vitro degradation of fluoranthene by bacteria isolated from petroleum sludge. Bioresource Technology, 102(4), 3709–3715.
Jin, X., Tian, W. J., Liu, Q., Qiao, K. L., Zhao, J., & Gong, X. X. (2017). Biodegradation of the benzo a pyrene-contaminated sediment of the Jiaozhou Bay wetland using Pseudomonas sp. immobilization. Marine Pollution Bulletin, 117(1-2), 283–290.
Ontanon, O. M., Gonzalez, P. S., Barros, G. G., & Agostini, E. (2017). Improvement of simultaneous Cr(VI) and phenol removal by an immobilised bacterial consortium and characterisation of biodegradation products. New Biotechnology, 37, 172–179.
Barreto, R. V. G., Hissa, D. C., Paes, F. A., Grangeiro, T. B., Nascimento, R. F., Rebelo, L. M., Craveiro, A. A., & Melo, V. M. M. (2010). New approach for petroleum hydrocarbon degradation using bacterial spores entrapped in chitosan beads. Bioresource Technology, 101(7), 2121–2125.
Deng, F. C., Liao, C. J., Yang, C., Guo, C. L., & Dang, Z. (2016). Enhanced biodegradation of pyrene by immobilized bacteria on modified biomass materials. International Biodeterioration and Biodegradation, 110, 46–52.
Costa, S. P., Angelim, A. L., Sousa, M., & Melo, V. M. M. (2014). Vegetative cells of Bacillus pumilus entrapped in chitosan beads as a product for hydrocarbon biodegradation. International Biodeterioration and Biodegradation, 87, 122–127.
Bayat, Z., Hassanshahian, M., & Capello, S. (2015). Immobilization of microbes for bioremediation of crude oil polluted environments: a mini review. Open Microbiology Journal, 9, 48–54.
Sedarati, M. R., Keshavarz, T., Leontievsky, A. A., & Evans, C. S. (2003). Transformation of high concentrations of chlorophenols by the white-rot basidiomycete Trametes versicolor immobilized on nylon mesh. Electronic Journal of Biotechnology, 6, 104–114.
Rocha, L. C., de Souza, A. L., Rodrigues Filho, U. P., Campana Filho, S. P., Sette, L. D., & Porto, A. L. M. (2012). Immobilization of marine fungi on silica gel, silica xerogel and chitosan for biocatalytic reduction of ketones. Journal of Molecular Catalysis B: Enzymatic, 84, 160–165.
Hoskeri, R. S., Mulla, S. I., & Ninnekar, H. Z. (2014). Biodegradation of chloroaromatic pollutants by bacterial consortium immobilized in polyurethene foam and other matrices. Biocatalysis and Agricultural Biotechnology, 3(4), 390–396.
Crini, G. (2005). Recent developments in polysaccharide-based materials used as adsorbents in wastewater treatment. Progress in Polymer Science, 30(1), 38–70.
Dash, M., Chiellini, F., Ottenbrite, R. M., & Chiellini, E. (2011). Chitosan-a versatile semi-synthetic polymer in biomedical applications. Progress in Polymer Science, 36(8), 981–1014.
Cadogan, E. I., Lee, C. H., & Popuri, S. R. (2015). Facile synthesis of chitosan derivatives and Arthrobacter sp. biomass for the removal of europium(III) ions from aqueous solution through biosorption. International Biodeterioration and Biodegradation, 102, 286–297.
Dumont, V. C., Mansur, H. S., Mansur, A. A., Carvalho, S. M., Capanema, N. S., & Barrioni, B. R. (2016). Glycol chitosan/nanohydroxyapatite biocomposites for potential bone tissue engineering and regenerative medicine. International Journal of Biological Macromolecules, 93(Pt B), 1465–1478.
Sotelo-Boyas, M. E., Correa-Pacheco, Z. N., Bautista-Banos, S., & Corona-Rangel, M. L. (2017). Physicochemical characterization of chitosan nanoparticles and nanocapsules incorporated with lime essential oil and their antibacterial activity against food-borne pathogens. LWT- Food Science and Technology, 77, 15–20.
Wangpradit, R., & Chitprasert, P. (2014). Chitosan-coated Lentinus polychrous Lev.: integrated biosorption and biodegradation systems for decolorization of anionic reactive dyes. International Biodeterioration and Biodegradation, 93, 168–176.
Mukta, J. A., Rahman, M., Sabir, A. A., Gupta, D. R., Surovy, M. Z., & Islam, M. T. (2017). Chitosan and plant probiotics application enhance growth and yield of strawberry. Biocatalysis and Agricultural Biotechnology, 11, 9–18.
Hsieh, F. M., Huang, C., Lin, T. F., Chen, Y. M., & Lin, J. C. (2008). Study of sodium tripolyphosphate-crosslinked chitosan beads entrapped with Pseudomonas putida for phenol degradation. Process Biochemistry, 43(1), 83–92.
Angelim, A. L., Costa, S. P., Farias, B. C. S., Aquino, L. F., & Melo, V. M. M. (2013). An innovative bioremediation strategy using a bacterial consortium entrapped in chitosan beads. Journal of Environmental Management, 127, 10–17.
Madueno, L., Coppotelli, B. M., Alvarez, H. M., & Morelli, I. S. (2011). Isolation and characterization of indigenous soil bacteria for bioaugmentation of PAH contaminated soil of semiarid Patagonia, Argentina. International Biodeterioration & Biodegradation, 65(2), 345–351.
Choi, Y. J., Kim, E. J., Piao, Z., Yun, Y. C., & Shin, Y. C. (2004). Purification and characterization of chitosanase from Bacillus sp. strain KCTC 0377BP and its application for the production of chitosan oligosaccharides. Applied and Environmental Microbiology, 70(8), 4522–4531.
Romao, L. P. C., Lead, J. R., Rocha, J. C., de Oliveira, L. C., Rosa, A. H., Mendonca, A. G. R., & Ribeiro, S. A. (2007). Structure and properties of Brazilian peat: analysis by spectroscopy and microscopy. Journal of the Brazilian Chemical Society, 18(4), 714–720.
de Carvalho, P. H. V., de Jesus, A. M. D., Prata, V. M., Bezerra, D. S. S., Romao, L. P. C., & Navickiene, S. (2010). Tropical peat as a versatile material for solid-phase extraction of pesticides from medicinal plant Cordia salicifolia. Journal of the Brazilian Chemical Society, 21(4), 659–664.
Costa, A. S., Romao, L. P. C., Araujo, B. R., Lucas, S. C. O., Maciel, S. T. A., Wisniewski, A., & Alexandre, M. R. (2012). Environmental strategies to remove volatile aromatic fractions (BTEX) from petroleum industry wastewater using biomass. Bioresource Technology, 105, 31–39.
Batista, A. P. S., Romao, L. P. C., Arguelho, M., Garcia, C. A. B., Alves, J. P. H., Passos, E. A., & Rosa, A. H. (2009). Biosorption of Cr(III) using in natura and chemically treated tropical peats. Journal of Hazardous Materials, 163(2-3), 517–523.
Cunha, G. D., Romao, L. P. C., Santos, M. C., Araujo, B. R., Navickiene, S., & de Padua, V. L. (2010). Adsorption of trihalomethanes by humin: batch and fixed bed column studies. Bioresource Technology, 101(10), 3345–3354.
Andersen, R., Chapman, S. J., & Artz, R. R. E. (2013). Microbial communities in natural and disturbed peatlands: a review. Soil Biology and Biochemistry, 57, 979–994.
Phang, I. R. K., Chan, Y. S., Wong, K. S., & Lau, S. Y. (2018). Isolation and characterization of urease-producing bacteria from tropical peat. Biocatalysis and Agricultural Biotechnology, 13, 168–175.
Ashelford, K. E., Fry, J. C., Bailey, M. J., & Day, M. J. (2002). Characterization of Serratia isolates from soil, ecological implications and transfer of Serratia proteamaculans subsp quinovora Grimont et al. 1983 to Serratia quinivorans corrig., sp nov. International Journal of Systematic and Evolutionary Microbiology, 52(Pt 6), 2281–2289.
Pakala, S. B., Gorla, P., Pinjari, A. B., Krovidi, R. K., Baru, R., Yanamandra, M., Merrick, M., & Siddavattam, D. (2007). Biodegradation of methyl parathion and p-nitrophenol: evidence for the presence of a p-nitrophenol 2-hydroxylase in a gram-negative Serratia sp. strain DS001. Applied Microbiology and Biotechnology, 73(6), 1452–1462.
Vaaje-Kolstad, G., Horn, S. J., van Aalten, D. M. F., Synstad, B., & Eijsink, V. G. H. (2005). The non-catalytic chitin-binding protein CBP21 from Serratia marcescens is essential for chitin degradation. The Journal of Biological Chemistry, 280(31), 28492–28497.
Gupta, A., & Thakur, I. S. (2015). Biodegradation of wastewater organic contaminants using Serratia sp. ISTVKR1 isolated from sewage sludge. Biochemical Engineering Journal, 102, 115–124.
Haq, I., Kumar, S., Kumari, V., Singh, S. K., & Raj, A. (2016). Evaluation of bioremediation potentiality of ligninolytic Serratia liquefaciens for detoxification of pulp and paper mill effluent. Journal of Hazardous Materials, 305, 190–199.
Machin-Ramirez, C., Morales, D., Martinez-Morales, F., Okoh, A. I., & Trejo-Hernandez, M. R. (2010). Benzo a pyrene removal by axenic- and co-cultures of some bacterial and fungal strains. International Biodeterioration and Biodegradation, 64(7), 538–544.
Gupta, R., Gigras, P., Mohapatra, H., Goswami, V. K., & Chauhan, B. (2003). Microbial alpha-amylases: a biotechnological perspective. Process Biochemistry, 38(11), 1599–1616.
Jobin, G., Couture, G. V., Goyer, C., Brzezinski, R., & Beaulieu, C. (2005). Streptomycete spores entrapped in chitosan beads as a novel biocontrol tool against common scab of potato. Applied Microbiology and Biotechnology, 68(1), 104–110.
Zheng, F., Cui, B. K., Wu, X. J., Meng, G., Liu, H. X., & Si, J. (2016). Immobilization of laccase onto chitosan beads to enhance its capability to degrade synthetic dyes. International Biodeterioration and Biodegradation, 110, 69–78.
Migneault, I., Dartiguenave, C., Bertrand, M. J., & Waldron, K. C. (2004). Glutaraldehyde: behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. Biotechniques, 37(5), 790–802.
Leite, F., Modesto, C. M. D., do Nascimento, R. F., & Dias, F. S. (2005). Adsorção de Cd(II) de soluções aquosas em microesferas de n-carboximetil-quitosana. Revista Iberoamericana de Polímeros, 6, 213–236.
Naumann, D. (2000). Infrared spectroscopy in microbiology. In R. A. Meyers (Ed.), Encyclopedia of Analytical Chemistry (pp. 102–131). Chichester: Wiley.
Grube, M., Gavare, M., Nescerecka, A., Tihomirova, K., Mezule, L., & Juhna, T. (2013). FT-IR spectroscopic analysis for studying Clostridium cell response to conversion of enzymatically hydrolyzed hay. Journal of Molecular Structure, 1044, 201–205.
Kasaal, M. R. (2008). A review of several reported procedures to determine the degree of N-acetylation for chitin and chitosan using infrared spectroscopy. Carbohydrate Polymers, 71(4), 497–508.
Kuyukina, M. S., Korshunova, I. O., Rubtsova, E. V., & Ivshina, I. B. (2014). Methods of microorganism immobilization for dynamic atomic-force studies (review). Applied Biochemistry and Microbiology, 50(1), 1–9.
Kafilzadeh, F., Amiril, P., Jahromil, A. R., & Mojoodi, N. (2013). Isolation and molecular identification of fluoranthene degrading bacteria from the mangrove sediments in south of Iran. International Journal of Biosciences, 3, 60–67.
Tao, X. Q., Lu, G. N., Liu, J. P., Li, T., & Yang, L. N. (2009). Rapid degradation of phenanthrene by using Sphingomonas sp. GY2B immobilized in calcium alginate gel beads. International Journal of Environmental Research and Public Health, 6(9), 2470–2480.
Xu, Y. H., & Lu, M. (2010). Bioremediation of crude oil-contaminated soil: comparison of different biostimulation and bioaugmentation treatments. Journal of Hazardous Materials, 183(1-3), 395–401.
Banerjee, I., Modak, J. M., Bandopadhyay, K., Das, D., & Maiti, B. R. (2001). Mathematical model for evaluation of mass transfer limitations in phenol biodegradation by immobilized Pseudomonas putida. Journal of Biotechnology, 87(3), 211–223.
Sarma, S. J., & Pakshirajan, K. (2011). Surfactant aided biodegradation of pyrene using immobilized cells of Mycobacterium frederiksbergense. International Biodeterioration and Biodegradation, 65(1), 73–77.
Covizzi, L. G., Giese, E. C., Gomes, E., Dekker, R. F. H., & Silva, R. (2007). Immobilization of microbial cells and their biotechnological applications. Semin Ciências Exatas e Tecnológicas, 28(2), 143–160.
Muxika, A., Etxabide, A., Uranga, J., Guerrero, P., & de la Caba, K. (2017). Chitosan as a bioactive polymer: processing, properties and applications. International Journal of Biological Macromolecules, 105, 1358–1368.
Xu, H. X., Wu, H. Y., Qiu, Y. P., Shi, X. Q., He, G. H., Zhang, J. F., & Wu, J. C. (2011). Degradation of fluoranthene by a newly isolated strain of Herbaspirillum chlorophenolicum from activated sludge. Biodegradation., 22(2), 335–345.
Funding
Financial support for this work was provided by the Brazilian agencies Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, process no. 307029/2013-1) and Fundação de Apoio a Pesquisa e Inovação Tecnológica do Estado de Sergipe (FAPITEC/SE, process no. 01920300723/201-1).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Garcia, A.C.F.S., Araújo, B.R., Birolli, W.G. et al. Fluoranthene Biodegradation by Serratia sp. AC-11 Immobilized into Chitosan Beads. Appl Biochem Biotechnol 188, 1168–1184 (2019). https://doi.org/10.1007/s12010-019-02980-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-019-02980-9