Abstract
A microbial consortium was directly taken from activated sludge and was used to solubilize rock phosphates (RPs) in a lab-scale bioreactor in this study. Results showed that the microbial consortium could efficiently release soluble phosphorus (P) from the RPs, and during 30-day incubation, it grew well in the bioreactor and reduced the pH of the solutions. The biosolubilization process was also illustrated by the observation of scanning electron microscopy combined with an energy dispersive X-ray spectroscopy (SEM–EDX), which showed an obvious corrosion on the ore surfaces, and most elements were removed from the ore samples. The analysis of microbial community structure by Illumina 16S ribosomal RNA (rRNA) gene and 18S rRNA gene MiSeq sequencing reflected different microbial diversity and richness in the solutions added with different ore samples. A lower richness and diversity of bacteria but a higher richness and diversity of fungi occurred in the solution added with ore sample 1 compared to that of in the solution added with ore sample 2. Alphaproteobacteria and Saccharomycetes were the dominating bacterial and fungal group, respectively, both in the solutions added with ore samples 1 and 2 at the class level. However, their abundances in the solution added with ore sample 1 were obviously lower than that in the solution added with ore sample 2. This study provides new insights into our understanding of the microbial community structure in the biosolubilization of RPs by a microbial consortium directly taken from activated sludge.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Phosphorus (P) is one of the most important macronutrients for plant growth [1]. Plant available P is relatively low in soils because the large portion of P is often bound by calcium (Ca) and magnesium (Mg) in calcareous soils and by iron (Fe) and aluminum (Al) in acidic soils, resulting in less available for the plant [2].
To reduce P deficiencies and ensure plant productivity, the use of natural rock phosphates (RPs) as an alternative source of P fertilizer has received significant interest in recent years and more particularly in China, the second-largest producer of RPs (behind the USA). Unfortunately, RPs is not plant available where the pH of the soils is greater than 5.5–6.0 and even when conditions are optimal, plant yields are lower than those obtained with soluble P [3].
Traditionally, RPs was chemically processed with sulfuric acid or phosphoric acid into P fertilizer. However, this strategy is not recommended to process mid-low grade RPs because of its high cost and inevitably environmental pollution [4].
With the current tendency for reduced process cost and environmental pollution, interest in biosolubilization of RPs through the action of P-solubilizing microorganisms and its use in agriculture are receiving greater attention recently. Naturally occurring P-solubilizing microorganisms can solubilize RPs and release a soluble form of P (HPO4 2− and H2PO4 −) into the soils solution where solubilized P can be absorbed by plant roots [5–7]. This process has been traditionally associated with chelation, ion exchange, and the production of organic acids [8, 9].
However, most current studies on biosolubilization of RPs were limited in only using single or few known strains. It was suggested that in bioleaching operations, multiple microorganisms assist bioleaching activities more effectively when presented as symbiotic consortia [10]. Hence, more researches are needed to understand the usage of various microbial consortia for the biosolubilization of RPs. In addition, most studies focused on the isolation of P-solubilizing microorganisms from soils, and seldom report is available so far in literature for biosolubilization of mid-low grade RPs by microorganisms collected from activated sludge. In this study, a microbial consortium enriched from activated sludge of a municipal wastewater treatment plant in Canada was used to solubilize mid-low grade RPs. Chemical analysis of the release of soluble P, pH, and microbial growth in the bioreactor was performed to follow the overall process. The analyses of ore surfaces by scanning electron microscopy and energy dispersive X-ray spectroscopy (SEM–EDX) to assess the corrosivity of the microbial consortium to the RPs were also examined. For a comprehensive understanding of the activity and distribution of the solution containing different RP samples, their microbial community structures were analyzed based on high-throughput Illumina MiSeq sequencing platform. The results in this study would provide theoretical guidance for the practical application of microbial consortium from activated sludge in the biosolubilization of mid-low grade RPs in a lab-scale bioreactor.
Materials and Methods
RP Samples
The samples of mid-low grade RPs used in this study were obtained from the Yunnan phosphate mines in China. They were originated from marine sedimentary phosphorite deposit. The main elements in the ore samples by X-ray fluorescence (XRF) analysis are shown in Table 1. X-ray diffraction (XRD) analysis showed that ore sample 1 was mainly composed of quartz and francolite, and ore sample 2 was mainly composed of quartz, dolomite, and francolite. Ore samples used inside the bioreactor were initially crushed and screened to a particle size in the range of 100–200 mesh before used.
Microorganisms
Activated sludge was collected from a municipal wastewater treatment plant in Edmonton, Canada. The microbial consortium in activated sludge were enriched by a modified Pikovskaya medium consisted of the following compounds (per liter): 10 g glucose, 0.5 g yeast extract, 0.5 g (NH4)2SO4, 0.2 g KCl, 0.1 g MgSO4·7H2O, 0.0001 g MnSO4·H2O, and 0.0001 g FeSO4·7H2O [11]. About 5 g/L Ca3(PO4)2 was added to the medium as a sole P source for selectively screening of P-solubilizing microorganisms. The enrichment was carried out in 2.5 L aeration tank aerated at a rate of 45 L/h of air at ambient temperature and initial pH 7 for 3 days, and subcultured at 2-week intervals.
Biosolubilization Experiments
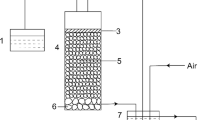
The biosolubilization experiments were performed in a 2.5 L completely mixed batch bioreactor agitated by stirring at 200 rpm and aerated at a flow of 1.2 L/min. Two liters of reaction solution including modified Pikovskaya medium inoculated with microbial consortium was added to each bioreactor. The concentration of the microbial consortium in reaction solution was adjusted to optical density (OD) at 0.6 under 600 nm by a spectrophotometer before used in this study. Autoclaved, uninoculated medium served as control. The two samples of RPs were then fed into the bioreactor at 2% (w/w), respectively. The experiment was entirely tested at ambient temperature and initial pH 7 for 30 days. During the experiment, 50 mL solution was taken daily from the bioreactor and analyzed to determine microbial growth, pH, and soluble P content. All experiments were performed in triplicate.
SEM–EDX Analysis
A small amount of ore residues were taken at random from the bioreactor on operation days 10 and 30. The residues were washed three times with P buffer and dehydrated by using a graded ethanol series, air-dried, and gold-coated for SEM–EDX observation. The analysis was performed using a VEGA3 (TESCAN, Czech) SEM equipped with an energy dispersive spectrometer at an accelerating voltage of 20 kV.
Microbial Community Analysis
After 30 days operation, microorganisms in the bioreactor were sampled to analyze the microbial community inside them via Illumina MiSeq sequencing. Genomic DNA was first extracted from the bioreactor using the PowerBiofilm™ DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA) according to its standard procedure.
Sequencing amplicon libraries were generated by PCR following the “16S Metagenomic Sequencing Library Preparation - Preparing 16S Ribosomal RNA Gene Amplicons for the Illumina MiSeq System” protocol (Illumina Part # 15044223 Rev. B, https://support.illumina.com). Internal parts of the 16S ribosomal RNA (rRNA) gene, covering variable regions V3 and V4, were PCR-amplified with the KAPA HiFi HotStart ReadyMix (KAPA Biosystems) and the primers 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′ and 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′ and purified with the Agencourt AMPure XP kit (Beckman Coulter Genomics). 18S ribosomal RNA primers are: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCAGCASCYGCGGTAATTCC-3′ and 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGACTTTCGTTCTTGATYRA-3′. The PCR process was conducted as follows: 95 °C for 3 min, followed by 25 cycles at 95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s, and a final extension at 72 °C for 5 min. PCR reactions were performed in triplicate. Each 25 μL PCR reaction mixture contained 12.5 ng of microbial DNA, 5 μL of each primer (1 μM), and 12.5 μL 2× KAPA HiFi HotStart ReadyMix. Purified amplicons were pooled in equimolar and sequenced on an Illumina MiSeq platform using the MiSeq Reagent Kit v3 in the 2 × 300 bp Paired-End mode. The sequencing data analysis was conducted by Mothur [12] following MiSeq SOP (http://www.mothur.org/wiki/MiSeq_SOP). Firstly, the raw paired-end reads were assembled to reduce sequencing and PCR errors of the reads, and then the reads containing ambiguous bases, incorrect barcode or primer sequences, or longer than 275 bp were excluded from further analysis. Putative chimeras were detected and excluded from previously treated sequences with UCHIME algorithm within Mothur. Taxonomy was assigned with Silva database using kmer searching method with cutoff of 80%. Reads were clustered into operational taxonomic units (OTUs) at 97% identity. Rarefaction and diversity statistics including library coverage, Chao value, and Shannon index were calculated for each sample after OTUs clustered. Finally, the raw reads were deposited into the National Center for Biotechnology Information Sequence Read Archive (SRA) database under accession SRP066037.
Analytical Methods
Microbial growth was measured as the optical density at 600 nm using a spectrophotometer (UV-3600; Shimadzu Corporation, Kyoto, Japan). The pH in the broth was recorded using water with a pH meter (AR 15, Accumet, Fisher Scientific) equipped with glass electrode (Cat. #13-620-108, Accumet, Fisher Scientific). Content of soluble P in the solution was determined by using the vanadium–ammonium molybdate colorimetric method by taking KH2PO4 as a standard according to the Standard Methods for Examination of Water and Wastewater [13]. The soluble P was expressed in terms of milligrams per liter P released in the solution. Values were given as mean ± standard deviation for triplicate samples. Data were analyzed by analysis of variance (ANOVA) and the means were compared with Duncan’s multiple range test (DMRT) at P < 0.05 level.
Results and Discussion
RP Biosolubilization by Microbial Consortium from Activated Sludge
A microbial consortium from activated sludge was used to solubilize mid-low grade RPs in a lab-scale bioreactor in this study. The concentration of the microbial consortium, pH, and content of soluble P in the solution with the two ore samples is illustrated in Fig. 1. Results showed that the microbial consortium could grow well during 30 days inoculation, although obvious fluctuation of the microbial concentration was observed during the experiment. The pH in the solution decreased sharply from the start of the experiment to the third day, and then the pH was fluctuant and kept at values of 3–5 for the duration of the experiment.
As a result of pH reduction, a significant increase of soluble P in the solution was observed during the experiment. The result showed that the microbial consortium could effectively solubilize the RPs in the lab-scale bioreactor. As for the two different ore samples, the microbial consortium could solubilize ore sample 2 (Fig. 1b) more effective than that of ore sample 1 (Fig. 1a). Moreover, ore sample 2 effected the larger reduction of pH than that of ore sample 1. However, there was no obvious difference of the microbial concentration in the solutions between with the two ore samples.
SEM–EDX Analysis
SEM observation revealed the morphological features of ore residue surfaces solubilized by the microbial consortium after 10 and 30 days (Fig. 2). As it can be seen from the images, the surfaces of ore samples without biosolubilization (Fig. 2a, b) were approximately smooth compared to surfaces in the presence of the microbial consortium, which presented as scraggly and formed many chasms (Fig. 2c, d, e, f). This suggests that the corrosion of the ore surface is due to the proton attack and thus made it scraggly and formed many chasms. In all cases, the microbial consortium could make different extents of corrosion of the residue surfaces with different ore samples, and the corrosion was becoming more serious when the action time was increased from 10 to 30 days.
SEM images of typical ore surface. a Ore sample 1 without biosolubilization (control). b Ore sample 2 without biosolubilization (control). c Ore sample 1 after 10 days of operation. d Ore sample 2 after 10 days of operation. e Ore sample 1 after 30 days of operation. f Ore sample 2 after 30 days of operation
EDX analyses were performed on all these features individually in order to find out if the amounts of the elements in the ore residues, especially P and Ca, had any changed. Results exhibit an obvious reduction of elements P and Ca in the residues of both ore sample 1 and ore sample 2 solubilized by the microbial consortium compared to the control (Fig. 3). It indicated that the main composition of RPs had undergone an obvious transformation, and it was solubilized successfully by the microbial consortium. Similarly, different extents of transformation were performed with different ore samples, and the amounts of P and Ca were reduced obviously when the action time increased from 10 to 30 days.
Typical EDX spectrums and elements content (embedded table) of the ore surface. a Ore sample 1 without biosolubilization (control). b Ore sample 2 without biosolubilization (control). c Ore sample 1 after 10 days of operation. d Ore sample 2 after 10 days of operation. e Ore sample 1 after 30 days of operation. f Ore sample 2 after 30 days of operation
Microbial Community Structures
Since the functions and behavior of microorganism influenced the microbial concentration and activity, further investigation by Illumina MiSeq sequencing was conducted to analyze the microbial community diversities and phylogenetic structures of the microorganisms in the solution added with different ore samples. Totally, 859,737 optimized sequences of the 16S rRNA gene and 726,392 optimized sequences of the 18S rRNA gene that widely represented the diversity of microbial community after removing low quality sequences and chimeras were generated from the microorganisms in the solutions added with ore sample 1 and ore sample 2, respectively. The sequence information of the samples and calculated microbial diversity index are listed in Table 2.
Illumina MiSeq sequencing results revealed the significant difference of microbial diversity between the solution added with ore sample 1 and ore sample 2. A total of 2582 bacterial OTUs and 4681 fungal OTUs from the solution added with ore sample 1 were obtained based on 97% similarity, while a total of 4826 bacterial OTUs and 3160 fungal OTUs from the solution added with ore sample 2 were obtained based on 97% similarity (Table 2).
The number of OTUs indicates the richness of the microbial community, whereas Shannon indexes represent the microbial diversity [14, 15]. The richness of the microorganisms was analyzed by number of OTUs through the rarefaction curve (Fig. 4). Results showed a lower richness of bacteria but a higher richness of fungi occurred in the solution added with ore sample 1 compared to that of in the solution added with ore sample 2. Although curves show similar patterns, none of the curves approached a plateau, suggesting that further sequencing would have resulted in more OTUs in each group [16].
Analysis of Shannon index revealed that the diversity of the bacterial community in the solution added with ore sample 1 was lower than that in the solution added with ore sample 2, whereas the diversity of the fungal community in the solution added with ore sample 1 was higher than that in the solution added with ore sample 2 (Table 2 and Fig. 2). A possible explanation for the differences of microbial community structure after application of activated sludge with different RPs could be related to the differences in the elemental composition of the two RPs. Several studies have indicated that mineral composition influences not only quantitative differences in the microbial concentration but also the microbial community structure [17–19].
All sequences were classified to the class level according to the Mothur software using the Silva classifier (Fig. 5). Results showed that multiple microorganisms existed in the microbial consortium from activated sludge, and they were jointly responsible for the biosolubilization of RPs. Many studies also indicated that activated sludge is a highly complex biological mixture with bacteria as the dominant group, but also contains eukaryotes, archaea, and viruses as determined by conventional molecular techniques and next-generation sequencing methods [20, 21]. These mixed microbial cultures can be used as seeds in biological conversion processes and enable them to better survive in the heterogeneous and challenging environment often encountered in the biosolubilization of mid-low grade RPs.
The relative bacterial community abundances at the class level of the microorganisms in the solutions added with ore sample 1 and ore sample 2 are illustrated in Fig. 5a. The relative abundances of dominant taxa varied between the microorganisms in the two solutions. Based on average abundance analysis, the bacteria in the solution added with ore sample 1 were mainly composed of TM7_class_incertae_sedis (34.52%), Alphaproteobacteria (21.76%), Gammaproteobacteria (19.84%), and Flavobacteria (9.91%). The most abundant bacterial species in the solution added with ore sample 2 belonged to Alphaproteobacteria, which abundances was 82.68%, obviously higher than that in the solution added with ore sample 1.
As is well known, Alphaproteobacteria was highly versatile in pollutant degradation capacities and detected in various biotreatment systems such as phenol-containing wastewater, domestic wastewater and coking wastewater [22]. In this study, Alphaproteobacteria was the dominant class, especially in the solution added with ore sample 2. This observation was similar to what was observed in studies on activated sludge using microarray analysis [23, 24].
The relative fungal community abundances at the class level are presented in Fig. 5b. Results showed that Saccharomycetes was the only dominant fungi at the class level in the solutions, especially in the solution added with ore sample 2, which abundances reached to 99.41%. However, its abundance in the solution added with ore sample 1 is 44.87%, which was obviously lower than that in the solution added with ore sample 2. Notably, 53.53% were not classified at the class level in the solution added with ore sample 1. The results of this study further indicated the existence of multiple organisms in the microbial consortium from activated sludge, and further studies are required in order to determine these unclassified organisms.
The diversity and abundance of both bacterial species and fungal species were different in the solutions added with ore samples 1 and 2. It indicated that the microbial composition can vary greatly with different ore samples. The microbial composition of activated sludge can vary greatly with source, temperature, pH, organic loading, environmental conditions, and operational parameters [25]. Therefore, the cause for the different structures of microbial community in the solutions added with different ore samples needs further studies.
Conclusions
A microbial consortium directly taken from activated sludge could efficiently solubilize mid-low grade RPs in a lab-scale bioreactor with modified Pikovskaya medium. During the 30-day incubation, the microbial consortium grew well in the bioreactor and reduced the pH in the solutions, which caused an acidic environment and thus facilitated the biosolubilization of the RPs. The biosolubilization process was also illustrated by SEM–EDX observations. Illumina MiSeq sequencing reflected different microbial community structures in the solutions added with different ore samples. A lower richness and diversity of bacteria but a higher richness and diversity of fungi occurred in the solution added with ore sample 1 compared to that in the solution added with ore sample 2. Multiple microorganisms existed in the microbial consortium from activated sludge, and the relative abundances of dominant taxa varied from different ore samples.
References
Sharpley, A. N. (1995). Soil phosphorus dynamics: agronomic and environmental impacts. Ecological Engineering, 5, 261–279.
Holford, I. C. R. (1997). Soil phosphorus: its measurement and its uptake by plants. Australian Journal of Soil Research, 35, 227–240.
Reddy, M. S., Kumar, S., & Khosla, B. (2002). Biosolubilization of poorly soluble rock phosphates by Aspergillus tubingensis and Aspergillus niger. Bioresource Technology, 84, 187–189.
Vaccari, D. A., & Strigul, N. (2011). Extrapolating phosphorus production to estimate resource reserves. Chemosphere, 84, 792–797.
Hafidi, M., Ouahmane, L., Thioulouse, J., Sanguin, H., Boumezzough, A., Prin, Y., Baudoin, E., Galiana, A., & Duponnois, R. (2013). Managing Mediterranean nurse plants-mediated effects on soil microbial functions to improve rock phosphate solubilization processes and early growth of Cupressus atlantica G. Ecological Engineering, 57, 57–64.
Liu, Z. G., Li, Y. C., Zhang, S. A., Fu, Y. Q., Fan, X. H., Patel, J. S., & Zhang, M. (2015). Characterization of phosphate-solubilizing bacteria isolated from calcareous soils. Applied Soil Ecology, 96, 217–224.
Ghosh, R., Barman, S., Mukherjee, R., & Mandal, N. C. (2016). Role of phosphate solubilizing Burkholderia spp. for successful colonization and growth promotion of Lycopodium cernuum L. (Lycopodiaceae) in lateritic belt of Birbhum district of West Bengal, India. Microbiological Research, 183, 80–91.
Vazquez, P., Holguin, G., Puente, M. E., Lopez-Cortes, A., & Bashan, Y. (2000). Phosphate-solubilizing microorganisms associated with the rhizosphere of mangroves in a semiarid coastal lagoon. Biology and Fertility of Soils, 30, 460–468.
Son, H. J., Park, G. T., Cha, M. S., & Heo, M. S. (2006). Solubilization of insoluble inorganic phosphates by a novel salt- and pH-tolerant Pantoea agglomerans R-42 isolated from soybean rhizosphere. Bioresource Technology, 97, 204–210.
Qiu, G. Z., Li, Q., Yu, R. L., Sun, Z. X., Liu, Y. J., Chen, M., Yin, H. Q., Zhang, Y. G., Liang, Y. L., Xu, L. L., Sun, L. M., & Liu, X. D. (2011). Column bioleaching of uranium embedded in granite porphyry by a mesophilic acidophilic consortium. Bioresource Technology, 102, 4697–4702.
Pikovskaya, R. I. (1948). Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Microbiología, 17, 362–370.
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., Lesniewski, R. A., Oakley, B. B., Parks, D. H., Robinson, C. J., Sahl, J. W., Stres, B., Thallinger, G. G., Van Horn, D. J., & Weber, C. F. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology, 75, 7537–7541.
Eaton, A. D., Clesceri, L. S., Rice, E. W., & Greenberg, A. E. (2005). Standard methods for the examination of water and wastewater. 21st Eds. Centennial Edition. American Public Health Association, Washington, DC.
Ma, J., Wang, Z., Yang, Y., Mei, X., & Wu, Z. (2013). Correlating microbial community structure and composition with aeration intensity in submerged membrane bioreactors by 454 high-throughput pyrosequencing. Water Research, 47, 859–869.
Liu, Z., Zhang, C., Wang, L., He, J., Li, B., Zhang, Y., & Xing, X. H. (2015). Effects of furan derivatives on biohydrogen fermentation from wet steam-exploded cornstalk and its microbial community. Bioresource Technology, 175, 152–159.
Li, Y. Y., Chen, L. Q., Wen, H. Y., Zhou, T. J., & Zhang, T. (2014). Pyrosequencing-based assessment of bacterial community structure in mine soils affected by mining subsidence. International Journal of Mining Science and Technology, 24, 701–706.
Carson, J. K., Rooney, D., Gleeson, D. B., & Clipson, N. (2007). Altering the mineral composition of soil causes a shift in microbial community structure. FEMS Microbiology Ecology, 61, 414–423.
Gleeson, D., Kennedy, N., Clipson, N., Melville, K., Gadd, G., & McDermott, F. (2006). Characterization of bacterial community structure on a weathered pegmatitic granite. Microbial Ecology, 51, 526–534.
Mechri, B., Attia, F., Tekaya, M., Cheheb, H., & Hammami, M. (2014). Agronomic application of olive mill wastewaters with rock phosphate increase the 10Me18:0 fatty acid marker of actinomycetes and change rhizosphere microbial functional groups under long-term field conditions. Soil Biology and Biochemistry, 70, 62–65.
Ju, F., Guo, F., Ye, L., Xia, Y., & Zhang, T. (2013). Metagenomic analysis on seasonal microbial variations of activated sludge from a full-scale wastewater treatment plant over 4 years. Environmental Microbiology Reports, 6, 80–89.
Wang, X., Hu, M., Xia, Y., Wen, X., & Ding, K. (2012). Pyrosequencing analysis of bacterial diversity in 14 wastewater treatment systems in China. Applied and Environmental Microbiology, 78, 7042–7047.
Figuerola, E. L. M., & Erijman, L. (2007). Bacterial taxa abundance pattern in an industrial wastewater treatment system determined by the full rRNA cycle approach. Environmental Microbiology, 9, 1780–1789.
Xia, S., Duan, L., Song, Y., Li, J., Piceno, Y. M., Andersen, G. L., Alvarez-Cohen, L., Moreno-Andrade, I., Huang, C. L., & Hermandowicz, S. W. (2010). Bacterial community structure in geographically distributed biological wastewater treatment reactors. Environmental Science and Technology, 44, 7391–7396.
Liang, S. B., Gliniewicz, K., Mendes-Soares, H., Settles, M. L., Forney, L. J., Coats, E. R., & McDonald, A. G. (2015). Comparative analysis of microbial community of novel lactic acid fermentation inoculated with different undefined mixed cultures. Bioresource Technology, 179, 268–274.
Zhang, T., Shao, M. F., & Ye, L. (2012). 454 pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME Journal, 6, 1137–1147.
Acknowledgements
This research work was kindly supported by the National Natural Science Foundation of China (51674178) and the Program for Excellent Young Scientific and Technological Innovation Team of Hubei Provincial Department of Education, China (T201506). The authors also thank Prof. Tong Yu, Prof. Zhenghe Xu, and Dr. Lei Zhu of the University of Alberta for their kindly support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xiao, C., Wu, X., Liu, T. et al. Microbial Community Structure of Activated Sludge for Biosolubilization of Two Different Rock Phosphates. Appl Biochem Biotechnol 182, 742–754 (2017). https://doi.org/10.1007/s12010-016-2358-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2358-3