Abstract
Halotolerant bacteria associated with Psoralea corylifolia L., a luxuriantly growing annual weed in salinity-affected semi-arid regions of western Maharashtra, India were evaluated for their plant growth-promoting activity in wheat. A total of 79 bacteria associated with different parts viz., root, shoot and nodule endophytes, rhizosphere, rhizoplane, and leaf epiphytes, were isolated and grouped based on their habitat. Twelve bacteria isolated for their potential in plant growth promotion were further selected for in vitro studies. Molecular identification showed the presence of the genera Bacillus, Pantoea, Marinobacterium, Acinetobacter, Enterobacter, Pseudomonas, Rhizobium, and Sinorhizobium (LC027447-53; LC027455; LC027457, LC027459, and LC128410). The phylogenetic studies along with carbon source utilization profiles using the Biolog® indicated the presence of novel species and the in planta studies revealed promising results under salinity stress. Whereas the nodule endophytes had minute plant growth-promoting (PGP) activity, the cell free culture filtrates of these strains enhanced seed germination of wheat (Triticum aestivum L). The maximum vigor index was monitored in isolate Y7 (Enterobacter sp strain NIASMVII). Indole acetic acid (IAA) production by the isolates ranged between 0.22 and 25.58 μg mL−1. This signifies the need of exploration of their individual metabolites for developing next-generation bio-inoculants through co-inoculation with other compatible microbes. This study has potential in utilization of the weed-associated microbiome in terms of alleviation of salinity stress in crop plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microorganisms, including fungi, bacteria, and archaea, are able to adapt to extremes of external temperatures, salinity/osmolarity, and other edaphic stresses in the natural soil environment. The adaptation of many wild plants and crop genotypes to highly saline conditions has been ascribed to their close interaction with microbes associated with their roots. However, these omnipresent and omnipotent microbes are less explored and under-utilized option for alleviation of salinity stress [1]. In fact, plant growth-promoting rhizobacteria (PGPR) are an attractive eco-friendly alternative to chemicals in agriculture and so far, the most attempts have concentrated on their screening from the rhizosphere of crop plants. Halophytic weeds thriving quite luxuriantly in relatively harsh environments with very limited nutrients play an important role in maintaining ecological balance, but these have largely been ignored. Salt-loving plants or halophytes of different habitats vary with respect to the composition of associated microbes. Hence, understanding the plant-specific composition of the root microbiome of halophytes is of utmost importance in order to explore the genetic and functional variation of microbes for the improvement of salinity tolerance in crop plants. Considering all these aspects, it is evident that the associated flora from wild plants and weeds may serve as a potent source for novel plant growth-promoting microbial resources that could most likely be implemented for alleviation of the majority of edaphic stresses in crop plants. In the present study, an extensive survey was conducted to target and explore Psoralea corylifolia L., a luxuriantly growing plant in the salt-affected soils of the Krishna river basin in the western part of Maharashtra, India, to harvest its associated bacteria, which have the potential as bio-inoculants for other agricultural crops. This plant has been mainly exploited for its multiple medicinal values, e.g., in treating skin diseases such as psoriasis, leprosy, leucoderma, etc. [2]; asthma; alopecia aretae; and diarrhea [3], and its action against diverse allergic responses and anti-tumor and anti-oxidant potential [4–6]. Biomolecules of medicinal importance include flavonoids, essential oil, alkaloids, coumarins, terpenoids, etc. However, the microbes associated with this plant and their PGP potential are yet to be explored. Therefore, attempts were made to harvest its associated microflora, which are beneficial for enhancing plant growth promotion under salinity stress. These were further evaluated in planta using wheat—a major cereal crop.
Materials and Methods
Collection of Samples
The weed (P. corylifolia L.) was collected from salt-affected barren soils distributed across the Krishna river basin from Sangli district (Maharashtra). Whole plants, along with the block of rhizosphere soil (covering the whole root system), were collected in pre-sterilized polyethylene bags and immediately brought to the laboratory at ambient conditions.
Processing of Samples and Isolation of Bacteria
The electrical conductivity (EC1:2) and pH values of the rhizosphere soil of the collected plants were measured using standard protocol [7]. The samples of the various parts of the plant were processed and subjected to isolation of associated microflora. The inhabitants of the plant parts—roots and root nodules—were isolated by treating the nodules and root fragments in 0.1 % HgCl2 for 5 min, followed by several times washing with sterile Milli Q water. The nodules and roots were then crushed in sterile saline (0.85 % NaCl) and streaked immediately on the Congo-red yeast extract mannitol agar (CRYEMA) amended with 5 % crude NaCl and pH 7.5 ± 0.2.The stem endophytes were also isolated in a similar manner. The leaf epiphytic flora was isolated using routine leaf impression technique on ammonium mineral salt (AMS) agar [8] and King’s B (KB) medium. The rhizoplanic microflora was isolated using the method described earlier [9]. The rhizosphere-inhabiting bacterial population was isolated by streaking the soil suspension on CRYEMA with 5 % salt and pH 8.2 ± 0.2.
Biochemical Characterization for PGP Traits and Salt Tolerance
The isolates were screened for salt tolerance ability using NaCl gradient ranging from 0 to 30 % by the steps of 1 %. The ability to fix atmospheric dinitrogen in vitro was determined using malate [10, 11] and mannitol (Ashby’s nitrogen-free mannitol agar) as carbon source. Siderophore production was determined on chrome azurol sulfonate (CAS) agar [12]. Exopolysaccharide production (Tryptic soy agar) and phosphate solubilization [13] abilities were tested at the respective salt concentrations (obtained during the salt tolerance experiment), allowing luxurious growth of the individual isolates.
Quantitative IAA production ability was determined by Salkowski’s method [14]. The isolates were grown at 30 ± 2 °C in yeast extract mannitol (YEM) medium with 0.1 % tryptophan under constant agitation at 150 rpm for 48 h. The cells were pelleted at 7000 rpm and 0.5 mL of the supernatant was mixed with 200 μL of orthophosphoric acid followed by 2.0 mL of Salkowski’s reagent (2 % 0.5 M FeCl3 mixed with 35 % HClO4); the tubes were kept in the dark for 30 min at room temperature and absorbance was recorded at 530 nm using UV–VIS 3000+ double beam spectrophotometer (LabIndia). The IAA (Fisher Scientific, USA) served as standard.
PCR Analysis of 16S rRNA Gene and Molecular Identification
The isolates were grown on YEM agar plates at 30 ± 2 °C for 24 and 48 h for fast- and slow-growing isolates, respectively. Well-isolated colonies were suspended in 1.5 mL of TE buffer (10 mM Tris–HCl; 1 mM Na-EDTA) and washed till the removal of the mucous. The genomic DNA of the purified isolates was isolated using an Ultra Clean DNA isolation kit (MoBio) following the manufacturer’s instructions. The 16S rRNA gene was PCR amplified using universal primers (Fwd 5′AGAGTTTGATCCTGGCTCAG3′ and Rev 5′ACGGCTACCTTGTTACGACTT3′). The 100-μL reaction mixture was composed of 100 ng of both primers, 75 ng of template DNA, 50 μL of 2× PCR master mix (Thermo Scientific) (Taq DNA polymerase, 0.5 U/μL; MgCl2, 4 mM; dNTPs (dATP, dCTP, dGTP, dTTP), 0.4 mM each) in the DNA Engine Tetrad2 thermal cycler (Biorad, USA), programmed for 94 °C, 4 min; 29 cycles of 94 °C, 45 s; 55 °C, 60 s; 72 °C, 90 s; and final extension at 72 °C, 10 min.
The resulting amplified product was electrophoresed on 1.2 % agarose gel. The PCR products were purified by the MinElute gel extraction kit (Qiagen). The purified DNA were sequenced using ABI 3130xlautomated genetic analyzer (Applied Biosystem, UK).The sequences were compared with available database using the NCBI-BLAST tool (http://www.blast.ncbi.nlm.nih.gov/Blast.cgi). The species level identification was done considering the 16S rRNA gene sequence similarity with that of the type sequences in the GenBank database. The sequences were aligned and compared using CLUSTAL W; the phylogenetic tree was constructed using the neighbor-joining method in MEGA6.0 [15]. The sequences were analyzed using NCBI database and submitted to DNA Data Bank of Japan (DDBJ).
Biolog-Based Biochemical Evaluation
The isolates were further subjected to detailed biochemical evaluation using Gen III plates and the inoculating fluid (IF) “A.” The log cultures of the isolates in single-colony form were suspended in the IF; the transmittance (97 %) was adjusted equivalent to that of a standard (provided by Biolog), followed by inoculation in 100-μL quantities in each well. The plates were incubated at 30 °C and read at every 12-h intervals till 48 h using BiologMicrostation™.
In Vitro Seed Germination Experiment
The fast- and slow-growing isolates with promising PGP traits were raised in YEM broth at 30 ± 2 °C for 24 and 48 h, respectively, with continuous shaking at 150 rpm. The cells were pelleted at 7000 rpm for 10 min, and the resulting supernatant were filtered using a 0.22-μm syringe filter and 100 μL of each filtrate was mixed with 20 ml molten (50 °C) water-agar (EC = 5.0dS/m) followed by immediate plating. These plates (n = 3) were used for the germination experiment. Sterile Milli-Q water, and uninoculated yeast extract mannitol broth served as the control.
The wheat seeds were surface sterilized by treatment with 5 % NaOCl solution [16], followed by several times washing with sterile Milli-Q water. The seeds were placed in the above plates and incubated in the dark till germination, followed by 12-h dark and light cycles. The germination percentage and root and shoot lengths were recorded after 7 days. The similar experiment was performed using YEM broth amended with 0.1 % tryptophan in order to detect the influence of the induced IAA on seed germination and development.
Statistical Analysis
Seed germination and IAA production data were statistically analyzed using analysis of variance (ANOVA) with subsequent Duncan’s multiple range test using SPSS (Windows 8.0). The differences at the 95 % confidence level were considered to be significant.
Results and Discussion
P. corylifolia L. is a well-known medicinal, annual weed capable of growing in diverse soil types across India [17]. It is principally distributed across the semi-arid area of northern India, including Uttarakhand, Rajasthan, Uttar Pradesh. and Punjab [18]; moreover, in this study, we have collected the same weed from the salinity-affected belt from the western regions of Maharashtra (Table S1). Seventy-nine halotolerant isolates with plant growth-promoting traits were obtained from the root, stem, leaf, and root surfaces and rhizosphere of P. corylifolia L. (Fig. 1). The weed being capable to grow luxuriantly in saline soil was thought to harbor the associated flora with the ability to tolerate elevated salt levels than the average. In order to evaluate the most predominant regions for the PGP bacteria, the isolates were grouped according to the origin of isolation (Fig. 1). The exclusive salt tolerance ability of the isolates confirmed the expectation regarding the presence of organisms tolerating elevated levels of salt (Fig. 1), where the reduced integer of culturable organisms with PGP traits in the rhizosphere and rhizoplane regions may be attributed to the higher concentration of the salt present in soil, which probably have restricted the proliferation of the salt-sensitive population. This fact was confirmed during the salt tolerance experiment, where the relative proliferation of the endophytic isolates seems restricted, in comparison with that of rhizosphere and rhizoplane at elevated salt levels.
Distribution of bacterial isolates associated with Psoralea corylifolia L., their average NaCl tolerance potential and average IAA production capacity group wise. The highest abundance of nodule endophytes is notable; whereas the rhizosphere and rhizoplane population was lowest. The rhizospheric population found topmost in salt tolerance ability. Note that the maximum average value of IAA was by the isolates which originated from rhizosphere; leaf inhabitants, in contrast, yielded a very low amount of IAA (Rh: rhizospheric; Rp: rhizoplanic; RE: root endophytes; NE: nodule endophytes; SE: stem endophytes; LE: leaf epiphytes)
The isolates originating from different plant parts exhibited varying PGP activities in vitro (Fig. 2). The siderophore and IAA production were the only traits that appeared among all the isolates. The EPS production can be attributed to the high levels of salt in the adjacent environment (Table S1), whereas the increased siderophore production among these isolates could be due to the constant iron-starved conditions due to the elevated pH, which increases the availability of insoluble ferric iron and vice versa [19, 20]. The induction or enhancement of siderophore production among the soil bacteria at pH beyond 7 has been frequent [21, 22]. The nodule inhabitants also exhibited higher siderophoregenic activity (almost all nodule-inhabiting isolates were positive for siderophore production), indicating the presence of an active iron-scavenging system to encounter the consistent iron deficiency; however, little count of the isolates found positive for EPS production, particularly due to the decreased levels of salt in the endophyte environment.
Based on colonial appearance and plant growth-promoting potential in terms of nitrogen fixation, siderophore production, phosphate solubilization, and as well as other in vitro PGP traits (Table S2), a total of 11 potent isolates were selected for detailed identification. The 16S rRNA gene sequences were submitted to DDBJ, with accession numbers LC027447-53, LC027455, LC027457, LC027459, and LC128410. Phylogenetic analysis (Fig. S1) of the same revealed a diverse group of organisms belonging to the following genera: Bacillus, Pantoea, Marinobacterium, Acinetobacter, Enterobacter, Pseudomonas, Rhizobium, and Sinorhizobium. Based on the biochemical characteristics (Table S3) and the phylogenetic positions, some of the isolates, viz, Marinobacterium sp., Pseudomonas sp., Rhizobium sp., and Sinorhizobium sp., highlighted the need for detailed identification. Surprisingly, none of the isolates selected for further characterization was found capable to thrive at 1 % NaCl, showing the need of higher salt concentration for optimum growth and metabolism. The presence of nodulating rhizobacteria in P. corylifolia L, from non-saline soils have also been demonstrated and classified earlier [18, 23]; this work, however, yielded significant knowledge regarding the P. corylifolia-associated halotolerant bacterial population and their PGP traits.
Plants secrete a variety of molecules predominantly comprising amino acids, vitamins, various sugars, polyamines, etc. which serve as signaling medium for the rhizosphere-inhabiting microbial community [24–26]. This comprises a complex phenomenon, including an array of inter- and intra-cellular processes, where the molecular signaling has vital role; unfortunately, the same is very susceptible to both biotic and abiotic factors [27]. The bacteria are known to induce changes in root exudate composition, principally in that of small molecules [28, 29] that are prerequisites for growth and development, typically for smooth organization and functioning of inter- and intra-cellular processes in plants; the use of bacterial consortia capable of overcoming the stress-susceptibility of plants therefore becomes crucial in such circumstances. The rhizosphere-microbial inhabitants secrete various phytohormones, growth factors, and a wide range of organic volatiles that play an indispensable role in plant growth and development [30].
The production of plant growth regulators is most common among the associated bacteria, which are principally intended to alter the physiology of the host for the ultimate benefit of the elaborator [31]. The IAA production ability of the rhizosphere isolates from this study was found predominant (Fig. 1), showing that there might be enhancement of germination under salt stress [32, 33] and help the plant sustain the stressful environment. The leaf-inhabiting isolates in contrast exhibited the least amount of IAA production. The IAA production ability of the microorganisms has been thought to be one of the most important criteria for screening of PGP due to its marked impact on the establishment and growth of a plant, particularly during seed germination and growth of the roots and the shoot [34, 35]. Moreover, in high salinity environments, plant growth is arrested due to higher salt concentration [36]; the physiological responses of the plants under salinity stress mainly include germination arrest, obstruction of the growth at seedling stage, root–shoot lengths, etc.; and the severity may vary depending upon the extent of salinity and the threshold tolerance of the plant. The germination experiment in wheat using the isolates in the present study also yielded linear results with that of the above (Fig. 3; Table S4), in which the impact of salinity is clearly evident in both the controls (CB and CW), respectively. However, the culture extracts of the isolates Y2, Y6, Y7, and Y10 showed a very significant enhancement in seed germination and seedling growth.
PGP activity of P. corylifolia-associated bacteria under salinity stress (EC = 5). CB (from left) represents the control with uninoculated culture broth; CW was the control with sterile Milli-Q water. Isolates Y1, Y3, Y5, and Y13 exhibited negligible enhancement effect on wheat germination, while that of the isolates Y2, Y6, Y7, Y10 was substantial
The isolates Y1, Y3, and Y5, though they exhibited good PGP ability in vitro, their reduced performance in the in planta studies (Fig. 3; Fig. 4) were quite unforeseen. This phenomenon can be attributed to the endophytic nature of these isolates, as all of them originated from root nodules, indicating the need for detailed endophytic studies including nodulation experiments. This also highlighted the requirement of the existence of complex interactions between the host and the endophytic microbe for optimal performance by the microorganisms; however, attempts towards the in vitro simulation of such environments could overcome the probable difficulties regarding the use of microbial metabolic products including those of endophytic microbes, instead of live cells for plant growth promotion in stress-prone environments.
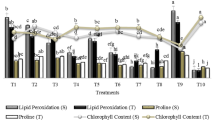
IAA production by the isolates vs. root/shoot development in wheat seedlings. Note the prominent fluctuations in root length throughout, where the root length seems to vary irrespective of IAA from bacterial origin. The shoot/root development clearly underlines the presence of some distinct factors that could impact the seedling development in wheat under salinity stress
The PGP abilities of the above isolates in particular need to be explored thoroughly; however, the probability regarding the inability of the culture extracts of the above isolates to induce the salinity stress tolerance in germinating seedlings also cannot be neglected, as the mechanism may perform at its peak in an endophyte environment. The isolates Y4, Y12, and Y14 were also from root nodules; however, they exhibited the PGP activity in planta, however, the results were quite fluctuating. It is therefore critical to evaluate the individual metabolites of these isolates for their ability to induce salinity stress tolerance in wheat and other crop plants. The isolates Y2 and Y10 dominated among all others in their PGP activity in wheat under salinity stress. The IAA production abilities of both the isolates differed noticeably; however, the culture extracts of both the isolates differed negligibly in the shoot/root ratio and root length in wheat, indicating the participation of multiple factors in stress alleviation that nevertheless require critical evaluation at the molecular level.
Auxins including IAA have been shown to play important roles in the regulation of growth and development of roots, leaves, and embryo [37, 38]. The IAA, though not an important hormone involved in seed germination, has role in the development of the seedlings [39]. Moreover the exogenous IAA has been shown to affect the process of germination and establishment by interacting with abscisic acid, gibberellins, and ethylene-mediated pathways [40, 41]. The tryptophan containing culture medium induced IAA production among the isolates; its impact on germination and growth parameters was studied in a separate experiment, in which similar results were obtained with the isolates except Y3, Y4, and Y6 (Fig. 5). Dramatic decrease in germination percentage was observed among the treatments, except for Y3, Y4, and Y6 where a slight increase in the germination percentage was noted. The most prominent downfall in germination percentage was recorded in the case of isolate Y8, the most potent IAA producer.
Similarly, impact on root length was also recorded, as IAA is one of the principal components influencing the root growth. Increase in root length was noted in the case of isolates Y1, Y5, Y6, Y12, and Y14; however, the extent of increase with reference to control appeared weaker (Fig. 6).
Influence of induced IAA on root length of wheat seedlings. CB: control—sterile culture broth; CW: control—sterile Milli-Q water. Figures in bold indicate the increased root length in the treatment with tryptophan containing culture filtrates/media (in case of CB). 1Values in the same data series represented by different letters are with significant difference (p = 0.05) in the Duncan’s multiple range test
Conversely, the seeds treated with the culture filtrate without tryptophan exhibited higher extent of root growth with reference to the control. Statistical analysis of the data revealed that isolates Y1 and Y14 belonged to the control group having sterile media containing tryptophan; however, there was a completely different pattern in the case of the treatment without tryptophan. The Y7 and Y8 isolates also showed a marked fall in root length in the presence of induced IAA.
Conclusion
The halotolerant bacteria accompanying the weed P. corylifolia L. promoted the growth of wheat under saline conditions. The impacts of isolates were independent of IAA. Some of the isolates have exceptional ability to produce IAA, indicating thereby their potential for large-scale usage. However, further studies are thus required on the application of bacterial cells for wheat seed germination, which will lead to the product development.
References
Seshadri, S., & Lakshminarasimhan, C. (2007). Population dynamics of P-solubilizers in the rhizosphere of major weed species from a tropical delta soil. In E. Velazquez & C. Rodriguez-Barrueco (Eds.), First international meeting on microbial phosphate solubilization (pp. 281–284). Berlin: Springer.
Kirtikar, K. R. & Basu, B. D. (1990). 2nd ed. Vol. 1. Dehradun, India: International Book Publisher; Indian Medicinal Plants; pp. 536.
Qiao, C. F., Han, Q. B., Song, J. Z., Mo, S. F., Kong, L. D., Kung, H. F., et al. (2007). Chemical fingerprint and quantitative analysis of FructusPsoraleae by HPLC. Journal of Separation Science, 30, 813–818.
Matsuda, H., Sugimoto, S., Morikawa, T., Matsuhira, K., Mizuguchi, E., Nakamura, S., & Yoshikawa, M. (2007). Bioactive constituents from Chinese natural medicines. XX. Inhibitors of antigen-induced degranulation in RBL-2H3 cells from the seeds of Psoralea corylifolia. Chemical and Pharmaceutical Bulletin, 55, 106–10.
Liu, R., Aifeng, L. I., Sun, A., & Kong, L. (2004). Preparative isolation and purification from Psoralea corylifolia by high speed counter current chromatography. Journal of Chromatography, 1057, 225–228.
Guo, J., Weng, X., Wu, H., Li, Q., & Bi, K. (2005). Antioxidants from Chinese medicinal herb—Psoralea corylifolia L. Food Chemistry, 91, 287–92.
Corwin, D. L., & Lesch, S. M. (2005). Apparent soil electrical conductivity measurements in agriculture. Computers and Electronics in Agriculture, 46, 11–43.
Zahra, S. O., Riccardo, T., & Berndt, G. (2004). Plant colonization by pink-pigmented facultative methylotrophic bacteria (PPFMs). FEMS Microbiology Ecology, 47, 319–332.
Supanekar, S. V., & Sorty, A. M. (2013). Siderophoregenic Klebsiella pneumoniaeSUP II from wheat (Triticum aestivum) rhizoplane. Indian Journal Research, 2, 243–45.
Reinhold, B., Hurek, T., Niemann, E. G., & Fendrik, L. (1986). Close association of Azospirillum and diazotrophic rods with different root zones of Kallar grass. Appliedand Environmental Microbiology, 52, 520.
Reinhold, B., Hurek, T., Fendrik, I., Pot, B., Gillis, M., Kersters, K., Thielemans, S., & de Ley, J. (1987). Azospirillumhalopraeferens sp. nov., a nitrogen fixing organism associated with roots of Kallar grass (Leptochloafusca (L.) Kunth)). lnternational Journal of Systematic Bacteriology, 37, 43.
Schwyn, B., & Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. AnalyticalBiochemistry, 160, 47–56.
Nautiyal, C. S. (1999). An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiology Letters, 170, 265–270.
Ehmann, A. (1977). The Van Urk-Salkowski reagent-a sensitive and specific chromogenic reagent for silica gel thin-layer chromatographic detection and identification of indole derivatives. Journal of Chromatography, 132, 267–276.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729.
Sauer, D. B., & Burroughs, R. (1986). Disinfection of seed surfaces with sodium hypochlorite. Phytopathology, 76, 745–749.
Sah, P., Agrawal, D., & Garg, S. P. (2006). Isolation and identification of furocoumarins from the seeds of Psoralea corylifolia L. Indian Journal of Pharmaceutical Science, 68, 768–71.
Prabha, C., Maheshwari, D. K., & Bajpai, V. K. (2013). Diverse role of fast growing rhizobia in growth promotion and enhancement of psoralen content in Psoralea corylifolia L. Pharmacognosy Magazine, 9, S57–S65.
Barak, M., & Ori, L. (2007). The effect of pH on the kinetics of spontaneous Fe (II) oxidation by O2 in aqueous solution—basic principles and a simple heuristic description. Chemosphere, 68, 2080–2084.
Sorty, A. M., & Shaikh, N. R. (2015). Novel co-enrichment method for isolation of magnetotactic bacteria. Journal of Basic Microbiology, 55, 520–526.
Gascoyne, D. J., Connor, J. A., & Bull, A. T. (1991). Isolation of bacteria producing siderophores under alkaline conditions. Applied Microbiology and Biotechnology, 36, 130–135.
Supanekar, S., Sorty, A., & Raut, A. (2013). Study of catechol siderophore from a newly isolated Azotobacter sp. SUP-III for its antimicrobial activity. Journal of Microbiology Biotechnology and Food Science, 3, 270–73.
Wang, Y. C., Wang, F., Hou, B. C., Wang, E. T., Chen, W. F., Sui, X. H., Chen, W. X., Li, Y., & Zhan, Y. B. (2013). Proposal of Ensiferpsoraleae sp. nov., Ensifersesbaniae sp. nov., Ensifermorelense comb. nov.andEnsiferamericanum comb. Nov. Systematic and Applied Microbiology, 36, 467–473.
Naz, I., Bano, A., & Ul Hassan, T. (2009). Isolation of phytohormones producing plant growth promoting rhizobacteria from weeds growing in Khewra salt range, Pakistan and their implication in providing salt tolerance to Glycine max. L. African Journal of Biotechnology, 8, 5762–5766.
Nelson, E. B. (2004). Microbial dynamics and interactions in the spermosphere. Annuual Review of Phytopathology, 42, 271–309.
Sutee, C., Chaum, S., & Sompornpailin, K. (2009). Differential accumulations of proline and flavonoids in indica rice varieties against salinity. Pakistan Journal of Botany, 41, 2497–2506.
Ortíz Castro, R., Contreras Cornejo, H. A., Macías Rodríguez, L., & LópezBucio, J. (2009). The role of microbial signals in plant growth and development. Plant Signaling and Behavior, 4, 701–712.
Badri, D. V., & Vivanco, J. M. (2009). Regulation and function of root exudates. Plant, Cell and Environment, 32, 666–681.
Bais, H. P., Park, S. W., Weir, T. L., Callaway, R. M., & Vivanco, J. M. (2004). How plants communicate using the underground information superhighway? Trends in Plant Science, 9, 26–32.
Castro, S. S., Herschkovitz, Y., Okon, Y., & Jurkevitch, E. (2007). Effects of inoculation with plant growth-promoting Rhizobacteria on resident rhizosphere microorganisms. FEMS Microbiology Letters, 276, 1–11.
Shih-Yung, H. (2010). IAA production by Streptomyces scabies and its role in plant microbe interaction. Masters thesis, Cornell University.
Nakbanpote, W., Panitlurtumpai, N., Sangdee, A., Sakulpone, N., Sirisom, P., & Pimthong, A. (2014). Salt-tolerant and plant growth-promoting bacteria isolated from Zn/Cd contaminated soil: identification and effect on rice under saline conditions. Journal of Plant Interactions, 9, 379–387.
Liu, Y., Shi, Z., Yao, L., Yue, H., Li, H., & Li, C. (2013). Effect of IAA produced by Klebsiellaoxytoca Rs-5 on cotton growth under salt stress. Journal of General and Applied Microbiology, 59, 59–65.
Fatima, Z., Saleemi, M., Zia, M., Sultan, T., Aslam, M., & Riaz-ur-Rehman Chaudhary, M. F. (2009). Antifungal activity of plant growth-promoting rhizobacteria isolates against Rhizoctoniasolani in wheat. African Journal of Biotechnology, 8, 219–225.
Wahyudi, A. T., Astuti, R. P., Widyawati, A., Meryandini, A., & Nawangsih, A. A. (2011). Characterization of Bacillus sp. strains isolated from rhizosphere of soybean plants for their use as potential plant growth for promoting Rhizobacteria. Journal of Microbiology and Antimicrobials, 3, 34–40.
Cuartero, J., & Fernandez-Munoz, R. (1999). Tomato and salinity. Scientia Horticulturae, 78, 83–125.
Popko, J., Hänsch, R., Mendel, R., Polle, A., & Teichmann, T. (2010). The role of abscisic acid and auxin in the response of poplar to abiotic stress. Plant Biology, 12, 242–258.
Rashotte, A. M., Brady, S. R., Reed, R. C., Ante, S. J., & Muday, G. K. (2000). Basipetal auxin transport is required for gravitropism in root of Arabidopsis. Plant Physiology, 122, 481–490.
Hentrich, M., Boettcher, C., & Duchting, P. (2013). ’The jasmonic acid signaling pathway is linked to auxin homeostasis through the modulation of YUCCA8 and YUCCA9gene expression’. Plant Journal, 74, 626–637.
Fu, X., & Harberd, N. P. (2003). Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature, 421, 740–743.
Liu, P. P., Montgomery, T. A., Fahlgren, N., Kasschau, K. D., Nonogaki, H., & Carrington, J. C. (2007). Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant Journal, 52, 133–146.
Acknowledgements
Financial assistance from Indian Council of Agricultural Research (ICAR), Govt. of India under Application of Microorganisms in Agriculture and Allied Sectors (AMAAS) scheme is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Highlights
Weed, Psoralea corylifolia L., growing in salinity-affected semi-arid regions, was explored for its associated plant growth-promoting bacteria.
The isolates exhibited potent plant growth-promoting properties in vitro. Culture filtrates containing metabolites significantly promoted seed germination in wheat under high salt environment.
This study has potential in exploitation of weed-associated microbes for development of salt stress resilient bio-inoculant in crop plants
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM. 1
(DOCX 236 kb)
Rights and permissions
About this article
Cite this article
Sorty, A.M., Meena, K.K., Choudhary, K. et al. Effect of Plant Growth Promoting Bacteria Associated with Halophytic Weed (Psoralea corylifolia L) on Germination and Seedling Growth of Wheat Under Saline Conditions. Appl Biochem Biotechnol 180, 872–882 (2016). https://doi.org/10.1007/s12010-016-2139-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2139-z