Abstract
The lipopeptide and its homologues are a kind of the five major biosurfactants with prominent interfacial and biological activities. A suite of mutagenesis method was adopted to expose a wild lipopeptide-producing strain Bacillus subtilis HSO121 to improve lipopeptide yield, and a stable mutant named R2-104 with a 2.0-fold production of lipopeptide was obtained. Compared to that of the wild strain HSO121, the lipopeptide produced by R2-104 showed a similar surface activity, but the course profiles of lipopeptide production during cultivation were different, with the peak yield of 500 mg at about 9 h by R2-104, and 400 mg at about 5 h by HSO121. The constituent abundance of the lipopeptide homologues produced by R2-104 was also different from that by HSO121. Combined methods of ESI-MS, GC-MS and MS-MS were applied for structural characterization of lipopeptide homologues, and it showed that the lipopeptides produced by R2-104 and HSO121 were attributed to a surfactin family with different constituents. The dominant constituent of the surfactin family produced by R2-104 was anteiso C15-surfactin with a relative content of 43.8 %, while the dominant one produced by HSO121was iso C14-surfactin with a relative content of 33.1 %.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The lipopeptide is an important part of biosurfactants produced by microorganisms such as genus Bacillus [1, 2] and Pseudomonas [3–5], and it has attracted much attention from both the scientific and industrial communities due to their prominent interfacial and biological activities [6, 7], and the great potentials in industrial fields, such as agriculture [8, 9], medicine [10, 11], food [12], cosmetics [6], environmental protection [13], and the petroleum industry to enhance oil recovery [2, 14]. During the last two decades, four main families of lipopeptides produced by microorganisms in laboratory, including surfactins [15], iturins [16], fengycins or plipastatins [17], and kurstakins [18], have been reported. However, the commercialization and the application in industrial practice of lipopeptides have not been fully achieved widely yet, and one of the reasons is the low yield and consequently the cost. The reported average yield of lipopeptides by wild strains under normal culture conditions was usually no more than 1 g/L [6, 15, 19, 20]. In order to improve the production performance of lipopeptides, except for screening wild lipopeptide-producing strains with high yield, fermentation engineering [16, 21, 22], and genetic engineering [23–25] were applied in the research of lipopeptides as well.
Mutagenesis was proved to be a cost-effective method to get forward mutants with higher production of microbial metabolites than that of the parent strains. Connor et al. [26] demonstrated a mutagenesis approach in developing a strain of Escherichia coli for production of 3-methyl-1-butanol by random mutagenesis and leveraging selective pressure toward l-leucine biosynthesis and screening for increased alcohol production. Lotfy et al. [27] used a stable mutant with an approximately 3.2-fold increase of citric acid by ultraviolet (UV) irradiation, ethyl methane sulfonate, and acridine orange mutagenesis to induce citric acid overproduction. Himabindu et al. [28] obtained a mutant with a 1.53-fold higher production of gentamicin than that of the parent strain after the gentamicin-producing strain Micromonospora echinospora was treated with chemical mutagens like EtBr and MNNG and physical mutagens such as ultraviolet. However, the research of using mutagenesis to obtain lipopeptide-producing mutants was not always reported. Kanda et al. [29] set up a strain-breeding and mutant selection system to improve the production of FR901379, a novel echinocandin type of lipopeptide antibiotic and obtained a variant strain M-7 showed 30.0 U/mL of production. Chen et al. [30] put forward the atmospheric pressure plasma jet as a novel mutagenic source for breeding high-yielding microbial mutant. A mutant strain designated as Bacillus subtilis B06 was successfully screened out which produced lipopeptides and showed higher antagonistic activity against F. graminearum. At the same time, mutagenesis could make more diverse in the growth of bacteria, gene information, and the category of products. Wu et al. [31] obtained an antitumor fungal mutant which produced seven novel and two known lipopeptides as well as five known polyketides by diethyl sulfate mutagenesis.
A series of structural analytical methods is key in characterization of the structural diversity of lipopeptides produced by mutants and to compare the similarity and difference with that of parent strains, since the microbial lipopeptides are a series of homologues. In general, a lipopeptide is composed of a fatty acid chain and a peptide chain with a specific sequence of amino acids [7]. The structural characterization of a lipopeptide was commonly distributed into two parts, including analysis of the fatty acid chain, and the composition and sequence of the peptide chain. The mass spectrometry (MS) was used preliminarily to identify the types of lipopeptides using the molecular weight (MW). Yang et al. [32] described a method to analyze the structure of fatty acid chains by using gas chromatography-mass spectrometry (GC-MS) after esterification. Comparison of mass spectrum of a lipopeptide before and after its hydrolysis was a feasible way to determinate the composition and sequence of the peptide chain [33]. Quadruple-time-of-flight tandem mass spectrum (Q-TOF MS/MS) could deduce the connection of amino acids in the peptide chain of a lipopeptide without hydrolysis directly. Analysis with the double hydrogen transfer (DHT) mechanism could also determine the C-terminal of the peptide chain [34].
In this paper, we present a stable B. subtilis mutant with enhanced lipopeptide production after composite random mutagenesis, and the course profile of lipopeptide production during cultivation, the composition and constituent abundance of lipopeptide homologues produced by the mutant was studied using combined methods of ESI-MS, GC-MS, and MS-MS.
Materials and Methods
Composite Mutagenesis
B. subtilis HSO121 [35] was grown in 100 mL sucrose culture (SC) medium containing: sucrose, 20.00 g/L; NH4NO3, 2.00 g/L; Na2HPO4 · 12H2O, 10.00 g/L; KH2PO4, 2.45 g/L; MgSO4, 0.10 g/L; yeast extract, 1.00 g/L; CaCl2, 0.78 mg/L; MnCl2, 1.26 mg/L in a 500-ml Erlenmeyer flask at 37 °C and 150 r/min for 12 h to reach the earlier stage of logarithmic phase.
The broth was diluted 100 times by normal saline and poured into petri dishes to make the thickness of the solution was 3 mm. The diluted broth was radiated by UV for 20 min with short wave by a UV light with the power of 20 W in super clean bench and the distance of irradiation was 50 cm. This dosage of UV light had been previously determined to give a survival rate of 10 %.
The resulted broth was subpackaged into 20-mL glass bottles with 5 mL in each bottle after UV radiation and then irradiated with gamma ray (Co-60, Institute of Nuclear Technology Application (INTA), East China University of Science and Technology) at the dose of 1000 Gy at room temperature. The dose rate was determined to be 2500 Gy/h by INTA. This dosage of gamma ray had been previously determined to give a survival rate of 10 %.
Screening Method
The resulted broth after composite mutagenesis was incubated at 37 °C for 20 h in the sucrose agar medium plates containing 1.8 % agar in the SC medium. Single colony was picked into 96-deep-well multiwell plates with 1.5 mL SC media in each well. After 12 h activated culture at 37 °C, mutants were incubated in new deep-well multiwell plates containing fresh medium for 48 h at 37 °C with the transfer volume of 100 μL.
Turbidometric method [36] was used to screen mutants which enhanced the lipopeptide production preliminarily. After 48 h of fermentation, cultures were centrifuged at 3000 × g for 20 min (11222, Sigma, Germany) to remove cells. A 200 μL cell-free supernatant of each mutant and 100 μL 10 % trichloroacetic acid were added into 96-well standard clrbtm; after 30 min standing in room temperature, turbidity was checked by Microplate Reader (M5, Molecular Devices, US) at 600 nm. Redistilled water of the same volume replaced 10 % trichloroacetic acid to mix with cell-free supernatant served as blank control. Mutants with much higher turbidity than B. subtilis HSO121 were purified through streaking on fresh agar plates and stored at −80 °C with 25 % (v/v) glycerol.
Determination of the Lipopeptide Production
Mutants with high turbidity were activated in 100 mL SC media in 500 ml erlenmeyer flask at 37 °C and 150 r/min for 20 h. The cultivation for 72 h at the same condition was proceed to produce lipopeptides with the transfer volume of 10 %. After centrifugation at 3000 × g for 20 min, 6 mol/L HCl was added to the cell-free supernatant until pH 2 was attained. The acidified cell-free broth was stored at 4 °C overnight for precipitation of lipopeptides. Crude lipopeptides were collected by centrifugation at 8000 × g for 30 min and dried in freeze dryer. After weighing the crude samples, lipopeptides were re-suspended in water and extracted 4 times with diethyl ether. The organic phases were collected, combined and evaporated with an air pump at room temperature, and dried in freeze dryer. The weight of production was recorded and compared with the parent strain to confirm the optimal mutant.
Fermentation and Monitoring of the Production of the Mutant
5 L fermentor (FMG-5 L, NCBIO, China) was used to cultivate the mutant continuously. After activated for 20 h, 150 mL bacterium solutions were inoculated into the fermentor containing 3000 mL SC medium. The temperature was constant at 37 °C and the pH was 7.20 in cultivation. The speed of agitator was 300 r/min and the ventilatory capacity was 1 vvm. Foam concentrate was collected every 2 h and the yield of lipopeptides was detected after acid precipitation by 6 mol/L HCl, centrifugation, and drying.
Stability of the Mutant
The mutant picked from the original agar plate was treated as the first generation and the production was determined. After stored at −20 °C overnight, the first generation was activated, transferred and cultivated by the same method, and the production was determined and recorded as the second generation. The production of the third, fourth and fifth generation was carried out by the same method.
Structural Analysis of the Lipopeptide from the Mutant
Determination of Molecular Mass
Electrospray ionization mass spectrometry (ESI-MS) (Thermo Finnigan, USA) was used to determinate the molecular weight of the lipopeptide. The instrument was operated in the positive mode and its capillary voltage, sample cone voltage and extraction cone voltages were 3 kV, 100 V and 6 V, respectively. The scanned area was 200–2000 m/z according to previous studies [37].
Analysis of Fatty Acid
Ten milligrams of extracted lipopeptide sample was dispersed in a 2-mL ampoule bottle with 1.0 mL of 6 mol/L HCl solution. The neck of the ampoule bottle was melted with alcohol blast burner and then sealed with a tweezers. The acid hydrolysis reaction was carried through at 90 °C for 24 h [38]. After hydrolysis, the whole product solution was transferred into a tube, and extracted four times with diethyl ether. The organic phases were collected, combined, and evaporated with an air pump at room temperature, and dried at 60 °C in vacuum oven for 2 h. Then 1.0 mL of 10 % (v/v) H2SO4-methanol solution was added into the residues to esterify at 55 °C for 6 h. After esterification, water and diethyl ether was added and extracted for four times. The organic phases containing fatty acid methyl esters were collected, and the solvent was removed with an air pump at room temperature. Finally, the esterified sample was dissolved with 0.5 mL of methanol.
To detect fatty acid methyl esters, GC-MS analysis was performed on a 6890 GC-7895 MS (Agilent,USA) equipped with a HP-5 MS capillary column, 30 m × 0.25 mm × 0.25 μm (Agilent, USA). The temperature of EI ion source was set at 230 °C with 70 eV of ionization energy. The carrier gas was helium (99.999 %), the flow rate was 1.0 mL/min, the injector temperature was 250 °C, the injection volume was 1.0 μL, and the split ratio was 10:1. The oven temperature was held initially at 60 °C for 3 min, then increased to 250 °C at 10 °C/min and finally kept at 250 °C for 10 min.
Amino Acid Analysis
The sample was hydrolyzed with the method mentioned above in the section of fatty acid analysis. After hydrolysis, the reactants were transferred into a vial. The solvent was removed with an air pump at room temperature and dried at 60 °C for 3 h in the vacuum oven, and the residues containing amino acids were obtained. Then the mixtures of 0.3 mL acetone and 0.2 mL N, O-bis (trimethylsilyl)-trifluoroacetamide (BSTFA; Fluka, USA) were added into the vial and heated at 60 °C for 20 min after being capped. The silylanized samples were analyzed with GC-MS.
GC-MS conditions were the same as for the analysis of fatty acid except for the programmed temperature conditions. The oven temperature was held initially at 80 °C for 3 min, then increased to 240 °C at 8 °C /min and finally kept at 240 °C for 10 min.
Identification of Connection of Amino Acids
The tandem mass spectrometry (Q-TOF Micro YA019, Micromass, UK) was utilized to determine the order of connection of amino acid residues in the biosurfactant with the method described by Yang et al. [34]. The electrospray mode was positive, the capillary voltage, sample cone voltage, ion energy and collision energy were 3.2 kV, 80 V, 1.6 V and 60 V, respectively.
Determination of the Surface Activity
Surface Tension
Lipopeptide samples were produced by R2-104 and obtained after acid precipitation and diethyl ether extraction. Aqueous solutions of lipopeptides were prepared at different concentrations from 10 to 400 mg/L by double distilled water. The surface tensions (SFT) of these solutions were measured with the plate method using a DCAT 21 tensiometer (DataPhysics, Germany) and the temperature was controlled at 25.0 ± 0.1 °C. The measurement was repeated three times and the average value was recorded. The surface tension between air and double distilled water was 71.8 mN/m at 25 °C.
Emulsification Activity
The cell-free broth obtained from the mutant was also checked for the emulsifying property. Equal volumes of cell-free broth and hexadecane were mixed by vortex at high speed for 5 min. The resulting mixture was incubated at 25 °C for 24 h and then the emulsification index (EI) value was calculated using the formula:
Results and Discussion
Selection of the Mutant and Lipopeptide Production Profile
An optimal forward mutant was screened and numbered as R2-104 after composite mutagenesis. The fermentative time course profile of lipopeptide production by the parent strain HSO121 and the mutant R2-104 were shown in Fig. 1.
Turbidometric method as a fast way to screen mutants was used based on the insoluble behavior of lipopeptides at low pH values [36]. A number of forward mutants with higher turbidity were selected and cultivated via the same method as the parent strain. After extraction and comparison of the production with that of the parent strain, an optimum forward mutant with higher production was selected and numbered as R2-104. The lipopeptides produced by R2-104 was extracted by acidic precipitation and diethyl ether extraction. The yield was 1747 mg/L (n = 3) after acidic precipitation and 866 mg/L (n = 3) after acidic precipitation and diethyl ether extraction. It was more than two times higher compared with the yield of the parent strain. Considering the yield of lipopeptides is not high in current studies, mutagenesis provided a feasible way to improve the yield of lipopeptides.
The differences of lipopeptide production between two strains were obvious compared with their growth conditions (Fig. 1). For HSO121, the output of lipopeptides mainly concentrated in the logarithmic growth phase especially the early and middle stages. Lipopeptides were produced after 2 h of incubation and showed a high level of output in the next 6 h. The lipopeptides production reached to its maximum of about 400 mg from 4 to 6 h, respectively, and the whole production phase was about 10 h. For R2-104, a continued lipopeptide production was detected from 2 to 20 h. The output was not very high at the start of fermentation but reached to a high level at 8 h to more than 500 mg. By comparison of the lipopeptide production, it is found that the mutant R2-104 showed a longer production life and a higher output at the whole fermentation process, though the production of lipopeptides was less than HSO121 at the beginning phase.
Meanwhile, the mutant R2-104 showed a good stability to produce lipopeptides. The yield of lipopeptides produced by 5 generations of the strain were 1747, 1860, 1609, 1788 and 1735 mg/L, respectively. After comparison of the yield of lipopeptides produced by different generations of the strain, it could be seen there was not any significant change between different generations, which indicated the mutant R2-104 could be used as a lipopeptide-producing strain reliably. The yield of lipopeptides after precipitation was proportional to that after precipitation and diethyl ether extraction. Moreover, precipitation was faster and convenient to implement. As a result, the weight of crude lipopeptides after precipitation was used to measure the production of lipopeptides unless otherwise noted.
Identification of the Lipopeptide
In consideration of the structural diversity of lipopeptide category and the change in mutagenesis, the molecular weights were determined and used for a primary confirmation the category of lipopeptides produced by R2-104. The result of ESI-MS is shown in Fig. 2.
The molecular weight of the lipopeptide produced by R2-104 was obtained by ESI-MS. Ionization ways of ESI-MS with positive ion modes were random with H+, Na+ and, K+. As shown in Fig. 2, the mass-to-charge ratios (m/z) of main peaks were m/z = 1008.6, 1022.6, 1030.7, 1036.6, 1044.6 and 1058.6, respectively. They were divided into two groups in which the difference value was 14: the first group contained m/z = 1008.6, 1022.6, and 1036.6 and the second group included m/z = 1030.7, 1044.6, and 1058.6. The difference value of 14 in the same group, the MW of CH2, indicated they were homologues with different carbon lengths. It was found that the difference of m/z in the two groups was 22, the same as the difference value of atomic weight between H and Na, which implied that the same molecule was ionized in two ways of H+ and Na+, respectively. In conclusion, the MW of the lipopeptides produced by R2-104 were mainly 1007, 1021, and 1035. It could be inferred the lipopeptide produced by R2-104 was surfactin homologues because they had the same MW as reported [20, 37].
Structure of the Lipopeptide
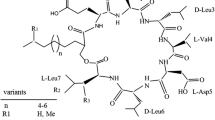
In consideration of the differences of the surfactin structure and composition of homologues, and the change brought by mutagenesis, we analyzed the structure of the surfactin produced by R2-104 by a systematic method and compared it with the parent strain, HSO121.
In order to confirm the structure of the fatty acid chain which formed the surfactin produced by R2-104 and the composition of surfactin homologues, the hydrolyzed sample was subjected to methyl esterification and analyzed by GC-MS. The GC result of the extracted ion chromatogram of m/z = 103 is shown in Fig. 3. The peak of m/z = 103 characterized as β-hydroxy fatty acids suggested the structure of [CH(OH)CH2COOCH3]+ [32]. Based on the analysis of MS information of each peak (S. 1), the whole composition of β-hydroxy fatty acids formed the surfactin produced by R2-104 was determined as in Fig. 3. It could be concluded that the eleven β-OH fatty acids were normal-C12, iso-C13, anteiso-C13, iso-C14, normal-C14, iso-C15, anteiso-C15, iso-C16, normal-C16, iso-C17 and anteiso-C17, respectively. The diverse molecular configurations made the difference in retention time between the same lengths of β-OH fatty acid methyl esters.
The analysis of the peptide part of the surfactin was conducted after hydrolysis and trimethylsilylation. The result of GC-MS was analyzed on the basis of the MS Agilent NIST 05 Chemical Structure Library (Fig. 4 and S. 2). The four peaks eluted at 8.5–16 min were identified as Val, Leu, Asp, and Glu, respectively, and the peak area ratio was about 1: 4: 1: 1. They were the components of the peptide chain in surfactins and had the same proportion as reported [15, 37].
In order to determine the sequences of amino acids in peptide chains, MS/MS was carried out according to the result of ESI-MS. Peaks with m/z = 1044.6, 1058.6, were chosen and subjected to MS/MS analysis (Fig. 5). As shown in Fig. 5 a, in accordance with a series of fraction ions of m/z = 1044.61, 931.52, 818.53, 703.50, 604.41 and 707.53, 594.38, 481.31, 382.06, 267.17, two sets of amino acid sequences Leu-Leu-Asp-Val and Leu-Leu-Val-Asp were received because of the sequential losses of amino acid and the composition information of amino acids from GC-MS. On the basis of double hydrogen transfer (DHT) mechanism [34], we recognized that an increase of 18 between m/z = 800.53 and 818.53 implied Leu-Leu-H2O was the C-terminal amino acid residues. Furthermore, the existence of H2O indicated the presence of a lactone ring which connected the -OH in fatty acid and the C-terminal of peptide. Since the C-terminal was occupied and there was no site to bind, the position of Glu was most probably at the N-terminal. This deduction was also in accordance with the molecular weight. Therefore, the amino acid sequence was determined to be N-Glu-Leu-Leu-Val-Asp-Leu-Leu-C. The difference between m/z 707.53 and 1044.61 was fitted with the fraction of C11H25C = CHCO-Glu, which indicated that the fraction of m/z = 1008.6 was the sodium adducts of C14-surfactin. According to the same method, the fractions of m/z = 1058.6 were recognized to be sodium adducts of C15-surfactin.
Structural Variation of the Lipopeptides Produced by the Parent Strain and R2-104
The lipopeptides produced by HSO121 were analyzed via the same method and proved to be of the surfactin family too. They had the same composition of fatty acids (S. 3) and amino acids (S. 4). Because the surfactin homologues were classified by the different length of β-OH fatty acids, the relative contents of surfactin homologues produced by R2-104 (Table 1) and HSO121 (Table 2) were determined by comparing the peak areas of every β-OH fatty acids.
It indicated that the composition of surfactin homologues produced by the mutant R2-104 was same as the parent strain HSO121. However, the relative contents of surfactin homologues produced by R2-104 were typically different from those by HSO121. The most dominant component in the homologues produced by HSO121 was the iso C14-surfactin, with the relative content of 33.1 %, followed by the anteiso C15-surfactin, with the relative content of 28.4 %. Howerer, the anteiso C15-surfactin came to be the most abundant component in the homologues produced by R2-104, with the relative content of 43.8 %, which, combined with iso C15-surfactin, made a major contribution to the high yield of surfactin family produced by the mutant R2-104.
Surface Activity of the Lipopeptide
The curve of surface tensions vs. the concentration of surfactin from R2-104 is shown is Fig. 6.
After linear fitting and calculating, the CMC of surfactin samples was confirmed to be 48.2 mg/L, the SFTCMC was 28.6 mN/m and the SFTmin was 27.8 mN/m. It indicated the surfactin produced by R2-104 possessed a low CMC and excellent surface property.
Meanwhile, hexadecane was emulsified by the R2-104 surfactin to high extent with an EI of 63.8 % (n = 3). The result could be seen in S. 5. Surface tension and emulsification activity were the main indicators to characterize the surface activity of the surfactants. It could be seen that lipopeptides produced by the mutant R2-104 has a prominent surface property and emulsifying property.
Conclusion
In this study, we obtained a stable B. subtilis mutant R2-104 with enhanced lipopeptide productivity after composite random mutagenesis. The lipopeptide produced by R2-104 showed a prominent surface property and emulsifying property. After fermentation, the condition of lipopeptide production of R2-104 and HSO121 were described and compared. R2-104 had a longer production process and a higher output. More than that, the composition and structure of the lipopeptide produced by the mutant was determined. It has been proved that the lipopeptide was a series surfactin homologues as same as HSO121. However, the proportion of surfactin homologues was changed and the main rise of output was caused by C15-surfactin.
References
Kim, P. I., Bai, H., Bai, D., Chae, H., Chung, S., Kim, Y., et al. (2004). Purification and characterization of a lipopeptide produced by Bacillus thuringiensis CMB26. Journal of Applied Microbiology, 97, 942–949.
Bezza, F. A., & Chirwa, E. M. N. (2015). Production and applications of lipopeptide biosurfactant for bioremediation and oil recovery by Bacillus subtilis CN2. Biochemical Engineering Journal, 101, 168–178.
Raaijmakers, J. M., Bruijn, I. D., & Kock, M. J. D. D. (2006). Cyclic lipopeptide production by plant-associated Pseudomonas spp.: diversity, activity, biosynthesis, and regulation. Molecular Plant-Microbe Interactions, 19(7), 699–710.
Trelita, D. S., & Saroj, B. (2012). Isolation and characterization of a lipopeptide bioemulsifier produced by Pseudomonas nitroreducens TSB.MJ10 isolated from a mangrove ecosystem. Bioresource Technology, 123, 256–262.
Souza, J. T. D., Boer, M. D., Waard, P. D., Beek, T. A. V., & Raaijmakers, J. M. (2003). Biochemical, genetic, and zoosporicidal properties of cyclic lipopeptide surfactants produced by Pseudomonas fluorescens. Applied and Environmental Microbiology, 69(12), 7161–7172.
Banat, I. M., Franzetti, A., Gandolfi, I., Bestetti, G., Martinotti, M. G., Fracchia, L., et al. (2010). Microbial biosurfactants production, applications and future potential. Applied Microbiology and Biotechnology, 87(2), 427–444.
Morikawa, M., Hirata, Y., & Imanaka, T. (2000). A study on the structure function relationship of lipopeptide biosurfactants. Biochimica et Biophysica Acta, 1488, 211–218.
Sachdev, D. P., & Cameotra, S. S. (2013). Biosurfactants in agriculture. Applied Microbiology and Biotechnology, 97(3), 1005–1016.
Chen, L. L., Wang, N., Wang, X. M., Hu, J. C., & Wang, S. J. (2010). Characterization of two anti-fungal lipopeptides produced by Bacillus amyloliquefaciens SH-B10. Bioresource Technology, 101(22), 8822–8827.
Weis, F., Beiras-Fernandez, A., & Schelling, G. (2008). Daptomycin, a lipopeptide antibiotic in clinical practice. Current Opinion in Investigational Drugs, 9(8), 879–884.
BenMohamed, L., Wechsler, S. L., Nesburn, A. B., BenMohamed, L., Wechsler, S. L., & Nesburn, A. B. (2002). Lipopeptide vaccines—yesterday, today, and tomorrow. The Lancet Infectious Diseases, 2(7), 425–431.
Nitschke, M., & Costa, S. G. V. A. O. (2007). Biosurfactants in food industry. Trends in Food Science & Technology, 18(5), 252–259.
Mulligan, C. N. (2009). Recent advances in the environmental applications of biosurfactants. Current Opinion in Colloid & Interface Science, 14(5), 372–378.
Pereira, J. F. B., Gudiña, E. J., Costa, R., Vitorino, R., Teixeira, J. A., Coutinho, J. A. P., et al. (2013). Optimization and characterization of biosurfactant production by Bacillus subtilis isolates towards microbial enhanced oil recovery applications. Fuel, 111, 259–268.
Peypoux, F., Bonmatin, J. M., & Wallach, J. (1999). Recent trends in the biochemistry of surfactin. Applied Microbiology and Biotechnology, 51(5), 553–563.
Mizumoto, S., Hirai, M., & Shoda, M. (2006). Production of lipopeptide antibiotic iturin A using soybean curd residue cultivated with Bacillus subtilis in solid-state fermentation. Applied Microbiology and Biotechnology, 72(5), 869–875.
Honma, M., Tanaka, K., Konno, K., Tsuge, K., Okuno, T., & Hashimoto, M. (2012). Termination of the structural confusion between plipastatin A1 and fengycin IX. Bioorganic & Medicinal Chemistry, 20(12), 3793–3798.
Abderrahmani, A., Tapi, A., Nateche, F., Chollet, M., Leclère, V., Wathelet, B., et al. (2011). Bioinformatics and molecular approaches to detect NRPS genes involved in the biosynthesis of kurstakin from Bacillus thuringiensis. Applied Microbiology and Biotechnology, 92(3), 571–581.
Hajfarajollah, H., Mokhtarani, B., & Noghabi, K. A. (2014). Newly antibacterial and antiadhesive lipopeptide biosurfactant secreted by a probiotic strain, Propionibacterium freudenreichii. Applied Biochemistry and Biotechnology, 174(8), 2725–2740.
Shaligram, N. S., & Singhal, R. S. (2010). Surfactin—a review on biosynthesis, fermentation, purification and applications. Food Technology and Biotechnology, 42(2), 119–134.
Das, K., & Mukherjee, A. K. (2007). Comparison of lipopeptide biosurfactants production by Bacillus subtilis strains in submerged and solid state fermentation systems using a cheap carbon source: Some industrial applications of biosurfactants. Process Biochemistry, 42(8), 1191–1199.
Piedrahita-Aguirre, C. A., Bastos, R. G., Carvalho, A. L., & Alegre, R. M. (2014). The influence of process parameters in production of lipopeptide iturin A using aerated packed bed bioreactors in solid-state fermentation. Bioprocess and Biosystems Engineering, 37(8), 1569–1576.
Miao, V., Coeffet-Le, G. M. F., Nguyen, K., Brian, P., Penn, J., Whiting, A., et al. (2006). Genetic engineering in Streptomyces roseosporus to produce hybrid lipopeptide antibiotics. Chemistry & Biology, 13(3), 269–276.
Li, Y. X., Li, Z. R., Yamanaka, K., Xu, Y., Zhang, W. P., Vlamakis, H., et al. (2015). Directed natural product biosynthesis gene cluster capture and expression in the model bacterium Bacillus subtilis. Scientific Reports, 5, 9383.
Qiu, Y. M., Xiao, F., Wei, X. T., Wen, Z. Y., & Chen, S. W. (2014). Improvement of lichenysin production in Bacillus licheniformis by replacement of native promoter of lichenysin biosynthesis operon and medium optimization. Applied Microbiology and Biotechnology, 98(21), 8895–8903.
Connor, M. R., Cann, A. F., & Liao, J. C. (2010). 3-Methyl-1-butanol production in Escherichia coli: random mutagenesis and two-phase fermentation. Applied Microbiology and Biotechnology, 86(4), 1155–1164.
Lotfy, W. A., Ghanem, K. M., & El-Helow, E. R. (2007). Citric acid production by a novel Aspergillus niger isolate: I. Mutagenesis and cost reduction studies. Bioresource Technology, 98(18), 3464–3469.
Himabindu, M., Potumarthi, R., & Jetty, A. (2007). Enhancement of gentamicin production by mutagenesis and non-nutritional stress conditions in Micromonospora echinospora. Process Biochemistry, 42(9), 1352–1356.
Kanda, M., Tsuboi, M., Sakamoto, K., Shimizu, S., Yamashita, M., & Honda, H. (2009). Improvement of FR901379 production by mutant selection and medium optimization. Journal of Bioscience and Bioengineering, 107(5), 530–534.
Chen, H., Chen, Z. J., Wu, M. B., & Deng, S. X. (2010). Screening the Fusarium graminearum inhibitory mutant strain from Bacillus subtilis by atmospheric-pressure plasma jet. Journal of Applied Microbiology, 108(1), 96–103.
Wu, C. J., Li, C. W., & Cui, C. B. (2014). Seven new and two known lipopeptides as well as five known polyketides: the activated production of silent metabolites in a marine-derived fungus by chemical mutagenesis strategy using diethyl sulphate. Marine Drugs, 12(4), 1815–1838.
Yang, S. Z., Wei, D. Z., & Mu, B. Z. (2007). Determination of the structure of the fatty acid chain in a cyclic lipopeptide using GC-MS. Journal of Biochemical and Biophysical Methods, 70(3), 519–523.
Yakimov, M. M., Abraham, W.-R., Meyer, H., Giuliano, L., & Golyshin, P. N. (1999). Structural characterization of lichenysin A components by fast atom bombardment tandem mass spectrometry. Biochimica et Biophysica Acta, 1438(2), 273–280.
Yang, S. Z., Wei, D. Z., & Mu, B. Z. (2006). Determination of the amino acid sequence in a cyclic lipopeptide using MS with DHT mechanism. Journal of Biochemical and Biophysical Methods, 68(1), 69–74.
Lv, Y. N., Yang, S. Z., & Mu, B. Z. (2005). Isolation and identification of a lipopeptide. Microbiology, 32(1), 67–73.
Mukherjee, S., Das, P., & Sen, R. (2009). Rapid quantification of a microbial surfactant by a simple turbidometric method. Journal of Microbiological Methods, 76, 38–42.
You, J., Yang, S. Z., & Mu, B. Z. (2015). Structural characterization of lipopeptides from Enterobacter sp. strain N18 reveals production of surfactin homologues. European Journal of Lipid Science and Technology, 117(6), 890–898.
Zhao, Y., Yang, S. Z., & Mu, B. Z. (2012). Quantitative analyses of the isoforms of surfactin produced by Bacillus subtilis HSO 121 using GC-MS. Analytical Sciences, 28, 789–793.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 21203063 and 51574125) and the 863 Program (Grant No. 2013AA064403).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 669 kb)
Rights and permissions
About this article
Cite this article
Meng, Y., Zhao, W., You, J. et al. Structural Analysis of the Lipopeptide Produced by the Bacillus subtilis Mutant R2-104 with Mutagenesis. Appl Biochem Biotechnol 179, 973–985 (2016). https://doi.org/10.1007/s12010-016-2044-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2044-5