Abstract
Plants are ubiquitously colonized by endophytic microorganisms which contribute significantly to plant health through production of plant growth regulators or disease suppression. In the present study, an endophytic bacterial isolate designated as BmB 1 with significant antifungal and plant growth promoting properties was isolated from the stem tissue of Bacopa monnieri (L.) Pennell. The isolate was studied in detail for the molecular and chemical basis of its bioactivity which proved it to have the presence of surfactin, iturin, and type I polyketide synthase (PKS) genes. For the analysis of the chemical basis of antifungal property, extract of the isolate was initially checked for its activity on test pathogens and LC-MS/MS based analysis further confirmed the presence of bacillomycin (m/z (M+H+) 1031.8) and surfactin (m/z (M+H+) 1008.6 and 1022.6) in the extract prepared. The light microscopic and SEM analysis of the treated and untreated mycelia of the pathogens clearly revealed the hypal destruction caused by the compounds produced by the selected isolate. This confirms the ability of the organism to directly inhibit the growth of the tested pathogens. The GC-MS analysis also confirmed the isolate to have the presence of volatile compounds with the expected role to induce induced systemic resistance (ISR) of the plant. Because of the multitargeted antifungal property, the isolate which was identified as Bacillus amyloliquefaciens can have potential biocontrol applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biodiversity richness and biosynthetic uniqueness are features of endophytic microorganisms that are highly impressive. They have been reported to possess the potential to produce an array of bioactive metabolites [1] with antifungal properties which enable the host plants to meet possible challenges generated by phytopathogenic fungi. So, identification of chemical basis of antifungal property of endophytes can have immense applications to explore them as biocontrol agents to prevent the plant diseases in an eco-friendly manner. Many endophytic Bacillus spp. have been reported to exhibit excellent natural product biosynthetic potential and hence much interest has been generated in recent years to develop Bacillus based commercial biocontrol formulations. The broad spectrum antiphytopathogenic properties of Bacillus spp. have been reported to be due to the production of wide range of secondary metabolites like lipopeptides, polypeptides, macrolactones, fatty acids, polyketides, lipoamides, and isocoumarins [2]. Antifungal compounds of peptide group that have been reported from Bacillus spp. mainly include mycobacillins, surfactins, mycosubtilins, and fungistatins [3]. In the case of Bacillus amyloliquefaciens, the peptide based as well as the volatile compounds trigger induced systemic resistance (ISR) [4]. This microbial induction of ISR enables the plant to resist of pathogenic microbes, viruses, and even nematodes which attack the plants. So, identification of various metabolites with antifungal properties from newly isolated Bacillus sp. deserves considerable importance because of the strain specific advantages expected from it. Also, presence of multitargeted antifungal traits among these isolates may act synergistically to provide effective protection from pathogens.
Medicinal plants can have highly efficient endophytic microflora to impart plant growth promoting and biocontrol properties and hence in the current study, the plant Bacopa monnieri was selected as the source for the isolation of endophytic bacteria. The plant is commonly known as “Brahmi” and is used in the preparation of many medicinal formulations. The major active compounds present in the plant include stigmastarol, alkaloids, flavonoids, betulicacid, beta-sitosterol, and saponins (bacoside A and B, bacopaside I and II, and bacopaside X, bacopasaponin C, bacopaside N2). In the present study, endophytic bacteria BmB 1 from B. monnieri (L.) was selected for studying the genomic and chemical basis of its antimicrobial property. PCR based genome mining confirmed the isolate to have the presence of genes for biosynthesis of surfactin, iturin, and type I polyketide synthase (PKS). The LC-MS/MS based analysis of the extract further confirmed the production of surfactin derivatives (m/z (M+H+)—1008.6, 1022.6) and bacillomycin (m/z (M+H+)—1031.5) by the selected bacterial isolate. The result of the GC-MS also suggests the isolate to have significant biosynthetic potential. The mechanistic basis of BmB 1 metabolites on cell wall of test fungi as studied by the light microscopy and SEM analysis confirmed its ability to disrupt cell wall with the resultant discharge of the cellular constituents. The plant growth promoting properties associated with BmB 1 further adds the multipotent application of BmB 1 as both biocontrol and phyto-enhancing agent.
Materials and Methods
Isolation of Endophytic Microorganisms and Screening for Antimicrobial Properties
The medicinal plant B. monnieri collected from Kerala Agricultural University was used as experimental material for the isolation of endophytic microorganisms. The surface sterilization and the isolation of endophytic bacteria were conducted using the procedures described by Aravind et al. [5]. Screening for antifungal activity of the isolates was done by dual-culture test against Rhizoctonia sp., Sclerotium sp., and Pythium sp. as per previously described methods by Shomura et al. [6]. The plates after incubation at 30 °C for 3–5 days were observed for the inhibition of the fungal mycelium.
Confirmation of Antifungal Activity
For confirming the activity of the selected endophytic organism, it was inoculated into 3 L of nutrient broth and was incubated at 30 °C for a period of 10 days with constant shaking in an incubator shaker. After incubation period, the cell free supernatant was collected by centrifugation at 10,000×g for 10 min at 4 °C. The collected supernatant was then acidified to pH 3.5 and was solvent extracted twice using equal volume of ethyl acetate. After extraction, the extract was made to dried powder and was then dissolved in methanol [7]. This was subjected to well diffusion method (20 mg/well) for the confirmation of the presence of antifungal compounds against Rhizoctonia sp., Sclerotium sp., and Pythium sp.
Molecular Screening for Natural Product Biosynthetic Gene Clusters
For screening the presence of known natural product biosynthetic gene clusters present in the isolate, PCR based screening method was done using primers specific for different biosynthetic gene clusters. For this, genomic DNA isolation was conducted using the genomic DNA isolation kit from Chromous Biotech Bacterial Genomic DNA Mini Spin Kit (RKT 17) as per the manufacturer’s procedure.
Primers used specifically to amplify different genes are summarized as Table S1. PCR was carried out in 50 μL reaction volume containing 50 ng of genomic DNA, 20 Pico moles of each primer (both forward and reverse), 1X EmeraldAmp GT PCR Master Mix (Takara Bio Inc., Japan). PCR was carried out for 35 cycles in a Mycycler™ (Bio-Rad, USA) with the initial denaturation at 94 °C for 5 min, cyclic denaturation at 94 °C for 30 s, annealing (temperature and time for specific primers are summarized in supplementary Table 1) and extension at 72 °C for 2 min with a final extension of 7 min at 72 °C. The amplified PCR product was visualized by electrophoresis in 1.5 % (w/v) agarose gel incorporated (10 mg/mL) for confirmation of amplification. The PCR product was then gel purified and sequenced using the Big Dye Terminator Sequence Reaction Ready Mix (Applied Biosystem, California, USA). The sequence data obtained was further analyzed by BLAST and detailed insilico phylogenetic analysis as per the previously described methods [7–9].
Identification of Bioactive Compounds by LC-MS/MS Analysis
The crude extract was subjected to LC-MS analysis to obtain the pseudomolecular ion of the active compound using Acquity H-Class (Waters, Milford, Massachusetts, USA) ultra performance liquid chromatography with BEH C18 column (1.7 μm, ɸ 2.1 × 50 mm), Xevo G2 (Waters, Milford, Massachusetts, USA) and Mass spectrometer. The electrospray ionization was used in both positive and negative mode with a scan range from m/z 50 to 1500, and the data acquirement time was 9 min. The pseudomolecular ion which was identified for possible bioactive compound from the LC-MS analysis was further confirmed by detailed LC-MS/MS based analysis using Acquity H-Class (Waters, Milford, Massachusetts, USA) ultra performance liquid chromatography with BEH C18 column (1.7 μm, ɸ 2.1 × 50 mm) and Xevo G2 (Waters, Milford, Massachusetts, USA) Quadruple Time of Flight (Q-TOF) Mass spectrometer.
GC-MS Analysis of Extract for Volatile Compounds
GC/MS analysis of the bacterial extract was performed on a 7890 GC system (Agilent Technology, Santa Clara, California, USA) 5975C inert MSD (mass selective detector) with an Agilent 190913-433, capillary column (30 m length, 250 μm i.d, 25 μm film thickness). Helium was used as carrier gas at a constant flow rate of 1.1 mL/min at an oven temperature programmed from 100 to 250 °C at the rate of 5 °C per minute. The mass spectra thus obtained were analyzed for major antifungal compounds by comparing with the database NIST 2011 GC/MS Library.
Effect of Compounds from Isolate BmB 1 on Fungal Morphology
For this, both light and electron microscopic analysis was done on treated (with 20 mg of extract/well) as well as control mycelia of Rhizoctonia sp. and Pythium sp. For light microscopy, the mycelial changes were observed under triocular microscope (Olympus®, Japan, model CX 41). For scanning electron microscopy (SEM), hyphal samples were fixed in 2.5 % glutaraldehyde at 4 °C for 2 h and washed 15 min in phosphate buffered saline (PBS) for four times. This was then dehydrated in a gradient of ethanol with each gradient treatment for 10 min and air-dried afterwards. SEM analysis was conducted by observing under JEOL USA SEM (JSM-6390) at 20 KV with a magnification of ×3700.
Optimization of Antifungal Activity of BmB 1 at Different Incubation Period
For optimizing the incubation period for production of antifungal compound, BmB1 was cultured on nutrient agar and the pathogens (Rhizoctonia sp. and Pythium sp.) were inoculated to this at different time intervals ranging from 0 h to 10 days. The growth of selected pathogen was then observed periodically.
Screening of BmB 1 for Plant Growth Promoting Properties
For screening BmB 1 for IAA production, the isolate was grown for 10 days at 28 °C in nutrient broth supplemented with 0.2 % (v/v) of L-tryptophan. After incubation, IAA production in the cell free supernatant was assayed by Salkowski’s method as per the methods described previously by Rahman et al. [10]. For screening the isolate for ammonia production, the organism was inoculated into peptone water and incubated for 2 days at room temperature. After incubation, 2–3 drops of Nessler’s reagent was added and formation of a brown color was observed as positive result [11]. The isolate was screened for the production of hydrogen cyanide was done using the method described by Jimtha et al. [12]. For this, the isolate was grown for 4 days in parafilm sealed nutrient agar plates containing 4.4 g/L (w/v) glycine with a Whatman filter paper no. 1 disc soaked in 2 % sodium carbonate in 0.5 % picric acid solution placed on the lid of the plate. The ACC deaminase production of the isolate was screened by using the methods described by Jimtha et al. [12]. For this, the isolated bacteria were grown on to DF salt minimal medium amended with 2 g/L ammonium sulfate. The bacterial isolate was screened for phosphate solubilizing property using the procedure described by Surange et al. [13]. For this, the isolate was inoculated on to the Pikovskaya medium containing 2.4 mg/mL bromophenol blue and was incubated for 48 h. The ability of the isolate for siderophore production was checked on Blue agar CAS medium containing chrome azurol S (CAS) and hexa decyl tri methyl ammonium bromide (HDTMA) as indicators [14] by incubating at 28 °C for 24 h. The BmB 1 was screened for nitrogen fixation using the method described by Jimtha et al. [12] on Jensen’s nitrogen free media, containing bromothymol blue as an indicator.
Identification of BmB 1
For this, genomic DNA previously isolated was used as the template for PCR amplification of 16S rDNA using the primers 16SF (5′–GAG TTT GAT CCT GGC TCA G–3′) and 16SR (5′–GAT ATT ACC GCG GCG CCT G–3′). The PCR product was further purified for its use as the template for sequencing PCR using Big Dye Terminator Sequence Reaction Ready Mix (Applied Biosystem, California, USA). The sequence data thus obtained was further subjected to BLAST analysis [8] and phylogenetic analysis [9].
Results
Screening of Antifungal Activity Against Selected Phytopathogens
Screening of antifungal property of BmB 1 has proved it to be highly active against Rhizoctonia sp. and Pythium sp. There was significant inhibition zone when the test fungi Rhizoctonia sp. and Pythium sp. were cultured along with BmB 1. However, the isolate was not found to have any inhibitory effect on the growth of Sclerotium sp. (Fig. S1). Also the crude extract from BmB 1 was also found to have activity against Rhizoctonia sp. and Pythium sp. as confirmed by well diffusion method. Inhibition of fungal growth near the well loaded with the crude extract was observed in both the cases (Fig. 1). This confirmed the presence of compounds with antifungal properties in the crude extract.
Molecular Screening for Natural Product Biosynthetic Gene Clusters
Among the different primers screened, BmB 1 was found to be positive for surfactin (srf) biosynthesis gene, iturin (Itu) biosynthesis gene, and gene for KS domain of type I polyketide synthase (PKS). The PCR product sizes were 645, 750, and 2000 bp for surfactin, type I PKS, and iturin, respectively (Fig. 2). The PCR products were further sequenced and BLAST analysis of aminoacid sequence of iturin revealed 73 % identity to that of iturin A synthetase A of B. subtilis (ABY89498). The aminoacid sequence of surfactin (srf) biosynthesis gene showed 100 % similarity towards 4′-phosphopantetheinyl transferase of B. amyloliquefaciens (CBI41443) by BLAST where as PKS gene showed only 91 % similarity to polyketide synthase gene of B. amyloliquefaciens (KMO08592) (Figs. 3, 4 and 5).
Identification of Antifungal Compound
For the identification of the antifungal compound, extract prepared from BmB 1 was subjected to LC–MS and LC–MS/MS analysis. From the detailed analysis of the LC-MS chromatogram with the reported mass, the presence of compounds identified were bacillomycin (m/z (M + H+) 1031.8), fengycin A (m/z (M + H+) 1463.6), iturin (m/z (M + H+) 1057.5, 1071.5, 1085.6), and surfactin (m/z (M + H+) 1008.6, 1022.6) (Fig. S2).
For further confirmation of the masses present, the pseudomolecular ion masses were subjected to fragmentation based analysis using tandem MS. The MS/MS based analysis confirmed the presence of bacillomycin (m/z (M+H+)1031.8) and surfactin (m/z (M+H+)1008.6 and 1022.6). The confirmation of the fragmented ions was done based on previous reports in the fragmentation of m/z 1031.5 which proved the presence of m/z 1018, 917, 905, 667, and 585 (Fig. 6). In the case of surfactin, the detailed LC-MS/MS analysis of m/z 1008.6 proved the presence of fragmented masses like m/z 990, 877, 685, 667, 554, and 441 which proved the presence of surfactin (Fig. 7). When the mass m/z 1022.6 was subjected to the fragmentation analysis, it yielded the pattern of fragmented masses m/z 1004, 909, 891, 685 667, 554, and 441 (Fig. 8). In case of both masses (m/z1008.6 and 1022.6), the presence of the base peak at m/z 685 also confirmed the presence of the compounds.
The extract was also analyzed for the presence of volatile compounds by GC-MS analysis. The volatile compounds identified and confirmed were benzeneacetic acid, pyrrolo[1, 2-a]pyrazine-1, 4-dione hexahydro-, pyrrolo[1, 2-a]pyrazine-1, 4-dione hexahydro-3-(2-methylpropyl), octadecanoic acid, 2, 5-piperazinedione, 3-benzyl-6-isopropyl, pyrrolo[1, 2-a]pyrazine-1, 4-dione hexahydro-3-(phenylmethyl), diisooctyl phthalate (Fig. S3 and Table 1).
Morphological Changes of Rhizoctonium and Pythium Induced by BmB 1
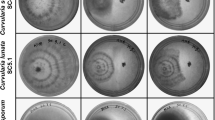
From the light microscopic analysis itself, severe mycelial destruction was observed in both Rhizoctonium sp. and Pythium sp. treated with isolate BmB 1. The shrinkage of the hyphal matrix of the fungi occurred and disintegration of mycelia and hyphae were observed. Lysis of hyphae and suppression of formation of new mycelia were also observed. The SEM images clearly explained the disruption of the fungal hyphae in the case of the treated samples and were clearly distinguishable from that of control where there was no visible disruption of the fungal cell wall (Fig. 9).
Microscopic analysis of the effect of isolate BmB 1 on the morphological alterations in the hyphal structure of phytopathogens. a Light microscopic image of untreated Rhizoctonia sp. b Light microscopic image of BmB 1 treated Rhizoctonia sp. c Light microscopic image of untreated Pythium sp. d Light microscopic image of BmB 1 treated Pythium sp. e SEM images of untreated Rhizoctonia sp. f SEM images of BmB 1 treated Rhizoctonia sp. g SEM images of untreated Pythium sp. h SEM images of BmB 1 treated Pythium sp.
Optimization of the Fermentation Time of the Isolate BmB 1
Time dependent antifungal activity of the BmB 1 against Rhizoctonia sp. and Pythium sp. was also examined. The isolate BmB 1 was found to exhibit maximum antifungal activity on the 4th day of incubation when co-inoculated with Rhizoctonia sp. and Pythium sp. (Figs. 10 and 11).
Optimization of antifungal activity against Rhizoctonia sp. at different incubation periods by BmB 1. a Pathogen inoculated at same day of inoculation. b Pathogen inoculated at 2nd day of inoculation. c Pathogen inoculated at 4th day of inoculation. d Pathogen inoculated at 6th day of inoculation. e Pathogen inoculated at 8th day of inoculation. f Pathogen inoculated at 10th day of inoculation
Optimization of antifungal activity against Pythium sp. at different incubation periods by BmB 1. a Pathogen inoculated at same day of inoculation. b Pathogen inoculated at 2nd day of inoculation. c Pathogen inoculated at 4th day of inoculation. d Pathogen inoculated at 6th day of inoculation. e Pathogen inoculated at 8th day of inoculation. f Pathogen inoculated at 10th day of inoculation
Plant Growth Promoting Properties
ACC deaminase production of BmB 1 was confirmed by its growth on Dworkin and Foster minimal salt medium. The isolate BmB 1 was also found to be ammonia - producing by the change of color of the broth to brown. The bacterial growth on Jensen’s medium showed its capability for nitrogen fixation. But, it was found not able to solubilize phosphate and produce HCN, and was also not producing indole acetic acid.
Identification of the Bacterial Isolate BmB 1
For the identification of the isolate BmB 1 molecular based method using 16SrDNA analysis was carried out. When the 16S rDNA gene sequence of BmB 1 obtained were analyzed with BLAST, it showed maximum identity of 99 % towards B. amyloliquefaciens (KF879305). The BLAST analysis result was further confirmed by phylogenetic analysis as it formed distinct clustering (Fig. 12).
Discussion
Endophytic microorganisms are considered to play key role in promotion of plant growth and protection from biotic and abiotic stresses. Due to the production of variety of bioactive metabolites, the endophytic microbiome of B. monnieri was also expected to be highly promising. Bacillus spp. with finely tuned chemical mechanisms which can be exploited for biocontrol applications, are well known for their ability to assign a large segment of its genome for the synthesis of diverse range of antimicrobial compounds including ribosomally synthesized compounds and nonribosomally synthesized cyclic peptide compounds like surfactins and iturins. Iturin producing B. amyloliquefaciens with anti-phytopathogenic activity against Rhizoctonia sp. has been reported previously as a safe alternative to chemical pesticides [15]. Experiments of Erlacher et al. [16] and Chowdhury et al. [17] have also suggested the use of Bacillus sp. as biocontrol agent against the phytopathogens affecting lettuce.
PCR based screening of the isolate BmB 1 proved it to have the presence of biosynthetic gene clusters for iturin, surfactin, and polyketides. BLAST analysis of the obtained sequences gave indirect evidence to the presence of lipopeptide based antifungal compounds in BmB1. The biosynthetic gene cluster of iturin has been reported to be encoded by iturin A operon composed of four open reading frames (orfs) ituD, ituA, ituB, and ituC [18]. However, the gene responsible for surfactin biosynthesis is present as part of srfA operon and the compound has multifaceted role in competency development and sporulation of bacteria. PCR based screening used in the current study was found to be highly helpful in the rapid selection of potential bacteria with multitargeted antifungal compounds.
LC/MS analysis showed BmB1 to have the presence of four compounds such as iturin, fengycin, bacillomycin, and surfactin. However, MS/MS based fragmentation analysis could confirm the presence of bacillomycin and surfactin only. The presence of bacillomycin was confirmed by the presence of masses m/z 1018, 917, 905, 667, and 585. At the same time, presence of surfactin derivative with mass m/z 1008.6 was confirmed by the presence of masses m/z 990, 877, 685, 667, 554 and 441 and with m/z 1022.6 confirmed by m/z 1004, 909, 891, 685 667, 554, and 441. The presence of the base peak at m/z 685 has been suggested as the final confirmation of the presence of the compounds as they are the precursor ion masses of the surfactin compound [19]. Even though the mode of action of the antagonistic volatile compounds produced by Bacillus spp. has not been studied in detail, some of them may be expected to trigger induced systemic resistance (ISR) in plants [4]. In the present study, the GC/MS analysis has identified a variety of compounds with possible potential for induced systemic resistance in plants.
The microscopical analysis of the BmB 1 treated mycelia of the Rhizoctonia sp. and Pythium sp. revealed its effect on the fungal morphology. The light microscopy itself showed the shrinkage of the hyphal matrix of the treated fungi. The disintegration and lysis of hyphae and suppression of formation of new mycelia were also observed. The SEM images clearly showed disruption of the fungal hyphae in the treated samples due to the mechanistic effect of compounds produced by the BmB 1 on fungal cell wall. There are many reports that suggest the mechanism of action of bioactive lipopeptide compounds to be targeting and binding directly to the cell membrane, which inturn has been suggested to cause the rapid depolarization of the membrane favoring the translocation of the peptide across the membrane to result in severe loss of the intracellular constituents and disruption of the cell wall. This direct binding mechanism also provides an additional advantage of reducing the development of resistance in the target organisms [20]. The ability of the BmB1 to suppress the growth of Rhizoctonia sp. and Pythium sp. in a very short span of 4 days as observed in the study is also indicative of its efficacy in treating the diseased plant without yield loss. Based on the genetic, metabolite, and morphological studies, it is suggested that the same mechanism might be the basis of the observed antifungal property of BmB 1. Apart from the antimicrobial production, the isolate BmB 1 was also showing significant plant growth promoting capacity. This makes the isolate to be fit for the use not only as a biocontrol agent but also as a bioenhancer in crop plants.
Rhizoctonia spp. are important soil borne fungal pathogens which causes sheath blight in corn, rice, cucumber; potato etc. [21]. Pythium spp. are also soil borne pathogens which causes damping off and root rot diseases on many plant species such as ginger, cabbage, broccoli, carrot, cucumber, melon, cotton, wheat etc. Both of them cause a severe reduction in the yields and sometimes total crop loss [22]. There are reports on the identification of microbes with biocontrol property against either Rhizoctonia sp. or Pythium sp. But, current result is significantly important as the isolate has broad activity against both Rhizoctonia sp. and Pythium sp. as confirmed by the LC-MS, GC-MS, and the SEM analysis. Hence, the result of this study will be of immense help for the development biocontrol formulation based on the ability of BmB 1 to control the fungal disease of wide range of crop plants. The endophytic origin of BmB 1 also makes it able to colonize inside the plants. This will also enable the long-term survival of the organism inside the plant. Organisms like B. amyloliquefaciens have been explored for plant probiotic potential due to their ability to interact with plant and also due to their excellent biocontrol mechanisms [17]. The organism in current study is remarkable and distinct because of its endophytic origin and the antifungal function that might have evolved in them as part of endophytic life.
In conclusion, the present study mainly focused on the bioactive potential of endophytic bacteria isolated from the stem tissue of B. monnieri. BmB1 isolated in the study was identified as B. amyloliquefaciens and was found to have genetic and biosynthetic make-up for production of surfactin and bacillomycin derivatives as identified by PCR and mass spectroscopic analysis. The antifungal properties of B. amyloliquefaciens were found to have mechanistic effect on cell wall of plant pathogens as observed by light and electron microscopic analysis. The occurrence of multi-targeted antifungal mechanisms of B. amyloliquefaciens makes it highly attractive to be exploited for biocontrol applications.
References
Jasim, B., John Jimtha, C., Jyothis, M., & Radhakrishnan, E. K. (2013). Plant growth promoting potential of endophytic bacteria isolated from Piper nigrum. Plant Growth Regulation, 71(1), 1–11. doi:10.1007/s10725-013-9802-y.
Ferreira, J. H. S., Matthee, F. N., & Thomas, A. C. (1991). Biological control of Eutypa lata on grapevine by an antagonistic strain of Bacillus subtilis. Ecology and Epidermiology, 81(3), 283–287.
Islam, M. R., Jeong, Y. T., Lee, Y. S., & Song, C. H. (2012). Isolation and identification of antifungal compounds from Bacillus subtilis C9 inhibiting the growth of plant pathogenic fungi. Mycobiology, 40(1), 59–66. doi:10.5941/MYCO.2012.40.1.059.
Chowdhury, S. P., Hartmann, A., Gao, X., & Borriss, R. (2015). Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42 - a review. Frontiers in Microbiology, 6, 780. doi:10.3389/fmicb.2015.00780.
Aravind, R., Kumar, A., Eapen, S. J., & Ramana, K. V. (2009). Endophytic bacterial flora in root and stem tissues of black pepper (Piper nigrum L.) genotype: isolation, identification and evaluation against Phytophthora capsici. Letters in Applied Microbiology, 48(1), 58–64. doi:10.1111/j.1472-765X.2008.02486.x.
Shomura, T., Omoto, S., Ohba, K., Ogino, H., Kojima, M., & Inouye, S. (1980). SF-1961, a new antibiotic related to bleomycin. The Journal of Antibiotics, 33(11), 1243–1248. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/6166601.
Jasim, B., Jimtha John, C., Shimil, V., Jyothis, M., & Radhakrishnan, E. K. (2014). Studies on the factors modulating indole-3-acetic acid production in endophytic bacterial isolates from Piper nigrum and molecular analysis of ipdc gene. Journal of Applied Microbiology, 117(3), 786–799. doi:10.1111/jam.12569.
Zhang, Z., Schwartz, S., Wagner, L., & Miller, W. (2000). A greedy algorithm for aligning DNA sequences. Journal of Computational Biology, 7(1–2), 203–214. doi:10.1089/10665270050081478.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., & Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28(10), 2731–2739. doi:10.1093/molbev/msr121.
Rahman, A., Sitepu, I. R., Tang, S.-Y., & Hashidoko, Y. (2010). Salkowski’s reagent test as a primary screening index for functionalities of rhizobacteria isolated from wild dipterocarp saplings growing naturally on medium-strongly acidic tropical peat soil. Bioscience, Biotechnology, and Biochemistry, 74(11), 2202–2208. doi:10.1271/bbb.100360.
Ahmad, F., Ahmad, I., & Khan, M. S. (2008). Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiological Research, 163(2), 173–181. doi:10.1016/j.micres.2006.04.001.
Jimtha, J., Smitha, P. V., Anisha, C., Deepthi, T., Meekha, G., Radhakrishnan, E. K., … Remakanthan, A. (2014). Isolation of endophytic bacteria from embryogenic suspension culture of banana and assessment of their plant growth promoting properties. Plant Cell, Tissue and Organ Culture (PCTOC), 118(1), 57–66. doi:10.1007/s11240-014-0461-0.
Surange, S., Wollum, A. G., II, Kumar, N., & Nautiyal, C. S. (1997). Characterization of Rhizobium from root nodules of leguminous trees growing in alkaline soils. Canadian Journal of Microbiology, 43(9), 891–894. doi:10.1139/m97-130.
Schwyn, B., & Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Analytical Biochemistry, 160(1), 47–56. doi:10.1016/0003-2697(87)90612-9.
Arrebola, E., Jacobs, R., & Korsten, L. (2010). Iturin A is the principal inhibitor in the biocontrol activity of Bacillus amyloliquefaciens PPCB004 against postharvest fungal pathogens. Journal of Applied Microbiology, 108(2), 386–395. doi:10.1111/j.1365-2672.2009.04438.x.
Erlacher, A., Cardinale, M., Grosch, R., Grube, M., & Berg, G. (2014). The impact of the pathogen Rhizoctonia solani and its beneficial counterpart Bacillus amyloliquefaciens on the indigenous lettuce microbiome. Frontiers in Microbiology, 5, 175. doi:10.3389/fmicb.2014.00175.
Chowdhury, S. P., Dietel, K., Rändler, M., Schmid, M., Junge, H., Borriss, R., … Grosch, R. (2013). Effects of Bacillus amyloliquefaciens FZB42 on lettuce growth and health under pathogen pressure and its impact on the rhizosphere bacterial community. PloS One, 8(7), e68818. doi:10.1371/journal.pone.0068818.
Tsuge, K., Inoue, S., Ano, T., Itaya, M., & Shoda, M. (2005). Horizontal transfer of iturin A operon, itu, to bacillus subtilis 168 and conversion into an iturin A producer. ANTIMICROBIAL AGENTS AND CHEMOTHERAPY, 49(11), 4641–4648. doi:10.1128/AAC.49.11.4641.
Tang, J.-S., Zhao, F., Gao, H., Dai, Y., Yao, Z.-H., Hong, K., … Yao, X.-S. (2010). Characterization and online detection of surfactin isomers based on HPLC-MS(n) analyses and their inhibitory effects on the overproduction of nitric oxide and the release of TNF-α and IL-6 in LPS-induced macrophages. Marine Drugs, 8(10), 2605–2618. doi:10.3390/md8102605.
Straus, S. K., & Hancock, R. E. W. (2006). Mode of action of the new antibiotic for gram-positive pathogens daptomycin: comparison with cationic antimicrobial peptides and lipopeptides. Biochimica et Biophysica Acta (BBA) - Biomembranes, 1758(9), 1215–1223. doi:10.1016/j.bbamem.2006.02.009.
Huang, X., Zhang, N., Yong, X., Yang, X., & Shen, Q. (2012). Biocontrol of Rhizoctonia solani damping-off disease in cucumber with Bacillus pumilus SQR-N43. Microbiological Research, 167(3), 135–143. doi:10.1016/j.micres.2011.06.002.
El-mohamedy, R. S. R. (2012). Biological control of pythium root rot of broccoli plants under. Journal of Agricultural Technology, 8(3), 1017–1028.
Acknowledgments
This study was supported by Department of Biotechnology (DBT), Government of India under DBT-RGYI and DBT-MSUB support scheme (BT/PR4800/INF/22/152/2012 Dtd 23.03.2012) and Kerala State Council Science Technology and Environment (KSCSTE) under KSCSTE-SARD Programme. The authors also acknowledge Prof. C. T. Aravindakumar, Hon. Director and Mr. Dineep D., Scientific Assistant of the Inter-University Instrumentation Centre, Mahatma Gandhi University, Kottayam for the help and support for the LC-MS/MS analysis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Compliance with Ethical Standards
We have not used any animal models for the experiments and thus do not require ethical committee clearance.
Conflict of Interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Jasim, B., Benny, R., Sabu, R. et al. Metabolite and Mechanistic Basis of Antifungal Property Exhibited by Endophytic Bacillus amyloliquefaciens BmB 1. Appl Biochem Biotechnol 179, 830–845 (2016). https://doi.org/10.1007/s12010-016-2034-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2034-7