Abstract
Jatropha curcas L. is a drought and salt-tolerant oil plant widely used for various purposes and has considerable potential as a diesel/kerosene substitute or extender. Understanding the molecular mechanisms underlie that the response to various biotic and abiotic stresses of this plant could be important to crop improvement efforts. Here, a new AP2/ERF-type transcription factor gene, named JcERF2, was isolated from the leaves of J. curcas. Sequence analysis showed that the JcERF2 gene contains a 759-bp open reading frame encoding a polypeptide of 252 amino acids. The predicted JcERF2 protein contained a conserved DNA-binding domain (the AP2/ERF domain) with 58 amino acids. The JcERF2 protein is highly homologous with other ERFs. JcERF2 was localized in the nucleus by analysis with a JcERF2-green fluorescent protein (GFP) fusion protein. Quantitative polymerase chain reaction (qPCR) analysis showed that JcERF2 was induced by drought, salt, abscisic acid, and ethylene. Overexpression of JcERF2 in transgenic tobacco plants enhanced the expression of biotic and abiotic stress-related genes, increased the accumulation of free proline and soluble carbohydrates, and conferred tolerance to drought and salt stresses compared to the wild type (WT). Taken together, the JcERF2 gene is a novel AP2/ERF transcription factor involved in plant response to environmental factors, which can be used as a potential candidate gene for genetic engineering of crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant growth and productivity are greatly limited by environmental stresses, of which drought, high salinity, and low temperature are the most serious threats in many areas of the world. Being sessile in nature, plants have evolved protective mechanisms to resist unfavorable conditions [8]. The protection is manifested by genes to modulate specific transcription factors via transduction cascades and transcriptional activation of series defense genes [45]. The AP2/ERF transcription family is a large family of plant transcription factors that includes four subfamilies called AP2, CBF/DREB, ERF, and RAV according to their types and numbers of DNA-binding domains [41]. The AP2 subfamily consists of two AP2/ERF domains that are mainly involved in the regulation of plant development [4, 9, 12]. The RAV subfamily, containing an AP2/ERF domain and a B3 domain, differs from the other transcription factor subfamilies, and some RAV transcription factors may cause transcriptional inhibition [23]. The CBF/DREB and ERF subfamily proteins contain one AP2/ERF domain each. The genes in the CBF/DREB subfamily primarily regulate the responses of plants to abiotic stress by recognizing the dehydration-responsive element or C-repeat (DRE/CRT). In contrast, the ERF subfamily regulates the responses of plants to biotic stress by recognizing the GCC box. Hundreds of transcription factor genes have been cloned from various plants since the first transcription factor was cloned from Zea mays by Paz-Ares [39].
The plant-specific ERF transcription factor family has numerous members among land plants and is involved in the signal transduction of the response to drought, salt, disease, and other biotic and abiotic stresses [6, 34]. The AP2/ERF transcription factor APETALA2 (AP2) gene was first isolated by Jofuku et al. [25] from Arabidopsis, after which the ERF transcription factors were successively isolated and studied in tomatoes [17], Arabidopsis thaliana [35], Nicotiana tabacum [13], pepper [49, 53], wheat [11], barley [26], rice [32], soybean [3, 7], cotton [24] and peanuts (Arachis hypogaea L.).
In rice, Cao et al. [5] showed that OsBIERF1, OsBIERF3, and OsBIERF4 were upregulated by salt, cold, drought, and wounding. The transcripts of TERF1 significantly accumulated under drought, cold, and abscisic acid (ABA) stress and involved in the ethylene and osmotic regulation pathways in tomato [56]. Overexpression of GmERF3 in tobacco increased their tolerance to salt, drought, and diseases [54]. Similarly, overexpression of RsERF1 in Arabidopsis exhibits improved expression of abiotic stress-related genes (e.g., ABF3, ABF4, ADH, Rab18, SUS1) and enhanced resistance to drought [3]. Studies have demonstated that ERF can increase tolerance to salt [3], drought [54], cold [53], high temperature, heavy metals, pathogens [49], and other biotic and abiotic stresses by activating or inhibiting expression of the target gene in ethylene signal transduction [18, 34]. ERF proteins may play pivotal roles in plant signal transduction and may be associated with different pathways [51]. Thus, the molecular mechanisms of ERF transcription factors should be investigated completely to contribute to agriculture and production.
Jatropha curcas L., belonging to the family of Euphorbiaceae, is a perennial, deciduous, drought-resistant shrub tree. It is native to Central America and widely distributed in tropical, subtropical, and hot, dry, valley areas, especially in Central and South America, Africa, India, and Southeast Asia [43]. J. curcas can be used to make biodiesel and biopesticides, in medical applications, to conserve soil, form biological hedges, provide animal feed, and other purposes [36]. The value of J. curcas as a biodiesel energy which may replace petroleum energy is receiving more attention because the depletion of traditional fossil fuels. As the genome of Jatropha being sequenced [21, 42] and genetic transformation technology being established [27, 29, 30, 37], the identification of a large number of agronomically important genes is now in progress. Nevertheless, few studies have focused on transcription factor genes in J. curcas. Only two papers have reported J. curcas AP2/EREBP transcription factors until now [47, 48].
In the present investigation, we cloned a new J. curcas ERF gene (JcERF2) by RACE PCR. Bioinformatics and expression analyses reveal that the predicted protein was located in the nucleus and induced by drought, salt, abscisic acid, and ethylene. Expression analysis was performed under stress conditions. Overexpression of JcERF2 transgenic tobacco plants was constructed to fully illustrated the molecular mechanism of JcERF2.
Materials and Methods
Plant Materials and Treatments
J. curcas seeds were collected from Xichang, Sichuan Province, China; germinated in moist vermiculite; and transplanted to a 1:1 mixture of nutrient soil and vermiculite under a 16-h light/8-h dark photoperiod at 28 °C. Seedlings grown to the five to six true-leaf stages were exposed to salt, drought, ABA, or ethylene. For salt and drought stress treatments, seedling roots were washed and immersed in 0.3 M NaCl and 20 % PEG6000, respectively [47]. For the ABA treatment, seedlings were sprayed with 200 μM ABA dissolved in 0.01 % alcohol solution [54]. Ethylene treatment was performed with 2 mL 40 % ethephon and 1 g NaHCO3 dissolved in 200 mL H2O [55]. To test the expression pattern of the JcERF2 gene, the treated materials were sampled at 0, 1, 3, 6, 12, 24, and 48 h, respectively, then frozen in liquid nitrogen and kept at −80 °C for further analysis.
Tobacco leaves were subjected to 70 % ethanol for 30 s, washed in sterile water three times, then disinfected in 0.1 % mercuric chloride for 8 min, and finally rinsed several time with sterile distilled water. The cotyledons were taken out from the seeds and placed in 40 mL Murashige and Skoog (MS) medium (Murashige and Skoog [33]) with 0.6 % (w/v) agar, pH 5.8, placed in the dark for 3 days, and then incubated at 26 °C in a 16-h light/8-h dark photoperiod.
Isolation and Sequence Analysis of the JcERF2 Gene
Total RNA was prepared from young leaves of J. curcas, using an RNA extraction kit (Tiangen Biotech, Beijing). Poly (A)+ RNA was isolated using an mRNA purification kit (Amersham Pharmacia Biotech, USA). The molecular cloning of the JcERF2 gene was accomplished by synthesizing first-strand complementary DNA (cDNA) from total RNA using a Primescrip RT reagent (TaKaRa, Dalian, China) and storing it at −20 °C. A 143-bp cDNA fragment was amplified from cDNA using AP2/ERF conserved domain degenerated primers (5′TGGGG(A/G)AAAT(T/G)(G/C)GC(T/G)GC3′ and 5′AA(A/G)TT(C/A/G)AC(T/C)TTAGCTTT3′). Degenerated primers were designed using the conserved sequence TRGVRQRPWGKVAEIRDP, commonly present in most of the ERF family.
The 143-bp cDNA fragment was cloned into pMD19-T easy vector for sequencing, and was confirmed to be an ERF gene. Next, 5′-RACE/3′-RACE methods were employed to extend the fragment into a full-length cDNA gene. The cDNA was obtained, denoted as JcERF2, and deposited in GenBank with the accession number of JF518881.1. The gene sequence of JcERF2 was analyzed by DNAMAN software, MEGA.5.1 software, a database search using Blastx (http://www.ncbi.nlm.nih.gov/), and the isoelectric point and molecular weight of JcERF2 were predicted by ExPASy (http://www.expasy.org/).
Expression of JcERF2 in Response to Stress Treatments
After selecting five to six true leaves from similarly sized seedlings, we exposed the seedlings to salt, drought, ABA, and ethylene and then sampled leaves at 0, 1, 3, 6, 12, 24, and 48 h. The expression of JcERF2 under these stress conditions was detected using real-time quantitative polymerase chain reaction (qPCR), and the 18sRNA gene of J. curcas was used as the reference gene. The primers were 5′ AGGAATTGACGGAAGGGCA3′ and 5′ GTGCGGCCCAGAACATCTAAG3′ for 18sRNA and 5′TGCTTCTGCCCCTTCTCG3′ and 5′TTGTTGGTGCTGCTGTGATG3′ for JcERF2.
Subcellular Localization Analysis of JcERF2
We constructed a transient expression vector by fusing JcERF2 to the N-terminal of green fluorescent protein (mGFP) under the control of CaMV 35S promoter (pBI221-JcERF2-mGFP). The recombinant vector was transferred into Arabidopsis protoplasts that were isolated from leaves as described by Abel and Theologis [1]. We cultured these for 24 h in the dark at 25 °C and then observed them using a confocal scanning laser microscope (Leica Microsystems, Heidelberg, Germany).
Generation of Transgenic Tobacco Plants
To produce overexpression tobacco, we amplified the open reading frame of JcERF2 using PCR. The product (759 bp) was cloned into the binary expression vector pBI121 (Invitrogen, USA) under the control of the CaMV 35S promoter. The transformation of tobacco was performed by the floral dipping method (Clough and Bent [10]) using Agrobacterium tumefaciens strain LBA4404. The transgenic plants were obtained through the Kan resistance screening of the pBI121 vector. Positive plants were obtained by RT-PCR analysis using total RNA that was extracted from transgenic tobacco as the template. T2 seeds of transgenic tobacco plants were harvested for use in later experiments.
Germination Experiment of JcERF2 Transgenic Tobacco
The seeds of transgenic and wild tobacco were sterilized as described previously and sown in ½ MS medium with different concentrations of NaCl (0, 50, 100, 150, 200 mM/L). They were maintained at 26 °C in the dark for 3 days and then 26 °C, in a 16-h light/8-h dark photoperiod for 7 days. We determined the germination rate and the root lengths of seedlings, each using 50 seeds, with three independent experiments. The data were analyzed with SPSS, version 13.0.
Tolerance of Overexpression Tobacco to Drought
The transgenic and wild tobacco seedlings were grown to the five to six true-leaf stages and treated with 30 % PEG600 for 15 days. We observed the phenotypic changes and detected the amounts of free proline [44], soluble carbohydrate [50], of the sample leaves from the control and stress treatment.
Expression Analysis of Putative Downstream Genes of JcERF2
Five putative downstream genes of JcERF2 were selected for analysis: NtPR1a (X06930.1), NtOsmotin (M29279.1), NtSAR8.2a (M97194.1), NtP5CS (HM854026.1), and NtSPSA (AF194022.1). Tobacco actin gene (JQ256516.1) was used as an internal control, and we explored the expression of these genes in wild and transgenic tobacco using quantitative real-time PCR. The primers that we used are shown in Table 1. Three replicates were performed.
Results
Molecular Cloning and Sequence Analysis of JcERF2
To understand the role of the ERF transcription factors from J. curcas, the novel gene JcERF2 was cloned from J. curcas leaves with degenerate primers based on the conserved amino acid domains of ERFs from other plants by RACE technology. Sequence analysis revealed that the full length of JcERF2 was 1367 bp, with an open reading frame (ORF) of 759 bp, a 5′-untranslated region (UTR) of 274 bp, a 3′UTR of 334 bp, and encodes a protein of 252 amino acids. According to ExPASy (http://web.expasy.org/), we predicted that the molecular weight of JcERF2 was 28.3 kDa with a isoelectric point (pI) of 5.83. The predicted JcERF2 protein has only one typical AP2 DNA-binding domain between 56 and 113 amino acid residues, suggesting that JcERF2 belongs to the ERF subfamily of the AP2/ERF family [19]. Subcellular localization signal analysis was conducted by http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_y.cgi, and it indicated that there was no nuclear localization signal in this protein.
Homology Analysis of JcERF2 Amino Acid Sequences
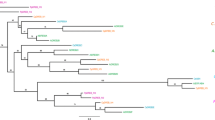
Blastx (http://www.ncbi.nlm.nih.gov/) analysis showed a high similarity between the predicted JcERF2 protein and other plant ERF proteins. Notably, JcERF2 shared a highest degree of similarity with Ricinus communis (80 %, XP002512669.1), as both J. curcas and R. communis are members of the family Euphorbiaceae. Homologous comparison also displayed that JcERF2 shared different degree of similarity with other ERFs from Morus notabilis (78 %, EXB64474.1), Cucumis sativus (78 %, XP004142046.1), Arabidopsis lyrata (74 %, XP002881295.1), Glycine max (74 %, XP003522452.1), and Citrus clementine (73 %, XP006436961.1) (Fig. 1a), which suggests that JcERF2 is a new member of AP2/ERF gene family. In order to investigate the evolutionary position of JcERF2 among the phylogenetic tree of various ERFs, a phylogenetic tree of ERFs was constructed using MEGA 5.1 software as shown in Fig. 1b. JcERF2 showed the closest relationship with R. communis.
Multiple sequence alignment and phylogenetic analysis of JcERF2. a Comparison of deduced amino acid sequences of AP2/ERF-related proteins that have high sequence similarity with JcERF2: Ricinus communis (XP002512669.1), Morus notabilis (EXB64474.1), Cucumis sativus (XP004142046.1), Arabidopsis lyrata (XP002881295.1), Glycine max (XP003522452.1), and Citrus clementina (XP006436961.1). Amino acid residues that are conserved in seven proteins are shown in dark gray. The sequence of JcERF2 between 56 and 113 bits is a conserved DNA-binding domain (ERF domain) marked by a transverse line. b The phylogenetic tree of JcERF2, showing the relationship between JcERF2 and other ERFs. It was generated using the unweighted pair-group method with arithmetic means (UPGMA) algorithm by MEGA4.1 software. The scale bar represents genetic distance
Expression Pattern of JcERF2 Gene Under Different Stress Conditions
As J. curcas can survive with high levels of salinity and dehydration, we have tested the expression level of JcERF2 in response to 0.3 M NaCl and 20 % PEG6000 in different time points. We also investigated the JcERF2 expression level when the seedlings were treated with ABA and ethylene since ERFs are involved in plant hormone signal transduction. The expression levels of JcERF2 were low under normal conditions but increased in response to salt, drought, ABA, and ethylene.
JcERF2 gene showed the highest level when exposed to 0.3 M NaCl for 1 h which indicates the irritability of plants, but in 20 % PEG6000, it appeared the highest level after a period time of 12 h stress, which may suggest that NaCl is a fast stimulator for JcERF2 without considering about the concentration of NaCl and PEG6000. When the seedlings were handled with ABA and ethylene, JcERF2 exhibit the highest expression level at 24 and 48 h. The expression level of JcERF2 first rise and then drop with the increasing of treatment duration. These phenomena may be explained by the irritability and adaptation of plant response to stress. Different stimulates caused by different expression level, from our data (Fig. 2), of the four treatments PEG6000 can lead to the highest expression level of JcERF2 much more than ABA, and followed by ethylene and NaCl. Thus, we can draw the conclusion that drought prompted the greatest increase expression of JcERF2. Further study still needed to explain the exact mechanism because our sampling time was not enough.
Expression of the JcERF2 gene in response to different stresses. Samples were taken at 0, 1, 3, 6, 12, 24, and 48 h. The expression of JcERF2 was detected by real-time qPCR, and the 18sRNA gene of Jatropha was used as the reference gene. a 300 mM NaCl, b 100 mM abscisic acid (ABA), c 20 % ethylene, d 30 % PEG6000. The mean value and standard deviation were obtained from three independent experiments
Subcellular Localization Analysis of JcERF2 Protein
Subcellular localization of proteins is important to elucidate their function in cells. Since JcERF2 is a transcription factor, it is expected to function in the nucleus, but bioinformatic analysis did not reveal a nuclear localization signal. Therefore, an in vivo targeting experiment was performed to fuse the JcERF2 coding region to the N-terminus of the mGFP gene under the control of the CaMV 35S promoter, transient expression vector transduction into Arabidopsis protoplasts [1]. Empty vector was used as a control. Localization of the fusion protein was then detected with a confocal laser fluorescence microscopy. As shown in Fig. 3, the JcERF2/mGFP fusion protein was exclusively located in the nucleus, whereas the control mGFP was uniformly distributed throughout the cell.
Subcellular localization of JcERF2 protein in Arabidopsis protoplasts. Arabidopsis protoplasts were transiently transformed with constructs containing either a control mGFP or JcERF2/mGFP under the control of the CaMV35S promoter. The subcellular location of the JcERF2/mGFP fusion protein or control mGFP alone were viewed with a confocal scanning laser microscope 24 h after transformation. Bright field images (left), fluorescence images (middle), and the corresponding overlay images (right) of representative cells expressing mGFP or JcERF2/mGFP fusion protein are shown
Overexpression of JcERF2 Enhanced Expression of Stress-Related Genes in Tobacco
In order to further study the function of JcERF2, we constructed the overexpression vector pBI121-JcERF2, in which JcERF2 was under the control of the CaMV 35S promoter. The vector was transformed into wild-type tobacco by the Agrobacterium-mediated leaf disc transformation method, seeds of the T2 generation harvested. Studies have demonstrated that PRs, osmotin, and SAR8.2 are GCC-box containing genes, their expression can be modulated by different ERF transcription factors [13, 17, 38]. According to the previous reports, P5CS mainly induced under salt, drought, ABA, and P5CS2 has a constitutive housekeeping gene [2]. So, we detected the expression levels of JcERF2, NtOsmotin, NtPR1a, NtSAR8.2a, NtP5CS, and NtSPSA by quantitative real-time PCR. As shown in Fig. 4, there was no expression of JcERF2 gene in wild-type tobacco, while in transgenic tobaccos, the expression of JcERF2 was significantly high. The expression of JcERF2 in transgenic tobacco implied that it can function well in tobacco, which suggests that JcERF2 can also express in other crops. The expression level of NtOsmotin, NtP5CS, NtPR1a, and NtSAR8.2a in transgenic tobacco is all higher than that in wild type (p < 0.05). But, the expression level of NtSPSA increased not so much as the others since the wild type also has a higher level of expression. The five selected gene showed a similarly expression level in the transgenic tobaccos suggested that those genes were modulated by the ERF subfamily and JcERF2 can regulate many genes involved in stress response.
Expression analysis of JcERF2 and putative downstream genes in wild-type and transgenic tobacco under normal conditions. Total RNA was isolated from the plants and analyzed by RT-qPCR using actin gene as a template control. OE1 to OE3 indicate JcERF2-transformed independent tobacco lines. The putative downstream genes are NtOsmotin (M29279.1), NtP5CS (HM854026.1), NtPR1a (X06930.1), NtSAR8.2a (M97194.1), and NtSPSA (AF194022.1). Error bars indicate SD from three independent experiments (*p < 0.05, **p < 0.01)
Seed Germination Assays
In order to determine the resistance of transgenic tobacco to salt, we conducted a germination experiment using the seeds of wild and transgenic tobacco. The seeds were sown in ½ MS medium containing different concentrations of NaCl. We determined the germination rate and root length of these seedlings. When treated with NaCl, the germination rate was significantly different; even at a low level of 50 mM NaCl stress, the transgenic tobacco seeds showed a higher rate of germination than wild type, and all of them showed a decrease germination rate with the increase of the NaCl concreation (p < 0.01) (Fig. 5a, b). The data also showed that few of the wild-type seeds can survive with 200 mM NaCl, and the germination ones showed growth retardation when compared with the transgenic tobacco seeds. The root length of transgenic and wild-type tobacco seedlings exhibits the same tendency, as shown in Fig. 5c, d. The wild-type and transgenic tobacco seedlings reached the same root length of 1.25 cm without NaCl, but the wild-type seedlings showed a more decrease in root length than the overexpression seedlings when the concentration of NaCl is increased. The length of wild-type seedlings is less than 1 cm with a 50 mM NaCl; the transgenic tobacco seedlings emerge this phenomenon when the NaCl concentration reached as much as 150 mM (p < 0.01) (Fig. 5c, d). These results demonstrated that overexpression of JcERF2 in tobacco improved seed germination rate and tolerance to NaCl.
Effect of salt stress on the seeding stage of wild-type (WT) and transgenic (OE1–OE3) tobaccos. a Tobacco seedlings grown for 7 days in Murashige and Skoog medium containing a 0 mM NaCl, b 50 mM NaCl, c 100 mM NaCl, d 150 mM NaCl, or e 200 mM NaCl. b Tobacco seedlings germination rate under salt stress. (Results are presented as mean ± SD from three independent experiments, and 50 seeds of each line were grown for each experiment.) c Tobacco seedlings grown for 7 days in Murashige and Skoog medium containing 0, 50, 100, 150, or 200 mM NaCl. d The root length of tobacco seedlings under salt stress. (Results are presented as mean ± SD from three independent experiments, and 20 seeds of each line were grown for each experiment.) The differences between the transgenic line and the WT plants are significant (p < 0.01) when salt stress is present
Defense Response is Enhanced in Transgenic Tobacco to Drought Stress
Soluble sugar and free proline are important physiological indicators of stress tolerance; these substances are positively correlated with response to stress in plants. In order to explore the functions of the JcERF2 gene in transgenic tobacco defensive reactions, seedlings of transgenic and wild-type tobaccos were irrigated with 30 % PEG6000 and leaves were collected after 15 days. The wild-type tobaccos were wilted slowly during the treatment, while the transgenic tobacco remain healthy and grow well (Fig. 6a).
Effects of drought stress on wild-type and transgenic tobacco plants. a The phenotypic changes in wild-type and transgenic plants 15 days after treatment with 30 % PEG, WT wild-type tobacco, OE transgenic tobacco. b Effects of drought stress on amounts of soluble carbohydrate. c Effects of drought stress on amounts of free proline. WT wild type, 1–3 different transgenic tobacco plants. Results are presented as mean ± SD from three independent experiments
Analysis of the amounts of free proline and soluble carbohydrates of the samples were performed. The wild-type and transgenic seedlings showed the same amounts of soluble carbohydrates in leaves before treatment, but the proline concentration is higher in transgenic tobacco seedlings than wild type (Fig. 6b, c), which implied that JcERF2 modulate the expression of the two substances by different mechanism. All the seedlings showed a higher concentration of proline and soluble carbohydrates when exposed to PEG6000, but the increase is significantly higher in transgenic tobacco seedlings. When compared the samples after treatment, we can find that transgenic tobacco seedlings have a higher concentration of proline and soluble carbohydrates than the wild type, which demonstrated that overexpression of JcERF2 in transgenic tobacco enhanced their tolerance to drought stress.
Discussion
J. curcas has great capability for environmental adaption, but the regulation mechanism of AP2/ERF genes in J. curcas is skimpy. Thus, the investigation of AP2/ERF genes in J. curcas is vital for understanding plant adaption to environmental stress. The expression of AP2/ERF transcription factors can be induced by a wide range of biotic and abiotic stresses, such as drought [14, 54], salt [54], hypoxia [15, 20, 31], pathogens [54], cold [41], submergence [14], freezing [53], potassium deficiency stress [28], and abscisic acid [46]. In our study, we observed that the transcript levels of JcERF2 could be enhanced by salt, drought, ABA, and ethylene. Previous reports indicated that most ERF transcription factors act as transcriptional activators, but some function in transcriptional inhibition [52]. Thus, we predicted that a cross-talk signal pathway exists in J. curcas and JcERF2 plays multiple roles in acquisition of plant stress tolerance. Investigation of the regulation and functional interactions, together with identification of new factors, is needed to understand the time-dependent action of JcERF2 in a wide response network to environment stresses.
Plant hormone signaling pathways are reported to participate in a variety of developmental processes and stress response of environmental stimuli [57]. The stress treatment of ABA and ethylene induced increased expression of JcERF2 at early stress period and then decreased, implicating that JcERF2 may be involved in ABA-dependent stress responses. The ABA signaling pathways are reported to comprise signal transducers and transcription factors [58], but the information of their exacted interaction mechanism is limited. Further studies should be done to identify the core sequence and element protein binding with and to explain the complex regulation mechanisms of gene expression in response to changes in external and internal environments by JcERF2 transcription factor. Besides, interactions between JcERF2 and other proteins are also important to clarify the regulatory steps on gene expression.
External stress stimulates the expression of JcERF2 and upregulates the expression of downstream genes involved in stress response, which improved the content of free proline and soluble sugar in plant [16, 22, 40]. The higher contents of proline in leaves of JcERF2-overexpressing lines under 30 % PEG conditions, suggesting the overexpression of JcERF2 gene, enhance the proline accumulation and further increase the drought tolerance of transgenic tobacco by controlling the osmotic pressure of cells. We found that the amounts of free proline and soluble sugar are expressed through different pathway in transgenic tobacco. Before treatment, proline in transgenic tobacco showed a higher content more than wild type, which suggest that JcERF2 may directly modulate the genes involved in the expression of proline, while JcERF2 may regulate the expression of soluble sugar with the help of other factors. From the qRT-PCR analysis, P5CS transcripts were higher in overexpression lines than wild type that confirmed that JcERF2 overexpression lines increased the content of proline may due to the upregulation of NtP5CS. In our study, we also found that overexpression of JcERF2 significantly upregulated the expression of a series of downstream genes (NtOsmotin, NtPR1a, NtSAR8.2a) in transgenic tobacco. Since the JcERF2 gene could not be found in wild-type tobacco, we can draw the conclusion that these genes were modulated by JcERF2, which may contribute to the fact that these genes contain GCC boxes. The transgenic line exhibited a higher level of NtSPSA gene than wild type, but the soluble sugar showed the same level in both lines before treatment, which demonstrated that JcERF2 affect sugar metabolism through different ways which include the expression of NtSPSA gene. The data presented in this report suggest that JcERF2 is a positive transcription factor function in cross-talk between different stress pathways in plants. The transgenic tobacco showed a higher tolerance to drought, and salt stress indicated that JcERF2 is an attractive engineering target in breeding plants for high antioxidant capacity to create multiresistant crops. However, the detail events involved in activating the defense response of JcERF2 still need to be further researched to completely understand its function, since transgenic crops being the most controversial topics in recent years. In order to improve our understanding of JcERF2, RNAi plants need to be constructed; molecular markers and genomic transcriptome analysis also exhibit great advantage in deeply understanding the mechanism involved in JcERFs.
In conclusion, the novel ERF transcription factor JcERF2 located in the nucleus and induced by exposure to salt, drought, ABA, and ethylene. Overexpression of JcERF2 in transgenic tobacco showed increased tolerance to salt and drought stress. JcERF2 upregulated the expression levels of JcERF2, NtOsmotin, NtPR1a, NtSAR8.2a, NtP5CS, and NtSPSA in transgenic tobacco, which indicated its multiple role in plant defense signal transduction networks.
References
Abel, S., & Theologis, A. (1994). Transient transformation of Arabidopsis leaf protoplasts: a versatile experimental system to study gene expression. The Plant Journal, 5, 421–427.
Archana, N. R., & Penna, S. (2013). Molecular evolution of plantP5CSgene involved in proline biosynthesis. Molecular Biology Reports, 40, 6429–6435.
Ayarpadikannan, S., Chung, E., Kim, K., So, H., Schraufnaggle, K. R., & Lee, J. (2014). RsERF1 derived from wild radish (Raphanus sativus) confers salt stress tolerance in Arabidopsis. Acta Physiologiae Plantarum, 36, 993–1008.
Boutilier, K., Offringa, R., Sharma, V. K., Kieft, H., Ouellet, T., Zhang, L. M., Hattori, J., Liu, C. M., van Lammeren, A. A., Miki, B. L. A., Custers, J. B. M., & van Lookeren Campagne, M. M. (2002). Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell, 14, 1737–1749.
Cao, Y. F., Song, F. M., Goodman, R. M., & Zheng, Z. (2006). Molecular characterization of four rice genes encoding ethylene-responsive transcriptional factors and their expressions in response to biotic and abiotic stress. Journal of Plant Physiology, 163, 1167–1178.
Cao, Y. F., Wu, Y. F., Zheng, Z., & Song, F. M. (2005). Overexpression of the rice EREBP-like gene OsBIERF3 enhances disease resistance and salt tolerance in transgenic tobacco. Physiological and Molecular Plant Pathology, 67, 202–211.
Chen, M., Xu, Z., Xia, L., Li, L., Cheng, X., Dong, J., Wang, Q., & Ma, Y. (2009). Cold-induced modulation and functional analyses of the DRE-binding transcription factor gene, GmDREB3, in soybean (Glycine max L.). Journal of Experimental Botany, 60, 121–135.
Chinnusamy, V., Schumaker, K., & Zhu, J. K. (2004). Molecular genetic perspectives on cross-talk and specificity in abiotic stress signaling in plants. Journal of Experimental Botany, 55, 225–236.
Chuck, G., Meeley, R. B., & Hake, S. (1998). The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet l. Genes & Development, 12, 1145–1154.
Clough, S. J., & Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal, 16, 735–743.
Egawa, C., Kobayashi, F., Ishibashi, M., Nakamura, T., Nakamura, C., & Takumi, S. (2006). Differential regulation of transcript accumulation and alternative splicing of a DREB2 homolog under abiotic stress conditions in common wheat. Genes & Genetic Systems, 81, 77–91.
Elliott, R. C., Betzner, A. S., Huttner, E., Oakes, M. P., Tucker, W. Q., Gerentes, D., Perez, P., & Smyth, D. R. (1996). AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell, 8, 155–168.
Fischer, U., & Dröge-Laser, W. (2004). Overexpression of NtERF5, a new member of the tobacco ethylene response transcription factor family enhances resistance to tobacco mosaic virus. Molecular Plant-Microbe Interactions, 17, 1162–1171.
Fukao, T., Yeung, E., & Bailey-Serres, J. (2011). The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell, 23, 412–427.
Gibbs, D. J., Lee, S. C., Isa, N. M., Gramuglia, S., Fukao, T., Bassel, G. W., Correia, C. S., Corbineau, F., Theodoulou, F. L., Bailey-Serres, J., & Holdsworth, M. J. (2011). Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature, 479, 415–418.
Gilmour, S. J., Sebolt, A. M., Salazar, M. P., Everard, J. D., & Thomashow, M. F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiology, 124, 1854–1865.
Gu, Y. Q., Wildermuth, M. C., Chakravarthy, S., Loh, Y. T., Yang, C., He, X. H., Han, Y., & Martin, G. B. (2002). Tomato transcription factors pti4, pti5 and pti6 activate defense responses when expressed in Arabidopsis. Plant Cell, 14, 817–831.
Gutterson, N., & Reuber, T. L. (2004). Regulation of disease resistance pathways by AP2/ERF transcription factors. Current Opinion in Plant Biology, 7, 465–471.
Hao, D. Y., Ohme-Takagi, M., & Sarai, A. (1998). Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. The Journal of Biological Chemistry, 273, 26857–26861.
Hinz, M., Wilson, I. W., Yang, J., Buerstenbinder, K., Llewellyn, D., Dennis, E. S., Sauter, M., & Dolferus, R. (2010). Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiology, 153, 757–772.
Hirakawa, H., Tsuchimoto, S., Sakai, H., Nakayama, S., Fujishiro, T., Kishida, Y., Kohara, M., Watanabe, A., Yamada, M., Aizu, T., et al. (2012). Upgraded genomic information of Jatropha curcas L. Plant Biotechnology, 29, 123–130.
Hongbo, S., Zongsuo, L., & Mingan, S. (2006). Osmotic regulation of 10 wheat (Triticum aestivum L.) genotypes at soil water deficits. Colloids and Surfaces. B, Biointerfaces, 47, 132–139.
Hu, Y. X., Wang, Y. H., Liu, X. F., & Li, J. Y. (2004). Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Research, 14, 8–15.
Jin, L., Huang, B., Li, H., & Liu, J. (2009). Expression profiles and transactivation analysis of a novel ethylene-responsive transcription factor gene GhERF5 from cotton. Progress in Natural Science, 19, 563–572.
Jofuku, K. D., den Boer, B. G., Montagu, M. V., & Okamuro, J. K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell, 6, 1211–1225.
Jung, J., Won, S. Y., Suh, S. C., Kim, H., Wing, R., Jeong, Y., Hwang, I., & Kim, M. (2007). The barley ERF-type transcription factor HvRAF confers enhanced pathogen resistance and salt tolerance in Arabidopsis. Planta, 225, 575–588.
Kajikawa, M., Morikawa, K., Inoue, M., Widyastuti, U., Suharsono, S., Yokota, A., & Akashi, K. (2012). Establishment of bispyribac selection protocols for Agrobacterium tumefaciens- and Agrobacterium rhizogenes-mediated transformation of the oil seed plant Jatropha curcas L. Plant Biotechnology, 29, 145–153.
Kim, W. K., Kim, J. H., Jeong, D. H., Chun, Y. H., Kim, S. H., Cho, K. J., & Chang, M. J. (2011). Radish (Raphanus sativus L. leaf) ethanol extract inhibits protein and mRNA expression of ErbB2 and ErbB3 in MDA-MB-231 human breast cancer cells. Nutrition Research Practice, 5, 288–293.
Kumar, N., Reddy, M., & Sujatha, M. (2013). Genetic transformation of Jatropha curcas: Current status and future prospects. In B. Bahadur, M. Sujatha, & N. Carels (Eds.), Jatropha, Challenges for a New Energy Crop. Volume 2 (pp. 535–546). New York: Springer.
Li, M. R., Li, H. Q., Jiang, H. W., Pan, X. P., & Wu, G. J. (2008). Establishment of an Agrobacteriuim-mediated cotyledon disc transformation method for Jatropha curcas. Plant Cell Tissue and Organ Culture, 92, 173–181.
Licausi, F., van Dongen, J. T., Giuntoli, B., Novi, G., Santaniello, A., Geigenberger, P., & Perata, P. (2010). HRE1 and HRE2, two hypoxiainducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. The Plant Journal, 62, 302–315.
Matsukura, S., Mizoi, J., Yoshida, T., Todaka, D., Ito, Y., Maruyama, K., Shinozaki, K., & Yamaguchi-Shinozaki, K. (2010). Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Molecular Genetics and Genomics, 283, 185–196.
Murashige, T., & Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum, 15, 473–497.
Ohme-Takagi, M., & Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell, 7, 173–182.
Oñate-Sánchez, L., & Singh, K. B. (2002). Identification of Arabidopsis ethylene-responsive element binding factors with distinct induction kinetics after pathogen infection. Plant Physiology, 128, 1313–1322.
Openshaw, K. (2000). A review of Jatropha curcas: an oil plant of unfulfilled promise. Biomass and Bioenergy, 19, 1–15.
Pan, J. L., Fu, Q. T., & Xu, Z. F. (2010). Agrobacterium tumefaciens-mediated transformation of biofuel plant Jatropha curcas using kanamycin selection. African Journal of Biotechnology, 9, 6477–6481.
Park, J. M., Park, C. J., Lee, S. B., Ham, B. K., Shin, R., & Paek, K. H. (2001). Overexpression of the tobaccoTsi1gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. The Plant Cell, 13, 1035–1046.
Paz-Ares, J., Ghosal, D., Wienand, U., Peterson, P. A., & Saedler, H. (1987). The regulatory C1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. The EMBO Journal, 6, 3553–3558.
Roosens, N. H., al Bitar, F. A., Loenders, K., Angenon, G., & Jacobs, M. (2002). Overexpression of ornithine-d-aminotransferase increases pro biosynthesis and confers osmotolerance in transgenic plants. Molecular Breeding, 9, 73–80.
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., & Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochemical and Biophysical Research Communications, 290, 998–1009.
Sato, S., Hirakawa, H., Isobe, S., Fukai, E., Watanabe, A., Kato, M., Kawashima, K., Minami, C., Muraki, A., Nakazaki, N., et al. (2011). Sequence analysis of the genome of an oil-bearing tree, Jatropha curcas L. DNA Research, 18, 65–76.
Schmook, B., & Serralta-Peraza, L. (1997). J. curcas: distribution and uses in the Yucatán Peninsula of Mexico. In G. M. Gübitz, M. Mittelbach, & M. Trabi (Eds.), Biofuels and industrial products from Jatropha curcas (pp. 53–57). Graz: DVB.
Shan, D. P., Huang, J. G., Yang, Y. T., Guo, Y. H., Wu, C. A., Yang, G. D., Gao, Z., & Zheng, C. C. (2007). Cotton GhDREB1 increases plant tolerance to low temperature and is negatively regulated by gibberellic acid. The New Phytologist, 176, 70–81.
Singh, K. B., Foley, R. C., & Oñate-Sánchez, L. (2002). Transcription factors in plant defense and stress responses. Current Opinion in Plant Biology, 5(5), 430–436.
Song, C., Agarwal, M., Ohta, M., Guo, Y., Halfter, U., Wang, P., & Zhu, J. (2005). Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell, 17, 2384–2396.
Tang, M. J., Sun, J. W., Liu, Y., Chen, F., & Shen, S. H. (2007). Isolation and functional characterization of the JcERF gene, a putative AP2/EREBP domain-containing transcription factor, in the woody oil plant Jatropha curcas. Plant Molecular Biology, 63, 419–428.
Tang, M. J., Liu, X. F., Deng, H. P., & Shen, S. H. (2011). Over-expression of JcDREB, a putative AP2/EREBP domain-containing transcription factor gene in woody biodiesel plant Jatropha curcas, enhances salt and freezing tolerance in transgenic Arabidopsis thaliana. Plant Science, 181, 623–631.
Tang, W., Charles, T. M., & Newton, R. J. (2005). Overexpression of the pepper transcription factor CaPF1 in transgenic Virginia pine (Pinus virginiana Mill.) confers multiple stress tolerance and enhances organ growth. Plant Molecular Biology, 59, 603–617.
Xiang, Y., Huang, Y., & Xiong, L. (2007). Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiology, 144, 1416–1428.
Xu, Z. S., Chen, M., Li, L. C., & Ma, Y. Z. (2011). Functions and application of the AP2/ERF transcription factor family in crop improvement. Journal of Integrative Plant Biology, 53, 570–585.
Yang, H. J., Shen, H., Chen, L., Xing, Y. Y., Wang, Z. Y., Zhang, J. L., & Hong, M. M. (2002). The OsEBP-89 gene of rice encodes a putative EREBP transcription factor and is temporally expressed in developing endosperm and intercalary meristem. Plant Molecular Biology, 50, 379–391.
Yi, S. Y., Kim, J. H., Joung, Y. H., Lee, S., Kim, W. T., Yu, S. H., & Choi, D. (2004). The pepper transcription factor CaPF1 confers pathogen and freezing tolerance in Arabidopsis. Plant Physiology, 136, 2862–2874.
Zhang, G. Y., Chen, M., Li, L. C., Xu, Z. S., Chen, X. P., Guo, J. M., & Ma, Y. Z. (2009). Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. Journal of Experimental Botany, 60, 3781–3796.
Zhang, H., Zhang, D., Chen, J., Yang, Y., Huang, Z., Huang, D., Wang, X. C., & Huang, R. (2004). Tomato stress-responsive factor TSRF1 interacts with ethylene-responsive element GCC box and regulates pathogen resistance to Ralstonia solanacearum. Plant Molecular Biology, 55, 825–834.
Zhang, X. L., Zhang, Z. J., Chen, J., Chen, Q., Wang, X. C., & Huang, R. F. (2005). Expressing TERF1 in tobacco enhances drought tolerance and abscisic acid sensitivity during seedling development. Planta, 222, 494–501.
Wang, S., Yao, W., Wei, H., Jiang, T., & Zhou, B. (2014). Expression Patterns of ERF Genes Underlying Abiotic Stresses in Di-Haploid Populus simonii × P. Nigra. The Scientific World Journal, 2014, 1–10.
Himmelbach, A., Yang, Y., & Grill, E. (2003). Relay and control of abscisic acid signaling. Current Opinion in Plant Biology, 6, 470–479.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (no. 31071448) and the Basic Research for Application project of Sichuan province (no. 2014JY0051).
Author information
Authors and Affiliations
Corresponding author
Additional information
Novelty Statement
• A new AP2/ERF-type transcription factor gene, JcERF2, was isolated from the leaves of Jatropha curcas.
• Experiments using a binary expression vector, pBI121-JcERF2, in tobacco suggested that JcERF2 plays a role in plant defense reactions.
• The overexpression of JcERF2 in tobacco enhanced the expression of some biotic and abiotic stress-related genes, increased the accumulation of free proline and soluble carbohydrates, and conferred tolerance to drought and salt.
Rights and permissions
About this article
Cite this article
Wang, X., Han, H., Yan, J. et al. A New AP2/ERF Transcription Factor from the Oil Plant Jatropha curcas Confers Salt and Drought Tolerance to Transgenic Tobacco. Appl Biochem Biotechnol 176, 582–597 (2015). https://doi.org/10.1007/s12010-015-1597-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-015-1597-z