Abstract
Polyhydroxyalkanoates (PHAs) are biodegradable and environmentally friendly natural polymers. In this study, we isolated a bacterium strain capable of synthesizing PHAs from the aerobic sludge of a sewage treatment plant. The bacterium was identified as Burkholderia cepacia via physiological and biochemical tests as well as 16S rDNA sequence analysis. Strain WN-H41 produced PHAs, which was identified as P3HB. These PHAs have a number average molecular weight of 2.6 × 104 Da, a polydispersity index (PDI) of 2.4, and its thermal properties include a glass transition temperature of 1 °C, a melting temperature of 171 °C, and a decomposition temperature of 280 °C. These properties indicate that P3HB produced by WN-H41 has a high purity and good thermal stability.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Plastic is widely used in the various fields because it is lightweight, noncorrosive, functionally diverse, and cold formed. Based on the report consumption of global plastic products, 240 million tons of plastic was produced in 2011 [1]. However, petrochemical-based plastics composed of polyethylene or polystyrene with a stable molecular structure and good anti-aging properties are difficult to degrade under natural state, thereby causing environmental pollution. Polyhydroxyalkanoates (PHAs) are a kind of water-insoluble polymer stored in the cell cytoplasm as carbon and energy storage materials [2]. In addition, PHAs have become a popular material used to solve the problem of “white pollution” [3]. PHAs can be degraded into H2O and CO2 in nature by microorganisms. To date, PHAs have been found to be incorporated by approximately 150 different monomers, such as 3-hydroxybutyrate, 4-hydroxybutyrate, 3-hydroxyvalerate, and 3-hydroxyhexanoate [4, 5]. Monomers can form all kinds of short-chain length (scl) and medium-chain length (mcl) PHA, including typical P3HB, polyhydalerate, poly(3-hydroxybutyrate-co-3-polyhydroxyvalerate), and poly(3-hydroxybutyrate-co-3-poly-hydro-xyhexanoate) via physical blending and block copolymerization. The use of physical blending and block copolymerization results in the formation all various PHAs with different molecular structures, which provide different properties and functionalities, such as biocompatibility, piezoelectricity, low oxygen permeability, and good physical properties. These properties are important in the application range of PHAs in the medical industry [6].
PHAs can be produced by wild type strains, recombinant strains, or plants. At present, pure bacterium fermentation is the major source of PHAs. Thus far, researchers have isolated PHAs produced by more than 300 bacteria strains, but only a few of them, such as Alcaligenes, Pseudomonas, Bacillus, and Ralstonia, can synthesize PHAs with a high concentration and high yield rate [7–9]. For most PHAs-producing microorganisms, the monomer structure of PHA depends on the use of the carbon source. Particular special structures of PHA share similar precursor structures, which require exogenous carbon sources [10]. As such, the cost of raw materials increases, which results in the limited commercialization of PHA.
Screening wild strains from various environments is one of the strategies to reduce the production cost of PHAs. Our study aims to screen PHA-producing strains from activated sludge and to study the growth characteristics of the screened strains. To determine the compositions and properties of the PHAs, Fourier-transform infrared (FTIR), nuclear magnetic resonance (NMR), gel permeation chromatography (GPC), and thermogravimetric analysis (TGA) were performed. This study will enrich our knowledge on PHA-synthesizing strains to lay the foundation for further research.
Materials and Methods
Isolation and Purification of Bacterial Strains
The strains were isolated from aerobic sludge that was collected from a sewage treatment plant at Shihezi. One gram of activated sludge with little sterile glass beads was combined with 20 mL of sterile normal saline. After shaking the culture at 100 rpm min-1 and 30 °C for 30 min, 0.5 mL of the homogeneous suspension was serially diluted and spread onto an isolation medium (glucose 20 g/L, beef extract 10 g/L, peptone 10 g/L, NaCl 5 g/L, agar 15 g/L, and Nile red (0.5 μg/mL) at pH 7.0 ± 0.2) to separate and screen the PHA-producing strains. After incubation for 48 h at 30 °C, the plates were observed under UV light (312 nm) [11]. Positive colonies with orange fluorescence were purified, and its extracts were further analyzed via FTIR. Finally, the target strain was obtained.
Morphological and Biochemical Characterization
The morphological identification of the strain was performed according to the procedure by Prasanna et al. [12] with minor modifications. The biochemical characterizations of the isolates were performed by the Guangdong Detection Center of Microbiology.
Amplification of 16S rDNA and Phylogenetic Analysis
The genomic DNA of the strain was isolated using a TIANamp Bacteria DNA Kit (Tiangen Biotech Co. Beijing). 16S rDNA gene was amplified via polymerase chain reaction (PCR) with universal primes 27 F [13] and 1492R [14]. PCR was carried out according to the procedure used by Kung et al. [15]. The PCR conditions were as follows: pre-denaturing DNA at 94 °C for 5 min, 30 thermal cycles of 94 °C for 30 s, 54 °C for 45 s, and final extension at 72 °C for 1 min.
Highly similar 16S rDNA sequences were obtained from the National Center for Biotechnology Information (NCBI) in the GenBank database by using the nucleotide BLAST command [16]. The phylogenetic tree was generated by the MEGA5.05 software, where a neighbor-joining program was used based on the bootstrap test of 1000 replicates [17].
Extraction and Characterization of PHAs
Polymer extraction was performed as described previously [18]. The cells were recovered via centrifugation (8000g, 10 min at 4 °C), washed twice with distilled water, and lyophilized (0.2 mbar, 12 h). PHAs were extracted from the lyophilized cell by using a methanol-chloroform mixture (v/v = 1/2) at 80 °C for 5 h in a Soxhlet apparatus. After filtration through a 0.45μm fiberglass filter, the filtrate was concentrated via rotary evaporation. The PHAs were precipitated with 10-fold volume quantities of cold ethanol (v/v) and centrifuged (5000g, 10 min at 10 °C). The precipitate was dried using a vacuum dryer until a constant weight.
The infrared spectra for the sample were obtained using a Nicolet Avatar 360 FTIR spectrometer (NICOLET, USA) with wavenumbers ranging from 4000 to 400 cm−1. All spectra were recorded with a total of 32 scans at room temperature [19]. For sample preparation, potassium bromide (KBr) was squeezed under pressure to form KBr pellets, and a polymer drip was dissolved in the pellet capillary.
About 20 mg of the samples were dissolved in 1 mL of deuterochloroform and combined with tetramethylsilane, which was used as the internal standard. 13C and 1H NMR spectra were obtained using Varian Inova-400 M (Varian, USA).
Weight average molecular weight (Mw), number average molecular weight (Mn), and polydispersity index (PDI) (PDI = Mw/Mn) were measured via GPC (Waters Alliance 2695, WATERS, USA). The operating temperature was 40 °C, and a PL gel 5 μm mixed-C column was used. THF was used as the eluent with a flow rate of 0.3 mL/min [5].
The thermal stability of the polymer was determined using an STA449F3 thermogravimetric analyzer (NETZSCH, Germany). The conditions were patterned to the study of Lau et al. [19]. Approximately 10 mg of the sample was heated at 30 to 900 °C at a heating rate of 10 °C/min in a nitrogen atmosphere of 30 mL/min. The temperature at 5 % weight loss of samples was defined as the decomposition temperature (Td) [20].
The glass transition temperature (Tg), melting temperature (Tm), and crystallization temperature (Tc) were determined via differential scanning calorimetry (DSC200F3, NETZSCH, Germany). Approximately 5 mg of the sample was placed in standard aluminum pans and heated at −60 °C to 180 °C at a heating rate of 20 °C/min under nitrogen atmosphere of 30 mL/min. The testing temperature was maintained at 180 °C for 1 min and quenched to −60 °C at a rate of 20 °C/min. Subsequently, the sample was reheated from −60 to 180 °C at a heating rate of 20 °C/min. Tg was taken as the midpoint of the total change in heat capacity. Tm and Tc were determined from the DSC spectrogram [5, 21].
Results and Discussion
Isolation and Identification of the Bacterial Strains
In this study, the strain was isolated from an aerobic sludge. The sludge sample was inoculated into an isolation medium containing Nile red (final concentration of 0.5 μg/mL), a kind of dye that can distinguish PHA accumulation in cells. Intense orange fluorescence can be observed in PHA-producing cells under UV light at 312 nm. Through fermentation, a bacterial strain that can synthesize PHA was extracted and named as WN-H41. Strain WN-H41 is gram-negative and produced small faint yellow colonies (0.2–0.5 mm) on nutrient agar after 24 h of incubation. The semitransparent colonies were smooth, moist, circular type, and edge tidy. The morphological and biochemical characteristics of WN-H41 are summarized in Table 1.
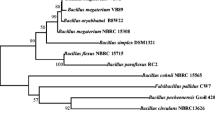
The 16S rDNA sequence (1436 bp of length) of strain WN-H41 was sent to NCBI, and a phylogenetic tree based on 16S rDNA was constructed (Fig. 1) using the MEGA5.05 software. The phylogenetic tree showed that strain WN-H41 together with B. cepacia 717, B. cepacia GG4, and Burkholderia diffusa R-15930 can be gathered into a cluster, wherein 16S rDNA homology are as high as 99.8 %. Combining all these morphological, physiological, and biochemical evidence, WN-H41 was identified as Burkholderia cepacia.
Chemical Characterization of PHAs
Infrared spectroscopy is a rapid screening tool for PHA-producing microorganisms [22]. The FTIR spectra of the polymer extracted from WN-H41 are illustrated in Fig. 2. Bands at 2935 cm-1 are attributed to the –CH3 asymmetric stretching vibrations; bands at 1735 cm-1 are attributed to the C = O stretching vibration peak, which is a characteristic peak of PHA [23]; bands at 1596 cm-1 is associated with the stretching vibration of C = C. Taking together the bands at 1284, 1183, 1135, 1052, and 975 cm-1, the polymer extracted from WN-H41 was confirmed to be PHA.
In addition, the peaks shown are consistent with those in the report of Hong et al. [24] about Poly(3-hydroxybutyrate) (PHB); thus, the preliminary polymer was identified as scl-PHA.
The 400 MHz 13C NMR spectrum (Fig. 3a) of the polymer shows that the polymer monomer is four carbon compounds. The chemical shifts at 169.27, 67.75, 40.94, and 19.91 ppm are assigned to carbonyl (C = O), methane (CH), methylene (CH2), and methyl (CH3) groups, respectively, which is in agreement with previous reports on PHB [25]. The 1H NMR spectrum (Fig. 3b) of the polymer isolated from strain WN-H41 shows the hydrogen signal on the methyl groups near 1.26 ppm. This peak is split into D peak, which means that its adjacent carbon only had single hydrogen atoms. Signals at 2.423 to 2.476 ppm are caused by the hydrogen in methylene, which is linked to the chiral carbon. As such, the two hydrogens on the methylene are asymmetrically split, thereby making the peak shape more complex. Signals at 5.210 to 5.289 ppm are assigned to hydrogen atoms in methane. Jan et al. [26] reported that the signal near 5.25 ppm is the characteristic peak of PHB. Based on the characterization of the polymer via FTIR and NMR, we finally confirmed that the PHA produced from strain WN-H41 is P(3HB).
Physical Characterization of PHAs
The GPC results showed that the polymer had a number average molecular weight (Mn) of 2.6 × 104 Da and a PDI of 2.4. The average molecular weights of particular PHB from wild type strains are 1 × 104 Da, whereas those of others are as high as 3 × 106 Da and PDI of 2 [27]. A low molecular weight PHB was obtained in this experiment. Studies have shown that PHB with a relatively low molecular weight has good biodegradability. For example, more quantity of PHB of 100 to 200 monomer polymerization was found in human blood cells [28]. Perhaps PHB in this size range can be applied to medical treatment as an embedding material of a slow-release drug.
The thermal stability and properties of P(3HB) were studied via TGA and DSC. Even the same kind of PHA produced from different microbes has different thermal properties. In general, Tg of PHB ranges from −5 to 5 °C [7], whereas the Tg of PHB in this study is 1 °C. The Tm of PHB from Strain WN-H41 is approximately 171 °C, which is very close to Tm (175 °C) of the PHB standard sample (Fig. 4a) [29].
Figure 4b shows that the Td of PHB from Strain WN-H41 is 280 °C, which is significantly higher compared with the PHB of Pseudomonas putida [18]. The significantly higher Td indicates that this polymer has a good thermal stability.
References
Zhang, B., & Kang, M. (2013). Life-cycle assessment for plastic waste recycling process. In A. Y. C. Nee, B. Song, S. K. Ong, & S. Y. Lim (Eds.), Based of the network evaluation framework (pp. 377–382). Singapore: Springer.
Tappel, R. C., Wang, Q., & Nomura, C. T. (2012). Journal of Bioscience and Bioengineering, 113, 480–486.
Legat, A., Gruber, C., Zangger, K., Wanner, G., & Stan-Lotter, H. (2010). Applied Microbiology and Biotechnology, 87, 1119–1127.
Park, S. J., & Lee, S. Y. (2004). Applied Biochemistry and Biotechnology, 114, 335–346.
Hu, D., Chung, A. L., Wu, L. P., Zhang, X., Wu, Q., Chen, J. C., & Chen, G. Q. (2011). Biomacromolecules, 12, 3166–3173.
Keshavarz, T., & Roy, I. (2010). Current Opinion in Microbiology, 13, 321–326.
Dias, J. M., Lemos, P. C., Serafim, L. S., Oliveira, C., Eiroa, M., Albuquerque, M. G., Ramos, A. M., Oliveira, R., & Reis, M. A. (2006). Macromolecular Bioscience, 6, 885–906.
Reinecke, F., & Steinbuechel, A. (2008). Journal of Molecular Microbiology and Biotechnology, 16, 91–108.
Chanprateep, S. (2010). Journal of Bioscience and Bioengineering, 110, 621–632.
Chen, G., Wu, Q., Xi, J., Yu, H. P., & Chan, A. (2000). Progress in Natural Science, 10, 843–850.
Spiekermann, P., Rehm, B. H., Kalscheuer, R., Baumeister, D., & Steinbüchel, A. (1999). Archives of Microbiology, 171, 73–80.
Prasanna, T., Babu, P. A., Lakshmi, P. D., Chakrapani, R., & Rao, C. R. (2011). Journal of Pharmaceutical Research and Review, 1, 15–18.
Reysenbach, A. L., Longnecker, K., & Kirshtein, J. (2000). Applied and Environmental Microbiology, 66, 3798–3806.
Miyoshi, T., Iwatsuki, T., & Naganuma, T. (2005). Applied and Environmental Microbiology, 71, 1084–1088.
Kung, S. S., Chuang, Y. C., Chen, C. H., & Chien, C. C. (2007). Letters in Applied Microbiology, 44, 364–371.
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., & Lipman, D. J. (1997). Nucleic Acids Research, 25, 3389–3402.
Kumar, S., Nei, M., Dudley, J., & Tamura, K. (2008). Briefings in Bioinformatics, 9, 299–306.
Wang, H. H., Zhou, X. R., Liu, Q., & Chen, G. Q. (2011). Applied Microbiology and Biotechnology, 89, 1497–1507.
Lau, N. S., Tsuge, T., & Sudesh, K. (2011). Applied Microbiology and Biotechnology, 89, 1599–1609.
Zhang, H. F., Ma, L., Wang, Z. H., & Chen, G. Q. (2009). Biotechnology and Bioengineering, 104, 582–589.
Bengtsson, S., Pisco, A. R., Johansson, P., Lemos, P. C., & Reis, M. A. (2010). Journal of Biotechnology, 147, 172–179.
Tripathi, A. D., & Srivastava, S. K. (2011). Journal of Polymers and the Environment, 19, 732–738.
Arcos-Hernandez, M. V., Gurieff, N., Pratt, S., Magnusson, P., Werker, A., Vargas, A., & Lant, P. (2010). Journal of Biotechnology, 150, 372–379.
Hong, K., Sun, S., Tian, W., Chen, G. Q., & Huang, W. (1999). Applied Microbiology and Biotechnology, 51, 523–526.
Doi, Y., Kunioka, M., Nakamura, Y., & Soga, K. (1986). Macromolecules, 19, 1274–1276.
Jan, S., Roblot, C., Courtois, J., Courtois, B., Barbotin, J. N., & Seguin, J. P. (1996). Enzyme and Microbial Technology, 18, 195–201.
Sudesh, K., Abe, H., & Doi, Y. (2000). Progress in Polymer Science, 25, 1503–1555.
Reusch, R. N., Sparrow, A. W., & Gardiner, J. (1992). Biochimica et Biophysica Acta, 1123, 33–40.
Bordes, P., Hablot, E., Pollet, E., & Avérous, L. (2009). Polymer Degradation and Stability, 94, 789–796.
Acknowledgments
The authors are grateful to the school of Chemistry and Chemical Engineering/Key Laboratory for Green Processing of Chemical Engineering of Xinjiang Bingtuan, Shihezi University, Shihezi, China for providing all the laboratory facilities.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zheng, B., Lu, J., Tong, Y. et al. Isolation and Characterization of Poly(3-hydroxybutyrate)-Producing Bacteria from Aerobic Sludge. Appl Biochem Biotechnol 175, 421–427 (2015). https://doi.org/10.1007/s12010-014-1271-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-014-1271-x