Abstract
Purpose of Review
The aim of this review was to compile a list of tools currently available to study bone cells and in particular osteocytes. As the interest (and importance) in osteocyte biology has greatly expanded over the past decade, new tools and techniques have become available to study these elusive cells,
Recent Findings
Osteocytes are the main orchestrators of bone remodeling. They control both osteoblasts and osteoclast activities via cell-to cell communication or through secreted factors. Osteocytes are also the mechanosensors of the bone and they orchestrate skeletal adaptation to loads. Recent discoveries have greatly expanded our knowledge and understanding of these cells and new models are now available to further uncover the functions of osteocytes.
Summary
Novel osteocytic cell lines, primary cultures, and 3D scaffolds are now available to investigators to further unravel the functions and roles of these cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The last decade has seen an expansion in the tools and models available to study skeletal processes and the establishment of new cell lines has proven fundamental in the discovery of novel therapeutics for skeletal diseases. Historically, osteocytes posed a technical challenge to biologists due to their location within the mineralized matrix and the associated difficulties in isolate and characterize these cells.
The extensive cell-to-cell communication and the architecture of the lacuna-canalicular network suggested two possible functions for these cells: to ensure communication between osteocytes deep within the matrix with the surrounding tissues and to provide an extensive surface for mineral exchanges [1]. It was postulated that osteocytes could mobilize calcium through a mechanism called “osteocytic osteolysis” and that this process was regulated by calciotropic hormones (parathyroid hormone (PTH) and 1,25-dihydroxy vitamin D3) [2, 3]. Histological observations that in various physiological and pathological states the size of the osteocytic lacunae was increased supported this hypothesis. The idea of “osteocytic osteolysis” was quickly abandoned when isolated chick osteocytes failed to reabsorb bone is a classical osteoclast pit-forming assay. Recent studies, however, have demonstrated that osteocyte-mediated calcium mobilization and remodeling of the perilacunar spaces are important for lactation [4, 5]. Nevertheless, the exact mechanism by which osteocytes control calcium homeostasis remains elusive.
In the late 1980s, Harold Frost initiated a series of investigations which led him to postulate that osteocytes were the skeletal “sensor” of mechanical forces, or “mechanostat,” capable of distinguishing between bone modeling and remodeling cues. In 1996, Bonewald’s group reported the isolation and characterization of the first osteocytic cell line, MLO-Y4, which allowed, for the first time, insights into the cellular and molecular mechanisms that control osteocytes biology. Prior to MLO-Y4, most of the research on osteocytes was done in calvaria chick embryos [1, 6,7,8,9,10,11,12,13], since these were the only osteocytes that could be isolated and purified, as described below. Over the past 20 years, the field of osteocyte biology has expanded exponentially; newly developed cell lines and tools have greatly enhanced our understanding of the functions and characteristics of these fascinating cells. In 2001, two independent papers described two rare skeletal diseases caused by a mutation of a protein, sclerostin [14], expressed predominantly, if not exclusively, in osteocytes [15,16,17]. This novel discovery brought a lot of interest in osteocytes and since then, a wealth of new biology has been revealed.
Cell Lines: New and Old
Osteocytes have been notoriously difficult to isolate. These cells are post-mitotic, i.e., they do not proliferate, and they reside in a mineralized matrix which is hard to remove and dissolve. The first isolation of a pure or “enriched” osteocyte population was done in chicken embryos using a monoclonal antibody OB7.3 capable of specifically binding to osteocytes [8] and avian osteocytes were, for several years, the only cells that could be analyzed in vitro [8]. MAb OB7.3 was specific for chicken and could not be used in mice or human preparations, limiting its application in osteocyte research. Interestingly, the identity of the antigen was revealed 15 years later when the same group demonstrated that MAb OB7.3 was targeted against the phosphate-regulated gene with homology to endopeptidase on the X chromosome (Phex), an endopeptidase highly express in osteocytes [13]. In humans, several diseases are associated with mutations in the PHEX gene, including X-linked hypophosphatemic rickets, suggesting that osteocytes might also play a role in phosphate homeostasis. Osteocytes are also the main source of yet another phosphatemic hormone, fibroblast growth factor 23 (FGF-23), but whether these cells indeed control phosphate homeostasis remains unclear.
MLO-Y4 and MLO-A5
The first “bona fide” murine osteocytic cell line to be isolated and characterized was MLO-Y4. These cells were derived from long bones of mice in which the immortalized SV40 T antigen (TAg) was driven by the 2.6 kb osteocalcin promoter [18••]. This pioneering study from Dr. Bonewald’s group started a wealth of in vitro investigations on osteocyte biology. Using MLO-Y4, it was demonstrated that fluid flow shear stress opens connexin 43 hemichannel and allows the secretion of PGE2 [19] and, more recently, that osteocytes influence muscle functions and vice versa [20, 21]. To this day, MLO-Y4 is still the most used cell model and probably the best characterized osteocytic cell line with almost 300 published reports [19, 20, 22,23,24,25,26,27,28]. These cells express many osteocytic markers, including Dentin Matrix Protein 1 (Dmp1), connexin 43 (Cx43), E11, MEPE, and PHEX but they express very low levels of Sost and FGF23, suggesting that they represent an early stage of osteocyte differentiation. MLO-Y4 cells, like primary osteocytes, express receptors for parathyroid hormone (PTH), prostaglandins, and irisin which binds to the αV/β5 integrin receptor and regulate both osteocyte survival and function [29]. Using similar approach, Bonewald’s group characterized an additional clone, MLO-A5 cells, which represent cells at the transition between osteoblasts to osteocytes [30]. Compared with osteoblasts, these cells express higher level of E11, PHEX, and MEPE and they rapidly mineralize the extracellular matrix even in the absence of osteogenic factors. Interestingly, they also display a subset of skeletal stem cell markers and, under the appropriate culture conditions, they differentiate into both adipogenic and chondrogenic cells, demonstrating multi-lineage plasticity [31].
HOB-01-C1
HOB-01-C1 was the first human osteocytic (or pre-osteocytic) cell line to be isolated and characterized [32]. These cells were derived from the femoral neck of an 82-year-old woman and immortalized using an adenovirus expressing the SV40TAg. Similar to MLO-Y4, the SV40TAg is constitutively expressed and these cells are permanently transformed. HOB-01-C1 cells display quite an exquisite regulation of osteocalcin expression in response of vitamin D treatment and an increase in cyclic AMP accumulation upon stimulation with PTH and forskolin. Surprisingly, despite being the only available human model, these cells were not further studied or analyzed, possibly due to the fast-paced progress of mouse genetics that shifted the interest of investigators from clinical to pre-clinical models and genetically modified animals. Recently, HOB-01-C1 cells have been used to study the interplay between multiple myeloma cells and osteocytes [33].
The identification of several specific osteocytic markers, such as dentin matrix protein 1(Dmp1) [34,35,36,37], matrix extracellular phosphoglycoprotein/osteocyte-factor 45 (MEPE/OF45), and sclerostin [15, 17, 38] allowed investigators to use these genes to specifically target osteocytes.
IDG-SW3 and Ocy454 Cells
As described above, MLO-Y4 and MLO-A5 cells have been used extensively to study osteocyte biology, although their validity as model for normal osteocyte is limited by their constitutively immortalized phenotype. Cell lines conditionally transformed with temperature-sensitive (ts) mutants of the SV40 TAg, which can be inactivated, in vitro, more faithfully reflect the functions of normal cells. Cells expressing the tsSV40 TAg proliferate indefinitely at temperature permissive for expression of functional tsTAg (i.e., 33 °C) but then revert to a non-transformed, non-prolifertaive phenotype when cultured at non-permissive temperature (i.e., 37–39 °C). IDG-SW3 and Ocy454 osteocytic cell lines were isolated using similar strategies. Both Bonewald’s and our group crossed mice in which the Dmp1 promoter was driving a Green Fluorescent Protein (8KbDmp1-GFP) [37] with mice in which tsSV40TAg was ubiquitously expressed [39••, 40]. IDG-SW3 cells were isolated by sequential digestions from the long bone of double transgenic mice and were then examined for expression of markers of mature osteocytes [40••]. These cells differentiate into mature osteocytes (as demonstrated by the expression of Sost/sclerostin) after 21 days in culture and they are an excellent model to study and analyze the transition from an osteoblast to an osteocyte. On the flip side, it takes more than 21 days in culture to detect Sost/sclerostin in these cells. Ocy454 cells were also isolated by sequential digestions from long bones of 4 weeks old female mouse and osteocytes were selected following two criteria: (1) sorted GFP-positive cells were required to have high levels of production of known osteocytic genes at early time point (10–14 days) in the absence of differentiation medium, and (2) respond to PTH by suppressing Sost and increasing RankL expression [39••]. Ocy454 cells express high level of Sost/Sclerostin after 10–14 days in culture, in the absence of mineralization medium, suggesting a more “mature” osteocytic phenotype than MLO-A5, MLO-Y4, and IDG-SW3 cells. Recently, we isolate and characterized a clonal derivative of Ocy454, namely Ocy454-12H cells which express high level of Sost/sclerostin after 5–7 days in culture in the absence of mineralizing medium and they are an ideal model to study Sost expression and regulation [41]. Interestingly, whereas markers of mature osteocytes like Sost, Dmp1, MEPE, and Phex highly correlate with osteocyte differentiation and maturation (i.e., they increase upon time in culture at non-permissive temperature), FGF-23 appears not to be directly regulated by the cell maturation. Both Ocy454 and Ocy454-12H are highly responsive to PTH and mechanical forces and they are an excellent model to study both hormonal responses and molecular mechanisms of mechano-transduction, as recently shown by several groups [42,43,44,45,46,47,48].
OmGFP66 and OmGFP10
The latest comers in the field of osteocyte biology are two cell lines derived from mice in which membrane-targeted GFP was driven by the Dmp1 promoter. When OmGFP66 are cultured for 14–21 days, they form highly organized 3D bone-like structures replicating the environment of osteocytes in vivo (i.e., cells embedded in the mineralized matrix within clearly defined lacuna) [49]. These cells offer an ideal in vitro model to study hormonal influence and further understand the molecular and cellular mechanisms by which osteocytes control their environment.
One advantage of using established cell lines over primary cells is the ability to quickly manipulate the genetic make-up of cell lines and to perform high throughput screening [48]. CRISPR/Cas9 or other genetic manipulations can be used effectively in all these cell lines to delete or increase the expression of genes of interests [39, 41, 50] and to introduce mutations in specific loci. These cell lines have been very useful in moving forward the current understanding of osteocyte biology, but they do have some limitations. First, osteocytes, in vivo, are post-mitotic, whereas these cell lines continue to proliferate, even when grown at non-permissive conditions. Second, osteocytes are a heterogenous population, expressing different subset of genes depending on their origins (weight-bearing vs non-weight-bearing bones) and localization (trabecular vs cortical bone). For example, osteocytes in the cortex of long bones express endothelial nitric oxide synthase (eNOS) and they release NO in response to loading, whereas osteocytes in non-load bearing calvaria do not [51]. It is plausible to speculate that the genetic make-up of osteocytes is site specific and future studies using single-cell RNA sequencing will delineate their transcriptomic profile.
Primary Cells
Immortalized cell lines, although very useful, do not fully recapitulate the phenotype of primary cells. To overcome this short fall, efforts have been devoted to develop and optimize methods to enrich for osteocytes both in pre-clinical and clinical models. Granted that homogeneous mature osteocyte population are still needed, great progress has been made to isolate primary cells which differentiate into mature osteocytes.
Pre-clinical
Several methods have been described to isolate osteocytes from murine bones. The first method to “enrich” for osteocytes was described by Gu et al. [52] by adding to the traditional sequential collagenase digestions few demineralization steps (EDTA digestion) and by increasing the number of digestions. Traditionally, the cells released from early digestions (1 and 2) are negative for alkaline phosphatase, do not form bone nodules, and express minimal osteoblastic markers. Whether these early populations are undifferentiated cells (maybe progenitors) or fibroblasts is unclear. Cells released between digestions 3 and 6 are enriched for osteoblastic features; they are highly positive for alkaline phosphatase (AP), they form bone nodules, and they express osteoblastic markers such as Runx2, osterix, and osteocalcin. Later digestions (7 and above) are enriched in osteocytes and they express markers such as Dmp1 and Sost and they are negative for AP. We took advantage of the 8KbDmp1-GFP mice to sequentially isolate calvarial cells and to show that the percentage of mature osteocytes (Dmp1-positive) present in digestion 7–9 was around 20% (unpublished observation), demonstrating the need for better enrichment protocols. Recently, several investigators [52,53,54,55] described new methods for isolating osteocytes from long bones and vertebra of adult mice. Stern et al. [53, 54] reported an increase in the percentage of osteocytes isolated from both young and old mice long bones by adding digestion and decalcification (EDTA treatment) steps allowing the release of embedded osteocytes. Although laborious and time consuming, the results are quite dramatic. Recently, Farr et al. [55] combined two collagenase digestions with immuno-depletion of hematopoietic and endothelial cells to rapidly isolate osteoblasts (enriched by selecting AP-positive cell) whereas the osteocytes were obtained from the remaining bone fragments. One note of caution in using these manipulations to isolate osteocytes is that they can change the transcriptomic make-up of the isolated cells. Ayturk et al. [56] reported that collagenase digestions, while removing contaminating non-skeletal cells, also altered the gene expressions profile of tibial diaphyseal bones.
Pre-clinical methods to isolate osteocytes can also take advantage of fluorescently labelled proteins. As described above, osteocytes can be isolated using fluorescently activated cell sorting (FACS) technique in transgenic animals in which these cells are fluorescently tagged, such as the 8Kb-Dmp1-GFP mouse model [37].
Moreover, new transcriptomic techniques, such as single-cell RNA sequencing, will further delineate the gene profile and characteristics of osteocytes.
Clinical
Over the last decade, a wealth of studies on genetically modified animals (and new osteocytic cell lines) has uncovered unexpected osteocyte functions. These cells not only control the skeletal responses to mechanical cues but they also secrete several hormones and cytokines that control bone homeostasis, muscle functions, and adipogenesis. In 2016, Prideaux et al. [57•] described a method to effectively isolate osteocytes from human surgical specimens, providing an important tool to study human osteocytes. This technique was recently optimized by Bernhardt et al. [58•, 59•] to increase the yield of primary cells by adding a prolonged culturing time (14 days of “resting” time) between the collagenase and EDTA digestions. This “resting” phase allows the proliferation, expansion, and differentiation of osteocytes. Using this technique to isolate osteocytes from human bone fragments, together with three-dimensional (3D) collagen gel culture, they reported an impressive enrichment for osteocytic genes such as DMP1 and SOST [58]. One limitation of this method, as pointed out by the authors, is the high donor variability. Whether this variability is intrinsic to the method or indeed reflects a real biological difference is currently unknown.
An important step for translating basic discoveries into therapeutics is to validate them in human cells prior to moving into human subjects. In the field of skeletal diseases, the efforts in translating pre-clinical findings into clinical outcomes are hampered by the lack of human osteocytic cell lines, with the exception of the HOB-01-C1 cells described above, and clinical models to study osteocytes. There is a pressing need in creating and establishing novel human osteocytic cell lines derived from different patients (gender and ages) and anatomical sites (load bearing or not) which will provide a highly needed platform to move from the bench to the bedside.
3D Cultures for Osteocytes
Most cells in the human body are organized in complex 3D structure and the cells secrete and are organized within an extracellular matrix (ECM) which defines the architectural geometry of the tissue and provides cytokines and growth factors. Each tissue has an ECM with unique composition and topology. In the bone and teeth, the ECM contains the mineral component (calcium and phosphate hydroxyapatite) which provides the strengths and rigidity needed for their functions. Despite the importance of 3D scaffolding, the classical 2D “in vitro” monolayer cell culture is still a dominant method in many biological studies. Monolayers provide some advantages over 3D cultures; reproducibility of seeding and recovery of cells is quite high in 2D culture whereas it poses come challenges when cells are embedded in 3D scaffolds.
Osteocytes “in vivo” are confined in their lacunae and connect with adjacent cells through their dendrites. Cell-to-cell contact, under these physiological conditions, is limited to the dendrites whereas the cell bodies are isolated from the rest of the cells. This “cell isolation” is lost when osteocytes are grown in 2D monolayers, and some of the “in vivo” functions and characteristics of osteocytes could be potentially affected by this non-physiological organization. Over the past decades, efforts have been made to study osteocytes under more physiological settings and several 3D scaffolds have been used to interrogate osteocyte biology.
3D structured scaffolds are fundamental for tissue engineering and regenerative medicine and current strategies involve the use of a variety of materials, such as natural and synthetic polymers, inorganic biomaterials (i.e., metals and ceramics), and their hybrid combinations [60]. The most commonly used biomaterials in skeletal tissue engineering are collagen and ceramics. Among the natural and synthetic polymers that are widely used as scaffolding materials, there are poly(lactic acid) (PLA), poly(glycolic acid) (PGA), and PLGA, that are chosen especially for their design flexibility, the surface modifiability, and the functional group availability. Composites, such as nanofibers, nanoparticles (NPs), and nanopores, combine polymers and scaffolding biomaterials and provide promising novel tools for bone regeneration. Below is the description of the most commonly used to date. For a comprehensive review of osteocytes and 3D cultures, see Zhang et al. [61].
Collagen Scaffolds
Bone matrix is comprised of 90% of collagen type 1 and collagen scaffolds have been widely used as 3D support for bone cell differentiation. Investigators have used various approaches to study bone cells (both osteoblasts and osteocytes) in this collagen-rich environment. Collagen scaffolds offer several advantages: type 1 collagen is widely available and biocompatible; it is easy to manipulate and can be used for 3D printing [62], or electrospinning and electrospray. Moreover, the pore size and stiffness of the scaffold can be adjusted by changing collagen concentration, by introducing chemical crosslinked compounds, or by adding hydroxyapatite.
Uchihashi et al. [63] reported that when seeded on top of collagen gels, osteoblasts (both MC3T3 and primary calvaria osteoblasts) migrate into and differentiated into osteocytes and resemble lacunae. Similarly, Bernhardt’s group [58, 64] reported differentiation of human osteocytes and high expression of DMP1, E11, PHEX, and other osteocytic markers when cells were cultured in 3D collagen gels.
Hydroxyapatite
The main inorganic component of bone is hydroxyapatite (HAp), a complex mineral comprising calcium and phosphorus. HAp has been used, either alone or in combination with other compounds, as a suitable scaffold for bone tissue engineering and studies have shown that HAp promotes osteoblastic cell differentiation. When human osteoblasts were grown on HAp/tricalcium phosphate (TCP) biphasic calcium phosphate (BCP) ceramic particle, they quickly differentiate into osteocytes, demonstrating the importance of substrates for cellular function and differentiation [65].
Microbeads and Nanoparticles
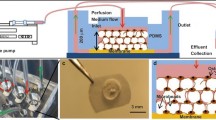
Biphasic calcium phosphate (BCP) microbeads have also been exploited to generate three-dimensional culture conditions that allow osteocyte differentiation. Studies have shown that when MLO-Y4 and HOB-01-C1cells and primary human bone–derived cells are cultured in BCP microbead (20–25 μm) under continuous perfusion, they differentiate into more mature osteocytes, as demonstrated by the expression of osteocytic markers [66,67,68].
Polystyrene
Synthetic material, such as polystyrene, offers the advantage to be easily manipulated and biologically inert. Indeed, the majority of culture vessels are made of either polystyrene or polypropylene. Scaffolds of various thickness or structural characteristics (pore sizes) can be designed and fabricated using these polymers. Cells easily attach to the polymer surface and they also migrate within the scaffold. We have used polystyrene scaffolds to grow Ocy454 cells in 3D conditions and demonstrated that, under these conditions, the cells can regulate FGF-23 expression in response to PTH treatment. Intriguingly, this response is lost when the cells are in 2D monolayer [39••], demonstrating the importance of complex structures for proper cellular responses.
Ex Vivo Models: Bone Explants
One limitation of engineered 3D scaffolds is that they only partially replicate the complexity of the tissue. Tissue organoids and “organ-on-chip” are being developed to address this shortcoming. In skeletal biology, investigators have been using ex vivo explants to study osteocyte functions, and, at the same time preserve the natural milieu for these cells. Several methods have been developed to enrich for osteocytes, and sequential collagenase digestions of bone marrow–deprived long bones have been shown to be effective to remove most of the contaminant hematopoietic and osteoblastic cells and provide explant highly enriched for osteocytes (i.e., osteocyte-enriched bone explants, or OEBEs) [69, 70]. Bone explants can be manipulated, subjected to mechanical loads, or used as a source of osteocyte-secreted factors. Ex vivo bone explants overcome the limitation of in vitro 2D culture and also provide a unique alternative to complex in vivo setting. For example, in vivo studies on the effects of hormones or mechanical forces on bones cannot discriminate between direct and indirect effects, possibly mediated by other organs and tissues. They represent an excellent intermediate model between in vitro and in vivo settings. For a complete review of models of organ culture available for skeletal research, see Bellido et al. [71••].
Conclusions and Looking Ahead
Over the past decade, a plethora of studies have shed light on the functions and characteristics of osteocytes and have propelled forward the field of skeletal biology. Neutralizing antibodies targeting sclerostin are now a powerful therapeutic option to treat skeletal disorders and few other novel therapeutics are in the pipelines.
Despite these great strides toward revealing the “secrets” of an osteocyte, few questions remain. First, there is an unmet need for a human-derived osteocytic cell line to validate (or investigate) any pre-clinical finding. Given the heterogeneity of bone cells, it will be important to create osteocytic cell lines from different anatomic sites and different patients. Nevertheless, there is an intrinsic limitation in using cell lines, since they represent only a specific population of cells. Ex vivo organ culture offers several advantages, including the maintenance of the osteocyte “native” environment. Novel models and tools should be developed to further replicate “in vitro” the complexity of bone. Some pioneer studies are now using new technologies such as biological 3D printing or “organ-on-chip” to generate and “in vitro” model that faithfully replicates the “in vivo’ conditions, which will provide further insights into the multifaceted functions of these cells.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Burger EH, Klein-Nulend J, van der Plas A, Nijweide PJ. Function of osteocytes in bone--their role in mechanotransduction. J Nutr. 1995;125(7 Suppl):2020S–3S.
Belanger L. Osteolysis: an outlook on its mechanism and causation. In: Gaillard PJ, Talmage R, Budy AM, editors. The parathyroid gland. Chicago: The University of Chicago Press; 1965. p. 137–43.
Bélanger LF. Osteocytic osteolysis. Calcif Tissue Res. 1969;4(1):1–12.
Qing H, Ardeshirpour L, Pajevic PD, Dusevich V, Jahn K, Kato S, et al. Demonstration of osteocytic perilacunar/canalicular remodeling in mice during lactation. J Bone Miner Res. 2012;27(5):1018–29.
Qing H, Bonewald LF. Osteocyte remodeling of the perilacunar and pericanalicular matrix. Int J Oral Sci. 2009;1(2):59–65.
Aarden EM, Burger EH, Nijweide PJ. Function of osteocytes in bone. J Cell Biochem. 1994;55(3):287–99.
Klein-Nulend J, van der Plas A, Semeins CM, Ajubi NE, Frangos JA, Nijweide PJ, et al. Sensitivity of osteocytes to biomechanical stress in vitro. FASEB J. 1995;9(5):441–5.
Nijweide PJ, Mulder RJ. Identification of osteocytes in osteoblast-like cell cultures using a monoclonal antibody specifically directed against osteocytes. Histochemistry. 1986;84(4–6):342–7.
Nijweide PJ, van der Plas A, Alblas MJ, Klein-Nulend J. Osteocyte isolation and culture. Methods Mol Med. 2003;80:41–50.
van der Plas A, Nijweide PJ. Isolation and purification of osteocytes. J Bone Miner Res. 1992;7(4):389–96.
van der Plas A, Aarden EM, Feijen JH, de Boer AH, Wiltink A, Alblas MJ, et al. Characteristics and properties of osteocytes in culture. J Bone Miner Res. 1994;9(11):1697–704.
van der Plas A, Nijweide PJ. JBMR anniversary classic. Isolation and purification of osteocytes. A van der Plas A, PJ Nijweide. Originally published in Volume 7, Number 4, pp 389–96 (1992). J Bone Miner Res. 2005;20(4):706–14.
Westbroek I, De Rooij KE, Nijweide PJ. Osteocyte-specific monoclonal antibody MAb OB7.3 is directed against Phex protein. J Bone Miner Res. 2002;17(5):845–53.
Balemans W, Patel N, Ebeling M, Van Hul E, Wuyts W, Lacza C, et al. Identification of a 52 kb deletion downstream of the SOST gene in patients with van Buchem disease. J Med Genet. 2002;39(2):91–7.
Van Bezooijen RL, Roelen BAJ, Visser A, Van Der Wee-pals L, De Wilt E, Karperien M, et al. Sclerostin is an osteocyte-expressed negative regulator of bone formation, but not a classical BMP antagonist. J Exp Med. 2004;199(6):805–14.
van Bezooijen RL, ten Dijke P, Papapoulos SE, Lowik CW. SOST/sclerostin, an osteocyte-derived negative regulator of bone formation. Cytokine Growth Factor Rev. 2005;16(3):319–27.
Winkler DG, Sutherland MK, Geoghegan JC, Yu C, Hayes T, Skonier JE, et al. Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. EMBO J. 2003;22(23):6267–76.
•• Kato Y, Windle JJ, Koop BA, Mundy GR, Bonewald LF. Establishment of an osteocyte-like cell line, MLO-Y4. J Bone Miner Res. 1997;12(12):2014–23 This article describes the isolation and characterization of MLO-Y4 cells.
Cherian PP, Siller-Jackson AJ, Gu S, Wang X, Bonewald LF, Sprague E, et al. Mechanical strain opens connexin 43 hemichannels in osteocytes: a novel mechanism for the release of prostaglandin. Mol Biol Cell. 2005;16(7):3100–6.
Huang J, Romero-Suarez S, Lara N, Mo C, Kaja S, Brotto L, et al. Crosstalk between MLO-Y4 osteocytes and C2C12 muscle cells is mediated by the Wnt/β-catenin pathway. JBMR Plus. 2017;1(2):86–100.
Kitase Y, Vallejo JA, Gutheil W, Vemula H, Jähn K, Yi J, et al. β-Aminoisobutyric acid, l-BAIBA, is a muscle-derived osteocyte survival factor. Cell Rep. 2018;22(6):1531–44.
Barragan-Adjemian C, Nicolella D, Dusevich V, Dallas MR, Eick JD, Bonewald LF. Mechanism by which MLO-A5 late osteoblasts/early osteocytes mineralize in culture: similarities with mineralization of lamellar bone. Calcif Tissue Int. 2006;79(5):340–53.
Guo D, Keightley A, Guthrie J, Veno PA, Harris SE, Bonewald LF. Identification of osteocyte-selective proteins. Proteomics. 2013;10(20):3688–98.
Dallas SL, Veno PA, Rosser JL, Barragan-Adjemian C, Rowe DW, Kalajzic I, et al. Time lapse imaging techniques for comparison of mineralization dynamics in primary murine osteoblasts and the late osteoblast/early osteocyte-like cell line MLO-A5. Cells Tissues Organs. 2009;189(1–4):6–11.
Kulkarni RN, Bakker AD, Everts V, Klein-Nulend J. Inhibition of osteoclastogenesis by mechanically loaded osteocytes: involvement of MEPE. Calcif Tissue Int. 2010;87(5):461–8.
Xiao Z, Zhang S, Mahlios J, Zhou G, Magenheimer BS, Guo D, et al. Cilia-like structures and polycystin-1 in osteoblasts/osteocytes and associated abnormalities in skeletogenesis and Runx2 expression. J Biol Chem. 2006;281(41):30884–95.
Zhang K, Barragan-Adjemian C, Ye L, Kotha S, Dallas M, Lu Y, et al. E11/gp38 selective expression in osteocytes: regulation by mechanical strain and role in dendrite elongation. Mol Cell Biol. 2006;26(12):4539–52.
Zhao S, Kato Y, Zhang Y, Harris S, Ahuja SS, Bonewald LF. MLO-Y4 osteocyte-like cells support osteoclast formation and activation. J Bone Miner Res. 2002;17(11):2068–79.
Kim H, Wrann CD, Jedrychowski M, Vidoni S, Kitase Y, Nagano K, et al. Irisin mediates effects on bone and fat via αV integrin receptors. Cell. 2018;175(7):1756–68.e17.
Kato Y, Boskey A, Spevak L, Dallas M, Hori M, Bonewald LF. Establishment of an osteoid preosteocyte-like cell MLO-A5 that spontaneously mineralizes in culture. J Bone Miner Res. 2001;16(9):1622–33.
Yang D, Gronthos S, Isenmann S, Morris HA, Atkins GJ. The late osteoblast/preosteocyte cell line MLO-A5 displays mesenchymal lineage plasticity. Stem Cells Int. 2019;2019:9838167.
Bodine PV, Vernon SK, Komm BS. Establishment and hormonal regulation of a conditionally transformed preosteocytic cell line from adult human bone. Endocrinology. 1996;137(11):4592–604.
Toscani D, Palumbo C, Dalla Palma B, Ferretti M, Bolzoni M, Marchica V, et al. The proteasome inhibitor bortezomib maintains osteocyte viability in multiple myeloma patients by reducing both apoptosis and autophagy: a new function for proteasome inhibitors. J Bone Miner Res. 2016;31(4):815–27.
Kalajzic I, Matthews BG, Torreggiani E, Harris MA, Divieti Pajevic P, Harris SE. In vitro and in vivo approaches to study osteocyte biology. Bone. 2013;54(2):296–306.
Kalajzic I, Staal A, Yang WP, Wu Y, Johnson SE, Feyen JH, et al. Expression profile of osteoblast lineage at defined stages of differentiation. J Biol Chem. 2005;280(26):24618–26.
Paic F, Igwe JC, Nori R, Kronenberg MS, Franceschetti T, Harrington P, et al. Identification of differentially expressed genes between osteoblasts and osteocytes. Bone. 2009;45(4):682–92.
Yang W, Lu Y, Kalajzic I, Guo D, Harris MA, Gluhak-Heinrich J, et al. Dentin matrix protein 1 gene cis-regulation: use in osteocytes to characterize local responses to mechanical loading in vitro and in vivo. J Biol Chem. 2005;280(21):20680–90.
van Bezooijen RL, ten Dijke P, Papapoulos SE, Löwik CW. SOST/sclerostin, an osteocyte-derived negative regulator of bone formation. Cytokine Growth Factor Rev. 2005;16(3):319–27.
•• Spatz JM, Wein MN, Gooi JH, Qu Y, Garr JL, Liu S, et al. The Wnt inhibitor sclerostin is up-regulated by mechanical unloading in osteocytes in vitro. J Biol Chem. 2015;290(27):16744–58 This article describes the isolation and characterization of Ocy454 cells.
•• Woo SM, Rosser J, Dusevich V, Kalajzic I, Bonewald LF. Cell line IDG-SW3 replicates osteoblast-to-late-osteocyte differentiation in vitro and accelerates bone formation in vivo. J Bone Miner Res. 2011;26(11):2634–46 This article describes the isolation and characterization of IDG-SW3 cells.
Shi C, Uda Y, Dedic C, Azab E, Sun N, Hussein AI, et al. Carbonic anhydrase III protects osteocytes from oxidative stress. FASEB J. 2017;32(1):440–52.
Lyons JS, Joca HC, Law RA, Williams KM, Kerr JP, Shi G, et al. Microtubules tune mechanotransduction through NOX2 and TRPV4 to decrease sclerostin abundance in osteocytes. Sci Signal. 2017;10(506):eaan5748. https://doi.org/10.1126/scisignal.aan5748.
He Q, Bouley R, Liu Z, Wein MN, Zhu Y, Spatz JM, et al. Large G protein α-subunit XLαs limits clathrin-mediated endocytosis and regulates tissue iron levels in vivo. Proc Natl Acad Sci U S A. 2017;114(45):E9559–E68.
He Q, Shumate LT, Matthias J, Aydin C, Wein MN, Spatz JM, et al. A G protein-coupled, IP3/protein kinase C pathway controlling the synthesis of phosphaturic hormone FGF23. JCI Insight. 2019;4(17):e125007. https://doi.org/10.1172/jci.insight.125007.
Ansari N, Ho PW, Crimeen-Irwin B, Poulton IJ, Brunt AR, Forwood MR, et al. Autocrine and paracrine regulation of the murine skeleton by osteocyte-derived parathyroid hormone-related protein. J Bone Miner Res. 2018;33(1):137–53.
Dobrosak C, Gooi JH. Increased sphingosine-1-phosphate production in response to osteocyte mechanotransduction. Bone Rep. 2017;7:114–20.
Gooi JH, Chia LY, Vrahnas C, Sims NA. Isolation, purification, generation, and culture of osteocytes. Methods Mol Biol. 1914;2019:39–51.
Wein MN, Liang Y, Goransson O, Sundberg TB, Wang J, Williams EA, et al. SIKs control osteocyte responses to parathyroid hormone. Nat Commun. 2016;7:13176. https://doi.org/10.1038/ncomms13176.
Wang K, Le L, Chun BM, Tiede-Lewis LM, Shiflett LA, Prideaux M, et al. A novel osteogenic cell line that differentiates into GFP-tagged osteocytes and forms mineral with a bone-like lacunocanalicular structure. J Bone Miner Res. 2019;34(6):979–95.
Sun N, Uda Y, Azab E, Kochen A, Santos RNCE, Shi C, et al. Effects of histone deacetylase inhibitor Scriptaid and parathyroid hormone on osteocyte functions and metabolism. J Biol Chem. 2019;294(25):9722–33. https://doi.org/10.1074/jbc.RA118.007312.
Zaman G, Pitsillides AA, Rawlinson SC, Suswillo RF, Mosley JR, Cheng MZ, et al. Mechanical strain stimulates nitric oxide production by rapid activation of endothelial nitric oxide synthase in osteocytes. J Bone Miner Res. 1999;14(7):1123–31.
Gu G, Nars M, Hentunen TA, Metsikko K, Vaananen HK. Isolated primary osteocytes express functional gap junctions in vitro. Cell Tissue Res. 2006;323(2):263–71.
Stern AR, Stern MM, Van Dyke ME, Jähn K, Prideaux M, Bonewald LF. Isolation and culture of primary osteocytes from the long bones of skeletally mature and aged mice. Biotechniques. 2012;52(6):361–73.
Stern AR, Bonewald LF. Isolation of osteocytes from mature and aged murine bone. Methods Mol Biol. 2015;1226:3–10.
Farr JN, Fraser DG, Wang H, Jaehn K, Ogrodnik MB, Weivoda MM, et al. Identification of senescent cells in the bone microenvironment. J Bone Miner Res. 2016;31(11):1920–9.
Ayturk UM, Jacobsen CM, Christodoulou DC, Gorham J, Seidman JG, Seidman CE, et al. An RNA-seq protocol to identify mRNA expression changes in mouse diaphyseal bone: applications in mice with bone property altering Lrp5 mutations. J Bone Miner Res. 2013;28(10):2081–93.
• Prideaux M, Schutz C, Wijenayaka AR, Findlay DM, Campbell DG, Solomon LB, et al. Isolation of osteocytes from human trabecular bone. Bone. 2016;88:64–72 This article describes the isolation of human osteocytes.
• Bernhardt A, Weiser E, Wolf S, Vater C, Gelinsky M. Primary human osteocyte networks in pure and modified collagen gels. Tissue Eng Part A. 2019;25(19-20):1347–55 This article describes the isolation of human osteocytes.
• Bernhardt A, Wolf S, Weiser E, Vater C, Gelinsky M. An improved method to isolate primary human osteocytes from bone. Biomed Tech (Berl). 2020;65(1):107–11 This article describes the isolation of human osteocytes.
Pina S, Ribeiro VP, Marques CF, Maia FR, Silva TH, Reis RL, et al. Scaffolding strategies for tissue engineering and regenerative medicine applications. Materials (Basel). 2019;12(11):1824. https://doi.org/10.3390/ma12111824.
Zhang C, Bakker AD, Klein-Nulend J, Bravenboer N. Studies on osteocytes in their 3D native matrix versus 2D in vitro models. Curr Osteoporos Rep. 2019;17(4):207–16.
Lee J, Kim G. Three-dimensional hierarchical nanofibrous collagen scaffold fabricated using fibrillated collagen and pluronic F-127 for regenerating bone tissue. ACS Appl Mater Interfaces. 2018;10(42):35801–11.
Uchihashi K, Aoki S, Matsunobu A, Toda S. Osteoblast migration into type I collagen gel and differentiation to osteocyte-like cells within a self-produced mineralized matrix: a novel system for analyzing differentiation from osteoblast to osteocyte. Bone. 2013;52(1):102–10.
Skottke J, Gelinsky M, Bernhardt A. In vitro co-culture model of primary human osteoblasts and osteocytes in collagen gels. Int J Mol Sci. 2019;20(8):1998. https://doi.org/10.3390/ijms20081998.
Boukhechba F, Balaguer T, Michiels JF, Ackermann K, Quincey D, Bouler JM, et al. Human primary osteocyte differentiation in a 3D culture system. J Bone Miner Res. 2009;24(11):1927–35.
Sun Q, Choudhary S, Mannion C, Kissin Y, Zilberberg J, Lee WY. Ex vivo construction of human primary 3D-networked osteocytes. Bone. 2017;105:245–52.
Sun Q, Choudhary S, Mannion C, Kissin Y, Zilberberg J, Lee WY. Ex vivo replication of phenotypic functions of osteocytes through biomimetic 3D bone tissue construction. Bone. 2018;106:148–55.
Sun Q, Gu Y, Zhang W, Dziopa L, Zilberberg J, Lee W. Ex vivo 3D osteocyte network construction with primary murine bone cells. Bone Res. 2015;3:15026.
Fulzele K, Krause DS, Panaroni C, Saini V, Barry KJ, Liu X, et al. Myelopoiesis is regulated by osteocytes through Gsα-dependent signaling. Blood. 2013;121(6):930–939. https://doi.org/10.1182/blood-2012-06-437160.
Fulzele K, Lai F, Dedic C, Saini V, Uda Y, Shi C, et al. Osteocyte-secreted Wnt signaling inhibitor sclerostin contributes to beige adipogenesis in peripheral fat depots. J Bone Miner Res. 2017;32(2):373–84.
•• Bellido T, Delgado-Calle J. Ex vivo organ cultures as models to study bone biology. JBMR Plus. 2020;4(3). https://doi.org/10.1002/jbm4.10345. This article reviews all techniques for organ culture and provide comprehensive overview of techniques and their applications.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Paola Divieti Pajevic declares no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Osteocytes
Rights and permissions
About this article
Cite this article
Divieti Pajevic, P. New and Old Osteocytic Cell Lines and 3D Models. Curr Osteoporos Rep 18, 551–558 (2020). https://doi.org/10.1007/s11914-020-00613-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11914-020-00613-3