Abstract
Purpose of review
In this review, we emphasize up-to-date practical cytogenetic and molecular aspects of chronic myeloid leukemia (CML) and summarize current knowledge on tyrosine kinase inhibitor (TKI) resistance and treatment response monitoring of CML.
Recent findings
The introduction of TKIs has changed the natural course of CML and markedly improved patient survival. Over the past decades, many research efforts were devoted to elucidating the leukemogenic mechanisms of BCR-ABL1 and developing novel TKIs. More recent studies have attempted to answer new questions that have emerged in the TKI era, such as the cytogenetic and molecular bases of treatment failure and disease progression, the clinical impact of genetic aberrations in Philadelphia chromosome (Ph)-positive and Ph-negative cells, and the biological significance of Ph secondarily acquired during therapy of other hematological neoplasms.
Summary
Recent progresses in the understanding of the cytogenetic and molecular mechanisms underlying therapeutic failure and disease progression have improved the risk stratification of CML and will be helpful in the design of novel therapeutic strategies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
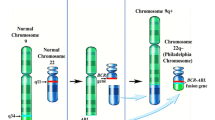
Chronic myeloid leukemia (CML) is a myeloproliferative neoplasm (MPN) characterized by a biphasic or triphasic natural history: an indolent chronic phase (CP) followed by an accelerated phase (AP) and then an aggressive blast phase (BP), or both. The BCR-ABL1 fusion, produced as a result of the t(9;22)(q34;q11.2), is a constitutively active tyrosine kinase that activates a network of downstream pathways and leads to leukemogenesis. The introduction of tyrosine kinase inhibitors (TKIs) has changed the natural course of CML and markedly improved patient survival. Despite the highly promising results, TKI resistance develops in ~ 13% of patients.
Ever since the first description of the Philadelphia chromosome (Ph)/der(22)t(9;22)(q34;q11.2), this translocation has served as a paradigm for how a single genetic abnormality causes malignant transformation and, most importantly, how a targeted therapy alters the disease course and dramatically improves patient outcome. However, many important questions remain unanswered. What are the mechanisms underlying TKI resistance? What are the cytogenetic and molecular bases of disease progression? What is the biological significance of chromosomal alterations in Ph-positive (Ph+) and Ph-negative (Ph-) cells? What is the importance of Ph acquired during therapy of other hematological malignancies? Herein we will try to synthesize recent advances on these important issues.
BCR-ABL1 Isoforms and Phenotypic Correlation
The normal ABL1 and BCR genes have 11 and 23 exons, respectively. In ABL1, the breakpoints are distributed in the intron between exons 1b and 1a, or in the intron between 1a and 2 (Fig. 1). Regardless of the ABL1 breakpoints, the two alternative first exons (1a and 1b) are always spliced out. The common exons 2–11 of ABL1 are then fused to different exon sets of BCR. In BCR, three breakpoint cluster regions are characterized: major (M-BCR), minor (m-BCR), and micro (μ-BCR), corresponding to three fusion proteins designated as P210, P190, and P230, respectively. All three BCR-ABL1 fusion proteins contain the common ABL1 domains. Depending on the BCR breakpoints, the fusion proteins may contain some or all of the BCR domains (Fig. 1) [1,2,3]. The e13a2 (b2a2) and e14a2 (b3a2) transcript subtypes, which encode P210, are seen in ~ 98% of all CML cases. The total frequency of other minor transcripts is ~ 2% [4, 5], including fusion transcripts lacking exon 2 of ABL1 (ela3, e13a3, and e14a3), e1a2+e13a2, e1a2+e14a2, e6a2, e12a1, e19a2, and “bizarre” insertions or breakpoints within exons [6].
Schematic representation of BCR and ABL1 gene structures and essential protein domains of BCR-ABL1 fusion proteins. a Gene structure and breakpoints of BCR and ABL1. In ABL1, the breakpoints (designated by parallel oblique lines) are distributed in the intron between exons 1b and 1a, or in the intron between exons 1a and 2. In BCR, most breakpoints in CML occur within the M-BCR region which encompasses exons 12–15. The m-BCR is located in the 3′ half of the first BCR intron. The μ-BCR is located further downstream between exons 19 and 21. b Essential domains of BCR-ABL1 fusion proteins. All three main BCR-ABL1 fusion proteins contain the common ABL1 domains, including the SRC homology domains SH2 and SH3, tyrosine kinase (Y-kinase) domain SH1, and DNA- and actin-binding domains (DBD and ABD). Depending on the BCR breakpoints, the fusion proteins may contain some or all of the following BCR domains: a coiled-coil (CC) oligomerization domain, a serine/threonine kinase (S/T-kinase) domain, a Rho guanine nucleotide exchange factor (Rho-GEF, also designated DBL-like) domain, a pleckstrin homology (PH) domain, a calcium-dependent lipid-binding domain (Cal-B), and a truncated RAC-GAP domain
There is a clear association of different BCR-ABL1 proteins with distinct disease phenotypes. The P210 form is reported in ~ 98% of CML but in < 20% of Ph+ B-lymphoblastic leukemia/lymphoma (B-ALL). In contrast, the P190 BCR-ABL1 is present in > 80% of Ph+ B-ALL but only rarely (~ 1%) observed in CML. CML patients with P190 tend to be older, present with monocytosis, and have a higher frequency of BP at initial presentation. Those who are not in BP initially have a higher risk of subsequent blastic transformation [7]. The rare e19a2 (P230) has been described in a small fraction of CML patients who often present with prominent neutrophilic maturation or thrombocytosis. Because of their low frequency, the clinical importance of other rare transcripts is unclear.
How different BCR-ABL1 proteins differentially drive lymphoid and myeloid leukemogenesis has not yet been completely elucidated. The three forms of BCR-ABL1 have been shown to be equally potent in inducing a CML-like MPN in a murine bone marrow transduction/transplantation model [8]. However, the expression of these three oncoproteins might be largely restricted to different hematopoietic cell types. Most studies suggest that P210 originates at the level of hematopoietic stem cells (HSCs) whereas P190 has a B cell progenitor origin without myeloid involvement [9]. It is important to keep in mind that HSCs and lineage-committed progenitors differ in many biological activities, such as metabolism, replication stress, chromatin condensation, transcriptional activity, and DNA damage response. These differences might play a role in inducing DNA breaks in different regions and generating different mutagenic patterns. In addition, P190 has a more potent lymphoid leukemogenic activity than P210 [10,11,12]. This may be associated with the increased intrinsic tyrosine kinase activity of P190 that allows elevated tyrosine phosphorylation of substrates critical for proliferation or transformation of lymphoid cells [13,14,15]. One key substrate is STAT6, a transcription factor implicated in lymphoid proliferative responses [16]. Its DNA binding activity is prominently activated via tyrosine phosphorylation by P190 but not P210 [14, 15]. How different domain compositions and overall structures of the two isoforms of BCR-ABL1 interact with different protein complexes and drive leukemogenesis remain elusive. One hypothesis is that differential subcellular localization causes the two isoforms to encounter different subsets of the proteome and consequently activates different signaling pathways [17,18,19].
Leukemogenic Effects of BCR-ABL1
The proto-oncoprotein ABL1 is a ubiquitously expressed non-receptor tyrosine kinase whereas BCR is a kinase with poorly understood functions. ABL1 shuttles continuously between the nucleus and cytoplasm but is predominantly localized in the nucleus in normal cells. However, fusion with BCR relocates ABL1 to the cytoplasm where it has the opportunities to interact with a number of proteins and exert its leukemogenic effect. Most importantly, the oligomerization domain encoded by the first exon of BCR constitutively activates the tyrosine kinase activity of ABL1 by promoting dimerization or tetramerization, which in turn autophosphorylates other sites on ABL1 and generates more binding sites for SH2 domain-containing proteins. Thus, BCR-ABL1 can recruit and activate multiple downstream signal transducers through the SH2 domains on these proteins.
The most clinically relevant and extensively-studied pathways include RAS/RAF1/MEK/ERK, PI3K/AKT, and SFKs/STAT1/STAT5 [15, 20,21,22,23,24]. In physiological conditions, these pathways are turned on by binding of growth factors or cytokines to their specific receptors. BCR-ABL1 abrogates this growth factor and cytokine dependence by activating essential downstream molecules through direct interaction or via the GRB2/SOS/GAB2 complex. Constitutive activation of these pathways induces endogenous reactive oxygen species and DNA double-strand breaks, causes unchecked cell cycle progression, impairs DNA repair, dysregulates cell adhesion, and inhibits apoptosis and autophagy [25,26,27]. BCR-ABL1 also activates MYC, which is related to drug resistance, aberrant DNA synthesis, and genomic instability [28]. The level of MYC in CML at diagnosis may predict treatment response and progression to BP [28]. Furthermore, the abovementioned events contribute to genomic instability and make CML cells more susceptible to developing further genomic abnormalities, likely a major driver of disease progression.
CML evolves through a partially understood multistep process. A hypothetical model considers the initiation and progression of CML being caused by a stepwise accumulation of cytogenetic or molecular aberrations. In this model, BCR-ABL1 is crucial in transforming HSCs and initiating CML. Further secondary cytogenetic, molecular, and epigenetic alterations, cooperating with increased expression levels of BCR-ABL1, provide a significant proliferative and survival advantage to a committed myeloid progenitor and lead to blastic transformation. The Ph is typically the sole cytogenetic abnormality in patients with CML-CP. With disease progression, additional chromosomal abnormalities (ACAs) increase (30% in AP and 70–80% in BP) [29••, 30]. The acquisition and accumulation of these abnormalities is due, at least in part, to genomic instability caused by BCR-ABL1. Both in vitro and in vivo studies have indicated a direct causal role of BCR-ABL1 in the development of karyotypic abnormalities and point mutations [31].
Monitoring CML Treatment Response
Ongoing assessment is required for early identification of patients who are not responding optimally so that alternative treatment strategies can be considered. Based on the time to reach various levels of responses, treatment responses can be categorized into three groups: optimal, failure, and warning (Table 1) [32••]. Optimal response is associated with the best long-term outcome, and patients are expected to live a similar lifespan to that of the general population. Continuing the current therapy is appropriate in this group. In contrast, “failure” means that a different treatment should be initiated to limit the risk of progression and death. “Warning” implies that although the current treatment may still be beneficial, the long-term outcome of treatment is less likely to be favorable. More frequent monitoring is required to permit timely changes in therapy in case of treatment failure.
Per the European LeukemiaNet (ELN) recommendations, response can be assessed with either cytogenetic analysis or quantitative reverse-transcription polymerase chain reaction alone; when local laboratory facilities permit, both cytogenetic and molecular tests are recommended until a complete cytogenetic remission (CCyR) and major molecular remission (MMR) are achieved [32••]. Following these landmarks, molecular tests alone may be sufficient (Table 2). However, in case of suspicion of warning, failure, or progression, cytogenetic analysis is mandated. In addition, mutation analysis is recommended in certain situations as following imatinib failure, up to 48% of cases with primary resistance and 68% of cases with secondary resistance show mutations in the BCR-ABL1 kinase domain [34]. Noteworthily, a single measurement of the BCR-ABL1 transcript level is not sufficient to define failure. Two tests at 3 and 6 months and supplementary tests in between should be performed before a decision is made to change the treatment. Furthermore, failures must be distinguished as either primary (a lack of initial response) or secondary (loss of an established response).
Patients with CML-CP who achieve CCyR have a favorable survival similar to that of the general population, and deeper treatment responses beyond CCyR provide no additional survival benefit [35]. However, a deeper response, such as MR4.5, may serve as an indicator of being one step closer to successful discontinuation of treatment [36]. In contrast, patients with CML-BP who achieve CCyR have a dismal survival similar to that of patients with CML-BP who achieve hematologic remission only [37]. Achieving molecularly undetectable leukemia is required for an optimal outcome, making precise and timely monitoring of BCR-ABL level essential in the management of CML-BP.
Mechanisms of TKI Resistance
The tyrosine kinase activity of BCR-ABL1 depends on the conformation of four highly conserved regions: (1) the ATP-binding loop (P-loop), which admits ATP that donates terminal phosphates to tyrosine residues on BCR-ABL1 and its substrates; (2) the direct binding site, which serves as a docking site for adapter proteins such as GRB2, CBL, and CRKL that further activate multiple downstream pathways; (3) the activation loop (A-loop), which switches between active (phosphorylated) and inactive (unphosphorylated) conformations. The inactive A-loop functions as a “pseudo-substrate” and is folded into and blocks the substrate binding site; and (4) the SH2 and SH3 domains, which assemble onto the catalytic domain of ABL1 and allosterically clamp it in an inactive stage. Importantly, imatinib only binds to ABL1 in the inactive state. Imatinib acts as a competitive inhibitor for ATPs. It binds to the catalytic domain locating close to the ATP binding pocket, displaces ATPs, and freezes the BCR-ABL1 kinase in the inactive non-ATP-binding conformation. This interaction blocks the tyrosine kinase activity and prevents BCR-ABL1-mediated autophosphorylation, and in turn substrate phosphorylation, thereby switching off downstream signaling pathways.
TKI resistance (~ 13% of patients) [38] can develop as a result of mechanisms involving BCR-ABL1-dependent and BCR-ABL1-independent pathways. The BCR-ABL1-dependent pathways include mutations and amplification of BCR-ABL1; the BCR-ABL1-independent pathways include resistance of CML stem cells, clonal evolution, activation of other tyrosine kinases, pharmacokinetic variability, and drug transport mechanisms.
Over 100 mutations in ABL1 have been identified that confer TKI resistance [39]. As the disease progresses, their frequency increases, with mutations detected in 27%, 52%, and 75% of patients with CML in CP, AP, and myeloid BP, respectively. Additionally, they are more common in acquired than primary resistance [40, 41]. These mutations contribute to TKI resistance through the following mechanisms: (1) interference with TKI binding by altering the three-dimensional structure of the TKI binding site (T315I mutation). This mutation is highly resistant to imatinib, nilotinib, dasatinib, and bosutinib, although ponatinib remains effective [32••, 42]; (2) preventing BCR-ABL1 from assuming the inactive conformation required for imatinib binding (e.g., mutations in position M351 and A-loop) or stabilizing the active conformation of BCR-ABL1 (e.g., P-loop mutations) [43]; and (3) changing the auto-inhibitory conformation by disrupting the SH3-SH2 domain clamp through destabilizing intramolecular interactions (SH2 domain mutations) [44]. Amplification of BCR-ABL1 can be caused by extra copies of Ph or ring chromosomes harboring multiple copies of BCR-ABL1. The level of BCR-ABL1 expression correlates with the speed at which resistance to imatinib develops, which provides further evidence that the BCR-ABL1 level serves as an excellent marker for TKI resistance [45].
TKIs eradicate CML progenitor cells but do not effectively target CML stem cells. Several possible mechanisms have been proposed to explain how CML stem cells escape the effects of TKIs, including low intracellular imatinib levels due to either inadequate active uptake or excessive drug efflux, elevated BCR-ABL1 expression, and a quiescent cell cycle status [46, 47]. Additionally, CML stem cell survival may not actually depend on BCR-ABL1 kinase [48,49,50]. Acquisition of ACAs in Ph+ cells is thought to drive disease progression. As will be discussed later, certain ACAs are associated with poor TKI response, particularly, 3q26.2 rearrangement [51, 52••]. Resistance can also be mediated through overexpression of other tyrosine kinases such as the SRC family kinases. Two SRC family kinases, LYN and HCK, are highly activated and are not suppressed by imatinib [53]. Moreover, TKI resistance can develop from constitutive activation of downstream signaling pathways such as the mTOR pathway [54]. Decreased responses to imatinib therapy might also be related to pharmacokinetic variability. These mechanisms may involve poor patient compliance, variation in metabolizing enzyme activity (most importantly CYP3A4), and drug-drug interaction [55]. Recent studies have shown that a lower level of expression or activity of hOCT1, an active influx transporter, is associated with a lower probability of achieving a cytogenetic or molecular remission [56].
ACAs in Disease Progression
Studies in the pre-TKI era stratified ACAs into “major” and “minor” routes based on their frequency. The abnormalities with a frequency of > 10% (of all cases with ACAs) were designated as major-route ACAs, including +Ph, +8, i(17q), and +19 [29••]. All others with a frequency of < 10% were designated as minor-route ACAs. In the 2017 update of the WHO Classification of Tumors of Hematopoietic and Lymphoid Tissue [57], the major-route but not minor-route ACAs, complex karyotype, 3q26.2 abnormalities at initial diagnosis, and ACAs emerging during therapy are considered defining criteria for CML-AP. Per ELN recommendations, major-route ACAs emerging during therapy, but not minor-route ACAs emerging during TKI therapy nor any ACAs detected at initial diagnosis define TKI failure and mandate a change of treatment [32••].
Most recent studies have established a four-tier risk stratification model based on the presence or absence of ACAs and the types of ACAs [58••, 59••]. These four different subgroups confer different risk of blastic transformation and patient outcome. This four-tier stratification remains somewhat valid for ACAs detected at initial diagnosis of CML, particularly for the high-risk ACAs, as follows:
-
High risk: 3q26.2 rearrangement, −7/7q- or i(17q), either as an isolated single ACA or as a component of a complex karyotype;
-
Intermediate risk-2: complex karyotypes without any of the three high-risk components;
-
Intermediate risk-1: single ACAs other than the three high-risk single ACAs;
-
Standard risk: no ACAs.
The 5-year cumulative probability of blastic transformation from initial diagnosis of CML for these four subgroups is 67%, 42%, 28%, and 10%, respectively, and the 8-year survival rate is 31%, 47%, 58%, and 80%, respectively. The pre-BP disease course in the high-risk patients treated with TKIs resembles that observed in CML patients treated in the pre-TKI era. These patients may benefit from timely hematopoietic stem cell transplantation before the onset of blastic transformation. Regardless of the risk of ACAs, interestingly, the three ACA subgroups have a similar latency from initial diagnosis of CML to their emergence (median: 13 months). In addition, the prognostic difference among the three ACA subgroups is minimized once the disease progresses to BP [52••, 59••].
The type of treatment received during CP seemingly influences the patterns of secondary abnormalities acquired in progression [29••]. Trisomy 8 is more commonly associated with treatment of busulfan than hydroxyurea. In patients treated with IFN-α, there is increased occurrence of unusual secondary abnormalities, such as del(7p) and del(13q), and cytogenetically divergent subclones and a higher incidence of cytogenetic evolution [60]. While aberrations following autologous stem cell transplantation seem to be similar to the ones seen in non-transplanted patients, the cytogenetic evolution after allogeneic transplant seems to be random, structurally complex, and sometimes transient. There are fewer major-route abnormalities but a significantly higher frequency of balanced translocations and divergent subclones. Cytogenetic changes in TKI-treated patients were originally thought to follow the same genetic evolution pattern as before the introduction of TKIs [61]. However, a recent study of a large cohort of CML-BP patients treated in the TKI era reveals a significant shift of ACA pattern, particularly the marked increase in the incidence of 3q26.2 rearrangement in the TKI era [52••]. The emergence of 3q26.2 rearrangement as a major-route change in the TKI era correlates with a high frequency of ABL1 mutations in patients harboring this rearrangement, supporting a role of TKI resistance in the changing cytogenetic landscape.
Molecular Genetic Changes in Disease Progression
The molecular changes in CML can be categorized, somewhat simplistically, into the following groups based on their functions:
-
1.
Increased level of BCR-ABL1 expression. Qualitative analyses have shown increased levels of the BCR-ABL1 mRNA and protein in CML-AP and BP compared with CP [62,63,64]. Furthermore, studies have supported a dose-dependent relationship for many of the leukemogenic effects of BCR-ABL1, including uncontrolled proliferation and escape from apoptosis [64, 65].
-
2.
Oncogenic activation or amplification of pro-oncogenes. These genes include mutation or amplification of RUNX1 [66], increased translation or amplification of MYC [67, 68], mutations of NRAS, KRAS [66], and the Wnt/beta-catenin pathway [69, 70], and gain-of-function mutation of GATA2 [71].
-
3.
Loss-of-function mutations of tumor suppressor genes. Mutations in the TP53 have been detected in approximately 20–30% of CML-BP [66, 72]. Deletions of CDKN2A (~ 50%) and RB1 (18%) are frequent in lymphoid BP [31, 73]. Mutations of IKZF1 are not detected in CML-CP [74], but are observed in 18–27% of CML-BP samples [66, 74], mainly of lymphoid BP.
-
4.
Suppressed expression or function of genes important for differentiation. Mutation of C/EPB alpha [75], t(3;21)(3q26.2;q22)/RUNX1-MECOM [76, 77], and translocations involving HOX genes are reported in myeloid BP [78, 79].
-
5.
Mutations in genes important for epigenetic regulation, such as ASXL (20%) and TET2 (8%), are seen in CML-BP [66]. CpG site methylation is significantly increased in CML-BP compared with CP [80]. Aberrant DNA methylation of multiple genes, such as the calcitonin gene, CDKN2B, PDLIM4, OSCP1, and the ABL1 promotor region, is associated with disease progression or TKI resistance [79,80,81,82].
As demonstrated in a recent study, the dynamics of additional mutations are associated with treatment outcomes [83]. Unsurprisingly, mutations acquired during TKI treatment are strongly correlated with treatment failure. ABL1 tyrosine kinase domain mutations exclusively follow this pattern. However, caution should be taken when interpreting clearance or persistence of pre-existing mutations as the prognostic impact depends on the timing of acquisition and cellular origin of these mutations. When mutations originate from a Ph− preleukemic clone, a durable satisfactory TKI response is usually achieved despite persistence of these mutations. Mutations acquired in preleukemic HSCs implied by the presence of these mutations in T cells at diagnosis and both Ph+ and Ph− clones may show persistence, significant reduction, or clearance following TKI therapy. No obvious association between treatment outcome and dynamics of these mutations following treatment is observed. These mutations are frequently seen in the genes associated with chromatin modification and DNA methylation. Although not currently widely used, assessment of the baseline mutational profile at diagnosis and determination of involvement of the T cell lineage may be of value in the future.
Clonal Cytogenetic Abnormalities in Ph-Negative Cells
About 3–9% of patients who receive imatinib develop clonal cytogenetic abnormalities in Ph− (CCA/Ph−) cells, most commonly −7, +8, −5, and −Y [84,85,86,87]. However, only a small subset of these patients (2–5%) develops clinically evident myelodysplastic syndrome (MDS) or acute myeloid leukemia (AML) [88, 89]. In the absence of dysplasia, the presence of CCA/Ph− does not adversely affect patient outcome, with the exception of −7. About 30% of patients with −7 develop MDS or AML [90]. According to the ELN recommendations, −7 and del(7q) detected during TKI treatment is a “warning” sign and requires more frequent cytogenetic and molecular genetic monitoring and long-term follow-up with the performance of bone marrow assessment [32••]. However, other CCA/Ph− do not necessitate immediate therapeutic intervention or more frequent monitoring. Bone marrow examination is reserved for cases with cytopenia or dysplastic morphology.
Ph Acquired Secondarily During Therapy of AML, MDS, and ALL
The acquisition of a secondary Ph during therapy of myeloid or lymphoid neoplasms is rare (summarized in [91•]). It has been reported in AML, MDS, B-ALL, and T-ALL. The emergence of secondary Ph is always associated with advanced stage of disease: during relapse or in refractory stage of de novo acute leukemia or after acute transformation from MDS. The transcript subtype is of overwhelmingly the e1a2 (P190) subtype (~ 74% of all cases). In the vast majority of patients, the diseases before and after the emergence of the Ph are clonally related and usually show similar morphological and immunophenotypic features. Features of CML are rare regardless of the size of Ph+ clones.
The emergence of a secondary Ph likely signifies terminal illness with profound genomic instability, in which the Ph is acquired randomly as a passenger event in the founder clone. Alternatively, in a subset of patients, the emergence of Ph could be due to the expansion of a minute Ph+ subclone that initially is not detectable by cytogenetic methods. Nonetheless, the emergence of Ph clones is not essential for the maintenance of the leukemic process. Incorporating TKIs into treatment may eradicate the Ph+ clones and potentially change the disease prognosis in a small subset of patients, but most patients have refractory disease despite the disappearance of the Ph+ clones. The outcome in these patients is extremely poor with a median survival of 4 months after the emergence of the Ph.
Ph Acquired During Therapy of MPN
JAK2 V617F (or rarely MPL W515L/K) and BCR-ABL1 were previously thought to be mutually exclusive. However, an increasing number of cases with coexistence of these two mutations have been described, acquired either simultaneously or sequentially. Herein, we only discuss the Ph arising in patients with a previous diagnosis of Ph− MPN. To date, < 40 cases have been reported in the literature [92,93,94,95], predominantly in patients with a history of polycythemia vera (~ 70%), followed by essential thrombocythemia and primary myelofibrosis. However, the mutational status of JAK2 and MPL was not examined in all cases of Ph− MPN. The Ph acquired after Ph− MPN is often the e13a2/e14a2 (P210) subtype (~ 70%) [93]. Additionally, the disease in most patients, if not all, is bona fide CML. These two features are in sharp contrast to the Ph acquired during therapy in MDS and acute leukemia, where the transcript subtype is of overwhelmingly the e1a2 (P190) subtype and features of CML are rare.
De Novo Ph+ Acute Leukemia
The Ph can also be seen in other types of leukemia, including de novo ALL, AML, and mixed phenotype acute leukemia (MPAL). Among all Ph+ acute leukemias, the overall frequencies of Ph+ ALL, Ph+ MPAL, and Ph+ AML are ~ 65%, ~ 30%, and < 5%, respectively [96]. The Ph is the most commonly detected cytogenetic abnormality in B-ALL patients: ~ 3% of pediatric patients, ~ 25% of adults, and up to 50% of patients older than 50 years [97]. The P190 form is documented in ~ 85% of cases. Patients with Ph+ B-ALL tend to have a high leukocyte count and a high frequency of central nervous system involvement. Microscopically, Ph+ B-ALL is similar to Ph− B-ALL. Ph+ B-ALL frequently expresses myeloid-associated antigens including CD13 and CD33, and CD25 expression is highly characteristic [98]. Very rare cases of Ph+ T-ALL have been also reported [99].
As is the case in ALL, the Ph is the most common recurrent genetic abnormality in MPAL and is found in ~ 30% of cases. The P190 form is detected in ~ 60% of cases. Patients with Ph+ MPAL tend to present with a high leukocyte count but are otherwise clinically similar to patients with Ph− MPAL. Most cases of MPAL have a B/myeloid phenotype, with few cases reported to show T/myeloid phenotype.
In the 2017 revised WHO classification [57], Ph+ AML is included as a new provisional entity. Ph+ AML is rare, comprising < 1% of all cases of AML. Although most Ph+ AML cases express the P210 form of BCR-ABL1, P190 is not uncommon and detected in 17–41% of cases [100,101,102]. Ph+ AML occurs primarily in adults and shows distinct hematological, morphological, and genetic characteristics from CML-BP, such as absence of basophilia, a slightly lower bone marrow cellularity, a lower myeloid-to-erythroid ratio, more prominent dysplasia with lack of significant myeloid maturation, and a significantly lower prevalence of major ACAs [100].
Conclusions
Although TKIs have revolutionized the treatment of CML patients and dramatically improved their outcome, therapy resistance remains a major problem. Ongoing assessment is required for early identification of patients who are not responding optimally so that alternative treatment strategies can be considered. Potential novel therapeutic strategies may include eradicating CML stem cells, inhibiting other tyrosine kinases that are overexpressed during progression, and simultaneously targeting both BCR-ABL1 and constitutively activated downstream molecules. Progress has been achieved in investigating the cytogenetic and molecular aberrations associated with disease progression. Based on the ACA-associated risk of blastic transformation, a four-tier risk stratification model has been established that would help identify high-risk patients who may need timely hematopoietic stem cell transplantation. Additionally, with the introduction of novel sequencing technology, patterns of molecular evolution will also likely be established, analogous with the identification of cytogenetic evolutionary routes in CML.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer. 2005;5(3):172–83.
Ahmed W, Van Etten RA. Alternative approaches to eradicating the malignant clone in chronic myeloid leukemia: tyrosine-kinase inhibitor combinations and beyond. Hematology Am Soc Hematol Educ Program. 2013;2013:189–200.
Cilloni D, Saglio G. Molecular pathways: BCR-ABL. Clin Cancer Res. 2012;18(4):930–7.
Goh HG, Hwang JY, Kim SH, Lee YH, Kim YL, Kim DW. Comprehensive analysis of BCR-ABL transcript types in Korean CML patients using a newly developed multiplex RT-PCR. Transl Res. 2006;148(5):249–56.
Arun AK, Senthamizhselvi A, Mani S, Vinodhini K, Janet NB, Lakshmi KM, et al. Frequency of rare BCR-ABL1 fusion transcripts in chronic myeloid leukemia patients. Int J Lab Hematol. 2017;39(3):235–42.
Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7(6):441–53.
Gong Z, Medeiros LJ, Cortes JE, Zheng L, Khoury JD, Wang W, et al. Clinical and prognostic significance of e1a2 BCR-ABL1 transcript subtype in chronic myeloid leukemia. Blood Cancer J. 2017;7(7):e583.
Li S, Ilaria RL Jr, Million RP, Daley GQ, van Etten R. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med. 1999;189(9):1399–412.
Castor A, Nilsson L, Åstrand-Grundström I, Buitenhuis M, Ramirez C, Anderson K, et al. Distinct patterns of hematopoietic stem cell involvement in acute lymphoblastic leukemia. Nat Med. 2005;11(6):630–7.
Lugo TG, Witte ON. The BCR-ABL oncogene transforms Rat-1 cells and cooperates with v-myc. Mol Cell Biol. 1989;9(3):1263–70.
Kelliher M, Knott A, McLaughlin J, Witte ON, Rosenberg N. Differences in oncogenic potency but not target cell specificity distinguish the two forms of the BCR/ABL oncogene. Mol Cell Biol. 1991;11(9):4710–6.
Voncken JW, Kaartinen V, Pattengale PK, Germeraad WT, Groffen J, Heisterkamp N. BCR/ABL P210 and P190 cause distinct leukemia in transgenic mice. Blood. 1995;86(12):4603–11.
Reckel S, Hamelin R, Georgeon S, Armand F, Jolliet Q, Chiappe D, et al. Differential signaling networks of Bcr-Abl p210 and p190 kinases in leukemia cells defined by functional proteomics. Leukemia. 2017;31(7):1502–12.
Danial NN, Pernis A, Rothman PB. Jak-STAT signaling induced by the v-abl oncogene. Science. 1995;269(5232):1875–7.
Ilaria RL Jr, van Etten RA. P210 and P190BCR/ABL induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J Biol Chem. 1996;271(49):31704–10.
Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4(3):313–9.
Telegeev GD, Dubrovska AN, Nadgorna VA, Dybkov MV, Zavelevich MP, Maliuta SS, et al. Immunocytochemical study of Bcr and Bcr-Abl localization in K562 cells. Exp Oncol. 2010;32(2):81–3.
Heisterkamp N, Voncken JW, Senadheera D, Gonzalez-Gomez I, Reichert A, Haataja L, et al. Reduced oncogenicity of p190 Bcr/Abl F-actin-binding domain mutants. Blood. 2000;96(6):2226–32.
Hantschel O, Wiesner S, Güttler T, Mackereth CD, Rix LL, Mikes Z, et al. Structural basis for the cytoskeletal association of Bcr-Abl/c-Abl. Mol Cell. 2005;19(4):461–73.
Wee P, Wang Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers (Basel). 2017;9(5):52.
Birge RB, et al. Crk and CrkL adaptor proteins: networks for physiological and pathological signaling. Cell Commun Signal. 2009;7:13.
Varticovski L, Daley GQ, Jackson P, Baltimore D, Cantley LC. Activation of phosphatidylinositol 3-kinase in cells expressing abl oncogene variants. Mol Cell Biol. 1991;11(2):1107–13.
Jain SK, Susa M, Keeler ML, Carlesso N, Druker B, Varticovski L. PI 3-kinase activation in BCR/abl-transformed hematopoietic cells does not require interaction of p85 SH2 domains with p210 BCR/abl. Blood. 1996;88(5):1542–50.
Shuai K, Halpern J, ten Hoeve J, Rao X, Sawyers CL. Constitutive activation of STAT5 by the BCR-ABL oncogene in chronic myelogenous leukemia. Oncogene. 1996;13(2):247–54.
Gaymes TJ, Mufti GJ, Rassool FV. Myeloid leukemias have increased activity of the nonhomologous end-joining pathway and concomitant DNA misrepair that is dependent on the Ku70/86 heterodimer. Cancer Res. 2002;62(10):2791–7.
Hoover RR, Gerlach MJ, Koh EY, Daley GQ. Cooperative and redundant effects of STAT5 and Ras signaling in BCR/ABL transformed hematopoietic cells. Oncogene. 2001;20(41):5826–35.
Nowicki MO, Falinski R, Koptyra M, Slupianek A, Stoklosa T, Gloc E, et al. BCR/ABL oncogenic kinase promotes unfaithful repair of the reactive oxygen species-dependent DNA double-strand breaks. Blood. 2004;104(12):3746–53.
Albajar M, Gomez-Casares MT, Llorca J, Mauleon I, Vaque JP, Acosta JC, et al. MYC in chronic myeloid leukemia: induction of aberrant DNA synthesis and association with poor response to imatinib. Mol Cancer Res. 2011;9(5):564–76.
•• Johansson B, Fioretos T, Mitelman F. Cytogenetic and molecular genetic evolution of chronic myeloid leukemia. Acta Haematol. 2002;107(2):76–94 Comprehensive and elegant review of cytogenetic changes in CML in pre-TKI era.
Mu Q, Ma Q, Wang Y, Chen Z, Tong X, Chen FF, et al. Cytogenetic profile of 1,863 Ph/BCR-ABL-positive chronic myelogenous leukemia patients from the Chinese population. Ann Hematol. 2012;91(7):1065–72.
Calabretta B, Perrotti D. The biology of CML blast crisis. Blood. 2004;103(11):4010–22.
•• Baccarani M, et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood. 2013;122(6):872–84 Updated recommendations for treatment and monitoring of CML.
Fioretos T. Chronic myeloid leukemia. In: Heim S, Mitelman F, editors. Cancer cytogenetics. Hoboken: Wiley; 2016. p. 153–74.
Soverini S, Branford S, Nicolini FE, Talpaz M, Deininger MW, Martinelli G, et al. Implications of BCR-ABL1 kinase domain-mediated resistance in chronic myeloid leukemia. Leuk Res. 2014;38(1):10–20.
Hanfstein B, Muller MC, Hochhaus A. Response-related predictors of survival in CML. Ann Hematol. 2015;94(Suppl 2):S227–39.
Hehlmann R, Hasford J, Pfirrmann M, Lauseker M, Saußele S, Hochhaus A, et al. Reply to H. Kantarjian et al. J Clin Oncol. 2014;32(27):3078–9.
Chen Z, Medeiros LJ, Kantajian HM, Zheng L, Gong Z, Patel KP, et al. Differential depth of treatment response required for optimal outcome in patients with blast phase versus chronic phase of chronic myeloid leukemia. Blood Cancer J. 2017;7(2):e521.
Chaitanya PK, Kumar KA, Stalin B, Sadashivudu G, Srinivas ML. The role of mutation testing in patients with chronic myeloid leukemia in chronic phase after imatinib failure and their outcomes after treatment modification: single-institutional experience over 13 years. Indian J Med Paediatr Oncol. 2017;38(3):328–33.
Quintas-Cardama A, Kantarjian HM, Cortes JE. Mechanisms of primary and secondary resistance to imatinib in chronic myeloid leukemia. Cancer Control. 2009;16(2):122–31.
Jabbour E, Kantarjian H, Jones D, Talpaz M, Bekele N, O'Brien S, et al. Frequency and clinical significance of BCR-ABL mutations in patients with chronic myeloid leukemia treated with imatinib mesylate. Leukemia. 2006;20(10):1767–73.
Soverini S, Colarossi S, Gnani A, Rosti G, Castagnetti F, Poerio A, et al. Contribution of ABL kinase domain mutations to imatinib resistance in different subsets of Philadelphia-positive patients: by the GIMEMA Working Party on Chronic Myeloid Leukemia. Clin Cancer Res. 2006;12(24):7374–9.
O'Hare T, Zabriskie MS, Eiring AM, Deininger MW. Pushing the limits of targeted therapy in chronic myeloid leukaemia. Nat Rev Cancer. 2012;12(8):513–26.
Apperley JF. Part I: Mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 2007;8(11):1018–29.
Sherbenou DW, Hantschel O, Kaupe I, Willis S, Bumm T, Turaga LP, et al. BCR-ABL SH3-SH2 domain mutations in chronic myeloid leukemia patients on imatinib. Blood. 2010;116(17):3278–85.
Barnes DJ, Palaiologou D, Panousopoulou E, Schultheis B, Yong ASM, Wong A, et al. Bcr-Abl expression levels determine the rate of development of resistance to imatinib mesylate in chronic myeloid leukemia. Cancer Res. 2005;65(19):8912–9.
Jiang X, Zhao Y, Smith C, Gasparetto M, Turhan A, Eaves A, et al. Chronic myeloid leukemia stem cells possess multiple unique features of resistance to BCR-ABL targeted therapies. Leukemia. 2007;21(5):926–35.
Thomas J, Wang L, Clark RE, Pirmohamed M. Active transport of imatinib into and out of cells: implications for drug resistance. Blood. 2004;104(12):3739–45.
Hamilton A, Helgason GV, Schemionek M, Zhang B, Myssina S, Allan EK, et al. Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood. 2012;119(6):1501–10.
Perl A, Carroll M. BCR-ABL kinase is dead; long live the CML stem cell. J Clin Invest. 2011;121(1):22–5.
Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest. 2011;121(1):396–409.
Wang W, Cortes JE, Lin P, Beaty MW, Ai D, Amin HM, et al. Clinical and prognostic significance of 3q26.2 and other chromosome 3 abnormalities in CML in the era of tyrosine kinase inhibitors. Blood. 2015;126(14):1699–706.
•• Chen Z, et al. Cytogenetic landscape and impact in blast phase of chronic myeloid leukemia in the era of tyrosine kinase inhibitor therapy. Leukemia. 2017;31(3):585–92 Demonstration of changes in ACA landscape, prognostic impact of ACAs, and relationship between ACAs in CML-BP in TKI era.
Wu J, Meng F, Kong LY, Peng Z, Ying Y, Bornmann WG, et al. Association between imatinib-resistant BCR-ABL mutation-negative leukemia and persistent activation of LYN kinase. J Natl Cancer Inst. 2008;100(13):926–39.
Quentmeier H, Eberth S, Romani J, Zaborski M, Drexler HG. BCR-ABL1-independent PI3Kinase activation causing imatinib-resistance. J Hematol Oncol. 2011;4:6.
Haouala A, Widmer N, Duchosal MA, Montemurro M, Buclin T, Decosterd LA. Drug interactions with the tyrosine kinase inhibitors imatinib, dasatinib, and nilotinib. Blood. 2011;117(8):e75–87.
White DL, Saunders VA, Dang P, Engler J, Venables A, Zrim S, et al. Most CML patients who have a suboptimal response to imatinib have low OCT-1 activity: higher doses of imatinib may overcome the negative impact of low OCT-1 activity. Blood. 2007;110(12):4064–72.
Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. Revised 4th ed. Switzerland: WHO Press; 2017.
• Wang W, et al. Risk stratification of chromosomal abnormalities in chronic myelogenous leukemia in the era of tyrosine kinase inhibitor therapy. Blood. 2016;127(22):2742–50 Stratification of CML patients based on ACA-associated patient survival in TKI era.
•• Gong Z, et al. Cytogenetics-based risk prediction of blastic transformation of chronic myeloid leukemia in the era of TKI therapy. Blood Adv. 2017;1(26):2541–52 Four-tier stratification of CML based on ACA-associated risk of blastic transformation.
Johansson B, Fioretos T, Billström R, Mitelman F. Aberrant cytogenetic evolution pattern of Philadelphia-positive chronic myeloid leukemia treated with interferon-alpha. Leukemia. 1996;10(7):1134–8.
Mitelman F, Johansson B, Mertens F. https://cgap.nci.nih.gov/Chromosomes/Mitelman. 2001. Accessed 1 Aug 2018.
Gaiger A, Henn T, Hörth E, Geissler K, Mitterbauer G, Maier-Dobersberger T, et al. Increase of BCR-ABL chimeric mRNA expression in tumor cells of patients with chronic myeloid leukemia precedes disease progression. Blood. 1995;86(6):2371–8.
Guo JQ, Wang JY, Arlinghaus RB. Detection of BCR-ABL proteins in blood cells of benign phase chronic myelogenous leukemia patients. Cancer Res. 1991;51(11):3048–51.
Barnes DJ, Schultheis B, Adedeji S, Melo JV. Dose-dependent effects of Bcr-Abl in cell line models of different stages of chronic myeloid leukemia. Oncogene. 2005;24(42):6432–40.
Cambier N, Chopra R, Strasser A, Metcalf D, Elefanty AG. BCR-ABL activates pathways mediating cytokine independence and protection against apoptosis in murine hematopoietic cells in a dose-dependent manner. Oncogene. 1998;16(3):335–48.
Grossmann V, Kohlmann A, Zenger M, Schindela S, Eder C, Weissmann S, et al. A deep-sequencing study of chronic myeloid leukemia patients in blast crisis (BC-CML) detects mutations in 76.9% of cases. Leukemia. 2011;25(3):557–60.
Notari M, Neviani P, Santhanam R, Blaser BW, Chang JS, Galietta A, et al. A MAPK/HNRPK pathway controls BCR/ABL oncogenic potential by regulating MYC mRNA translation. Blood. 2006;107(6):2507–16.
Jennings BA, Mills KI. c-myc locus amplification and the acquisition of trisomy 8 in the evolution of chronic myeloid leukaemia. Leuk Res. 1998;22(10):899–903.
Jamieson CH, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351(7):657–67.
Radich JP, Dai H, Mao M, Oehler V, Schelter J, Druker B, et al. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc Natl Acad Sci U S A. 2006;103(8):2794–9.
Zhang SJ, Ma LY, Huang QH, Li G, Gu BW, Gao XD, et al. Gain-of-function mutation of GATA-2 in acute myeloid transformation of chronic myeloid leukemia. Proc Natl Acad Sci U S A. 2008;105(6):2076–81.
Hehlmann R. How I treat CML blast crisis. Blood. 2012;120(4):737–47.
Sill H, Goldman JM, Cross NC. Homozygous deletions of the p16 tumor-suppressor gene are associated with lymphoid transformation of chronic myeloid leukemia. Blood. 1995;85(8):2013–6.
Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453(7191):110–4.
Yin Y, Li J, Yan W, Cheng Z, Sun N, Zhang G. CEBPA mutation in a case of chronic myeloid leukemia presenting in myeloid blast crisis. Leuk Lymphoma. 2017;58(3):708–10.
Dash AB, Williams IR, Kutok JL, Tomasson MH, Anastasiadou E, Lindahl K, et al. A murine model of CML blast crisis induced by cooperation between BCR/ABL and NUP98/HOXA9. Proc Natl Acad Sci U S A. 2002;99(11):7622–7.
Nucifora G, Birn DJ, Espinosa R 3rd, Erickson P, LeBeau M, Roulston D, et al. Involvement of the AML1 gene in the t(3;21) in therapy-related leukemia and in chronic myeloid leukemia in blast crisis. Blood. 1993;81(10):2728–34.
Di Giacomo D, et al. Blast crisis Ph+ chronic myeloid leukemia with NUP98/HOXA13 up-regulating MSI2. Mol Cytogenet. 2014;7:42.
Shteper PJ, Ben-Yehuda D. Molecular evolution of chronic myeloid leukaemia. Semin Cancer Biol. 2001;11(4):313–23.
Heller G, Topakian T, Altenberger C, Cerny-Reiterer S, Herndlhofer S, Ziegler B, et al. Next-generation sequencing identifies major DNA methylation changes during progression of Ph+ chronic myeloid leukemia. Leukemia. 2016;30(9):1861–8.
Jelinek J, Gharibyan V, Estecio MRH, Kondo K, He R, Chung W, et al. Aberrant DNA methylation is associated with disease progression, resistance to imatinib and shortened survival in chronic myelogenous leukemia. PLoS One. 2011;6(7):e22110.
Issa JP, Kantarjian H, Mohan A, O'Brien S, Cortes J, Pierce S, et al. Methylation of the ABL1 promoter in chronic myelogenous leukemia: lack of prognostic significance. Blood. 1999;93(6):2075–80.
Kim T, Tyndel MS, Kim HJ, Ahn JS, Choi SH, Park HJ, et al. Spectrum of somatic mutation dynamics in chronic myeloid leukemia following tyrosine kinase inhibitor therapy. Blood. 2017;129(1):38–47.
Terre C, et al. Report of 34 patients with clonal chromosomal abnormalities in Philadelphia-negative cells during imatinib treatment of Philadelphia-positive chronic myeloid leukemia. Leukemia. 2004;18(8):1340–6.
O'Dwyer ME, Gatter KM, Loriaux M, Druker BJ, Olson SB, Magenis RE, et al. Demonstration of Philadelphia chromosome negative abnormal clones in patients with chronic myelogenous leukemia during major cytogenetic responses induced by imatinib mesylate. Leukemia. 2003;17(3):481–7.
Jabbour E, Kantarjian HM, Abruzzo LV, O'Brien S, Garcia-Manero G, Verstovsek S, et al. Chromosomal abnormalities in Philadelphia chromosome negative metaphases appearing during imatinib mesylate therapy in patients with newly diagnosed chronic myeloid leukemia in chronic phase. Blood. 2007;110(8):2991–5.
Medina J, Kantarjian H, Talpaz M, O'Brien S, Garcia-Manero G, Giles F, et al. Chromosomal abnormalities in Philadelphia chromosome-negative metaphases appearing during imatinib mesylate therapy in patients with Philadelphia chromosome-positive chronic myelogenous leukemia in chronic phase. Cancer. 2003;98(9):1905–11.
Deininger MW, Cortes J, Paquette R, Park B, Hochhaus A, Baccarani M, et al. The prognosis for patients with chronic myeloid leukemia who have clonal cytogenetic abnormalities in Philadelphia chromosome-negative cells. Cancer. 2007;110(7):1509–19.
Kovitz C, Kantarjian H, Garcia-Manero G, Abruzzo LV, Cortes J. Myelodysplastic syndromes and acute leukemia developing after imatinib mesylate therapy for chronic myeloid leukemia. Blood. 2006;108(8):2811–3.
Groves MJ, Sales M, Baker L, Griffiths M, Pratt N, Tauro S. Factors influencing a second myeloid malignancy in patients with Philadelphia-negative −7 or del(7q) clones during tyrosine kinase inhibitor therapy for chronic myeloid leukemia. Cancer Genet. 2011;204(1):39–44.
• Kurt H, Zheng L, Kantarjian HM, Tang G, Ravandi-Kashani F, Garcia-Manero G, et al. Secondary Philadelphia chromosome acquired during therapy of acute leukemia and myelodysplastic syndrome. Mod Pathol. 2018. Comprehensive study of emergence of secondary Ph and its significance.;31:1141–54.
Chen Z, Wang W, Verstovsek S, Cortes JE, Medeiros LJ, Hu S. Chronic myelogenous leukemia in patients with MPL or JAK2 mutation-positive myeloproliferative neoplasm. Int J Lab Hematol. 2015;37(6):e150–2.
Soderquist CR, Ewalt MD, Czuchlewski DR, Geyer JT, Rogers HJ, Hsi ED, et al. Myeloproliferative neoplasms with concurrent BCR-ABL1 translocation and JAK2 V617F mutation: a multi-institutional study from the bone marrow pathology group. Mod Pathol. 2018;31(5):690–704.
Pingali SR, et al. Emergence of chronic myelogenous leukemia from a background of myeloproliferative disorder: JAK2V617F as a potential risk factor for BCR-ABL translocation. Clin Lymphoma Myeloma. 2009;9(5):E25–9.
Mirza I, Frantz C, Clarke G, Voth AJ, Turner R. Transformation of polycythemia vera to chronic myelogenous leukemia. Arch Pathol Lab Med. 2007;131(11):1719–24.
Shimizu H, Yokohama A, Hatsumi N, Takada S, Handa H, Sakura T, et al. Philadelphia chromosome-positive mixed phenotype acute leukemia in the imatinib era. Eur J Haematol. 2014;93(4):297–301.
de Franca Azevedo I, et al. Frequency of p190 and p210 BCR-ABL rearrangements and survival in Brazilian adult patients with acute lymphoblastic leukemia. Rev Bras Hematol Hemoter. 2014;36(5):351–5.
Jaso J, Thomas DA, Cunningham K, Jorgensen JL, Kantarjian HM, Medeiros LJ, et al. Prognostic significance of immunophenotypic and karyotypic features of Philadelphia positive B-lymphoblastic leukemia in the era of tyrosine kinase inhibitors. Cancer. 2011;117(17):4009–17.
Verrma SP, Dutta TK, Vinod KV, Dubashi B, Ariga KK. Philadelphia chromosome positive pre-T cell acute lymphoblastic leukemia: a rare case report and short review. Indian J Hematol Blood Transfus. 2014;30(Suppl 1):177–9.
Soupir CP, Vergilio JA, Cin PD, Muzikansky A, Kantarjian H, Jones D, et al. Philadelphia chromosome-positive acute myeloid leukemia: a rare aggressive leukemia with clinicopathologic features distinct from chronic myeloid leukemia in myeloid blast crisis. Am J Clin Pathol. 2007;127(4):642–50.
Konopleva M, et al. Molecular biology and cytogenetics of chronic myeloid leukemia. In: P.H. Wiemik, J.P. Dutcher, and M.A. Gertz, eds. Neoplastic Disease of the Blood. Springer. 2018:29-47.
Reboursiere E, Chantepie S, Gac AC, Reman O. Rare but authentic Philadelphia-positive acute myeloblastic leukemia: two case reports and a literature review of characteristics, treatment and outcome. Hematol Oncol Stem Cell Ther. 2015;8(1):28–33.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Molecular Testing and Diagnostics
Rights and permissions
About this article
Cite this article
Zhou, T., Medeiros, L.J. & Hu, S. Chronic Myeloid Leukemia: Beyond BCR-ABL1. Curr Hematol Malig Rep 13, 435–445 (2018). https://doi.org/10.1007/s11899-018-0474-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11899-018-0474-6