Abstract
Purpose of Review
Following cardiac injury, the heart has limited ability to regenerate leading to decreased efficiency and function. Cardiac reprogramming offers a promising treatment to ameliorate the damage caused by ischemia through conversion of cardiac fibroblasts to induced cardiomyocytes (iCMs). Here, we aim to highlight the recent advancements of the last 5 years by discussing the various aspects of cardiac reprogramming including characterization of the cardiac fibroblast, the endogenous environment of the heart, the molecular mechanisms during reprogramming, the epigenetic landscape, and the mechanics of delivering reprogramming factors.
Recent Findings
Due to generally low efficiency of direct cardiac reprogramming, many researchers have continued to improve the efficiency of iCM induction and continued exploration of the basic science behind the technique. The field is continuing to optimize individual aspects of reprogramming that can be leveraged together to improve overall effectiveness.
Summary
Over the last several years, knowledge regarding the process of direct cardiac reprogramming and the many factors that affect its efficiency has increased significantly. Individual aspects have continued to be optimized, and it will be essential going forward to synthesize this information. Cardiac reprogramming continues to advance towards clinical translatability.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
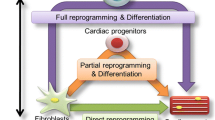
Cardiovascular disease is the leading cause of death worldwide [1]. Ischemic events, such as myocardial infarction (MI), are significant contributors to this statistic due to resultant cardiomyocyte death. Following an MI, the myocardium undergoes significant remodeling in an attempt to maintain the integrity and functionality of the heart through the injury [2]. After the initial inflammatory response, cardiac fibroblasts activate and proliferate to produce extracellular matrix proteins to form a collagen scar [2]. While initially protective, the fibrotic scar results in decreased efficiency and function of the heart. Unfortunately, cardiomyocytes, the primary workforce of the heart, retain minimal regenerative properties into adulthood leaving them unable to repair damage or recover from the injury [3]. The ability to capitalize on the abundance of other cell types within the heart to replenish cardiomyocytes offers a promising treatment for cardiac disease and has led to the growing field of direct cardiac reprogramming.
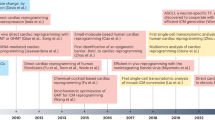
In 2010, Takahashi and Yamanaka et al. demonstrated the ability to induce pluripotency through the expression of four factors, Oct 3/4, Sox2, c-Myc, and Klf4, within adult and embryonic fibroblasts [4]. Attaining a pluripotent state then permits differentiation into a different cell type of interest, such as cardiomyocytes [5]. While very promising, this technique also has several limitations including the risk of tumorigenesis, the heterogeneity of the resulting tissue, and the immaturity of the cardiomyocytes. This approach was further optimized in the method of direct cardiac reprogramming which allows for the conversion of cardiac fibroblasts directly to induced cardiomyocytes (iCMs) in vitro and in vivo through the ectopic expression of transcription factors, such as MEF2C, GATA4, and TBX5 (MGT), without returning to a progenitor state [6,7,8]. These techniques have significant potential for improving cardiac function and restoring non-functional heart tissue, but they still require significant optimization due to low efficiency and incomplete resemblance of iCMs to endogenous cardiomyocytes [6,7,8]. Soon after, many began to explore ways to improve reprogramming through testing new combinations of factors or microRNAs (miRNAs) [9,10,11,12,13,14,15,16,17]. While the field has continued to explore different reprogramming cocktails, it has additionally expanded through the characterization of other aspects of reprogramming such as investigation of the cardiac fibroblast, the endogenous environment of the heart, the molecular mechanisms behind reprogramming, the epigenetic landscape, and the delivery method of reprogramming factors which have all resulted in a greater understanding of direct cardiac reprogramming and improved efficiency of conversion. This review will highlight the last 5 years of advancements in direct cardiac reprogramming.
Characterization of Cardiac Fibroblasts
The heart is a complex tissue made of many different cell types including cardiomyocytes, fibroblasts, endothelial cells, and immune cells [18]. The percentage of cardiomyocytes can vary from 30 to 50% depending on the region of the heart, while the remaining cells are non-myocytes [18]. Cardiac fibroblasts are targeted for reprogramming due to their abundance in the heart and for their increased role during and after injury [6]. Following an MI, fibroblasts transdifferentiate into myofibroblasts, their activated form, which synthesize and deposit extracellular matrix and collagen to provide contractile support to the damaged heart tissue [19]. While this role is initially essential for the maintenance of heart function, the scar tissue is unable to resolve and can become pathologic resulting in decreased heart function. An overview of this process is reviewed here [20].
Fibroblasts have been utilized as the target for reprogramming since the initial development of the technique [6]. At that time, it was established that fibroblasts do not return to a progenitor state while transitioning to an induced cardiomyocyte, but other information regarding how fibroblasts are affected throughout the process remained unexplored. In vitro and in vivo fibroblasts are constantly progressing to myofibroblasts due to environmental and mechanical stress [21]. Despite this predisposition, lineage tracing has shown that iCMs are derived directly from fibroblasts and do not pass through an activated myofibroblast state [21]. iCMs undergo a change in the actin isoforms. Isoforms associated with a cardiomyocyte gene program are induced, while those related to a myofibroblast state are downregulated [21]. Cells undergoing reprogramming must overcome the previously formed structure of the fibroblast that acts as a scaffold for fibroblast activation into myofibroblasts [21]. Altering this gene program could be a significant barrier particularly within in vitro experiments where fibroblasts are transdifferentiated by the mechanical stress and tension applied by a cell culture dish. The necessity of altering the actin isoforms may explain the low efficiency of reprogramming and expounds on the limitations of reprogramming in a dish. Within the heart, each cell type has been further characterized using single-cell transcriptomics which revealed heterogeneity and led to the identification of several subpopulations by their differential gene expression [18, 22•]. Three subpopulations of cardiac fibroblasts were identified by their transcriptome, each with a unique functional state including cellular response, immune response, and cytoskeleton organization [22•]. Based on their distinct morphologies, transcriptomes, and preliminary experiments, it is believed that each subpopulation is responsible for executing different functions [22•]. Developing an understanding of how each of these populations contribute and function during tissue maintenance and scar formation could offer significant insight into the process of fibrosis in the heart. The continued characterization and understanding of fibroblasts will optimize their use and targeting for cardiac reprogramming.
Exploring the Endogenous Environment of the Heart
It has previously been observed that reprogramming is more biologically successful in vivo than it is in vitro likely due to contributions from the environment [7]. A two-dimensional (2D) culture does not accurately recapitulate the in vivo process. As previously mentioned, plating fibroblasts in a cell culture dish can lead fibroblasts to transdifferentiate, likely impeding their ability to be reprogrammed. Exploration of the mechanical stress placed on fibroblasts by their environment was explored by testing varying matrix stiffness [23]. Utilizing a soft matrix that is comparable to the myocardium not only results in a higher number of cardiac troponin T positive cells, but also additionally appears to promote a high level of maturation, as demonstrated by increased beating of the iCMs [23]. Yes-associated protein (YAP) and transcriptional coactivator with PDZ-binding domain (TAZ) signaling, known for its role in sensing the cellular environment, are suppressed which results in decreased fibroblast gene program contributing to enhanced reprogramming [23]. To continue to explore the role of the environment, a cylindrical three-dimensional (3D) hydrogel was developed and used to encapsulate fibroblasts for reprogramming targeting [24]. Utilizing the previously proven miRNA cocktail, the efficiency of reprogramming was increased in the 3D model [24]. This model led to the increased expression of several matrix metalloproteinase which have previously been shown to play a role in cardiac differentiation and were proven to be responsible for the increased efficiency [24]. These studies clearly suggested that the environment is essential for successful in vitro reprogramming, but still lacked an understanding of the local environment and stimuli within the heart. With the knowledge that typical culture medium lacks many of the signals present within the heart, several variations of cardiogenic compounds were tested [25]. The screen indicated that the addition of fibroblast growth factor 2 and 10 and vascular endothelial growth factor to the medium activated several transcriptional regulators and converted partially reprogrammed cells to functional iCMs through the p38MAPK and PI3K/AKT pathways and likely others [25]. AKT has separately been identified in a screen of protein kinases to accelerate reprogramming [11]. To further explore the environment within the heart, an in vitro biomatrix was developed utilizing human adult cardiac fibroblasts [26•]. After decellularization, it contained typical components of the cardiac basement membrane such as fibronectin, laminin, and collagen IV [26•]. They utilized the biomatrix within 3D hydrogels, leading to an even more improved reprogramming efficiency [26•]. Continuing to explore the differences between in vitro and in vivo reprogramming offers information to improve the in vitro model, but more importantly begins to elucidate previously unexplored contributors to cardiac reprogramming in vivo.
The Underlying Molecular Mechanisms of iCM Reprogramming
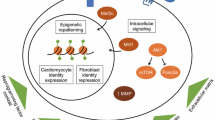
Direct cardiac reprogramming relies heavily upon transcription factors and their downstream effects. As researchers have continued to explore the variety of factors that can improve efficiency, many have additionally identified the signaling pathways and how they may be contributing at the molecular level. Shortly after the exploration of reprogramming began, several pathways immediately came to attention including Akt1/protein kinase B, TGFβ, and Wnt [11, 27, 28]. In contrast, the role of Notch signaling in cardiac cell differentiation and heart development has long been debated with some showing evidence that it is essential, while others show it is inhibitory to cardiac cell fate [29,30,31,32]. DAPT treatment, causing inhibition of Notch signaling, has been shown to increase efficiency of reprogramming by enhancing MEF2C binding [33]. Additionally, TLR3, which had previously been shown to play a role in iPS and endothelial cell reprogramming, was determined to significantly contribute to cardiomyocyte maturation through activation of NFκB [34]. It can cause changes in the expression of various epigenetic modifiers that improves DNA accessibility for reprogramming [34]. This information was leveraged to identify natural RNA sensing receptor ligands and to develop synthetic RNA oligonucleotides that are capable of similarly enhancing cardiomyocyte maturation [35]. Many previous studies have performed factor screening and exploration, but always within healthy cardiac fibroblasts. A more recent a study utilized cardiac fibroblasts post-MI as they may act as a better model for the starting cell should reprogramming be utilized translationally for cardiovascular disease treatment [36]. Through a screen of 22 candidate transcription factors, selected for their role in heart development, they found that MGT with two additional transcription factors, Sall4 and Myocd, induced more iCMs [36]. They further characterized the role of these genes and found that Sall4 plays a role in promoting the functional, beating properties of the cardiomyocyte, while Myocd contributes to expression of cardiac sarcomeric proteins [36]. While these studies have identified factors to activate cardiomyocyte programming, surprisingly autophagy is also activated during reprogramming, likely to assist in disposing of fibroblast structures and cell content [37]. Despite autophagy’s significant role, the knockdown of Beclin1, a known regulator of autophagy, actually increases the efficiency of reprogramming by activating the Wnt/β-catenin pathway through Lef1 and the PI3K complex [37]. While the continued identification of additional factors and other contributing gene programs to improve reprogramming remains of importance, there is significant value in further understanding the function of transcription factors during the process of reprogramming. By understanding how transcription factors act the field may increasingly be able to incorporate the information to optimize and simplify the reprogramming cocktail to improve chances of translational use.
The Epigenetic Landscape of iCM Reprogramming
Epigenetics plays a major role in normal cell fate determination and cellular reprogramming due to its direct effects on gene expression. Early on it was determined that iCMs gain a chromatin status similar to endogenous cardiomyocytes in at least some cardiac specific genes, even more so than cardiomyocytes derived from induced pluripotent stem cells [6, 38]. During direct cardiac reprogramming, the cells must first undergo a rapid activation of the cardiac loci through reduced H3K27me3 and increased H3K4me3, meanwhile the opposite occurs in fibroblast loci leading to slow suppression of the fibroblast pathways [39]. Additionally, upon investigating the promoters of several cardiac genes, it was found that certain CpGs within these loci played a significant role in total demethylation and therefore transcriptional activation [39]. To further understand the role that epigenetics plays in reprogramming, a screen was performed searching for epigenetic regulators. Bmi1 was identified to have an inhibitory effect on several genes related to cell proliferation and also plays a role in repressing endogenous Gata4 expression [40]. By knocking down Bmi1 and removing these functions, reprogramming was enhanced [40]. From the screen, several RNA splicing factors and several genes were identified that were also inhibitory to reprogramming [41]. Outside of understanding the role of epigenetics, there was also interest in understanding how to manipulate epigenetics to benefit reprogramming. miRNAs have been utilized to alter chromatin status. miR-1, miR-133, miR-208, and miR-499 are able to alter histone methyltransferases and demethylases to cause reduction of H3K27me3 [42]. More recently, it was also determined that the expression of Mef2c and Tbx5 of the original reprogramming cocktail, partially function through promoting chromatin remodeling to an accessible state [43]. Through the utilization of a screen for epigenetic modifiers, the histone reader PHF7 was identified and proven to be a strong activator of cardiac reprogramming [44]. Previously it had been found that it binds directly to several different histone marks, but it also cooperates with the SWI/SNF complex at cardiac gene super enhancers to increase chromatin accessibility and transcription factor binding [44]. Epigenetics has additionally been explored and compared in cardiac, hepatic and neuronal direct reprogramming, which identified Ascl1’s unexpected potential for cross lineage reprogramming [45••]. When combined with MEF2C, they act cooperatively to alter epigenetics and transcriptional activity [45••]. Optimizing chromatin accessibility and manipulation of epigenetics will continue to play a vital role in improving the efficiency of reprogramming.
The Mechanics of Delivering Reprogramming Factors
Delivering reprogramming factors to fibroblasts in a safe and effective manner continues to be a significant barrier for future therapeutic use. Traditionally, reprogramming factors have been delivered through a viral construct, but despite high transfection efficiencies, the rate of reprogramming remains fairly low [46]. Additionally, the use of viral vectors can come with a safety concern for use in clinic due to their ability to integrate into DNA [46]. This has led to a recent expansion in research focused on safer, more efficient delivery methods. Rather than the typical lentivirus or retrovirus, Sendai virus has been successfully utilized as a non-integrating, non-pathogenic vector to induce both in vitro and in vivo direct reprogramming while also preventing worsening of cardiac function [47]. Not only was this new vector promising due to its lack of integration, but it also led to increased efficiency generating 100-fold more beating iCMs and a shorter time to induce beating cells from mouse fibroblasts [47]. In the last several years, researchers have sought a non-viral option for reprogramming. The first evidence of an alternative method was the use of a modified mRNA gene delivery platform that results in prompt gene expression for only 10 days [48]. The ability to transiently express genes following heart injury while limiting the extent to which gene expression is altered offers a unique opportunity for a translatable, short-term treatment. Unfortunately, the CM-like cells were not functional [48]. In the last year, lipoplexes were utilized to deliver miRNAs to adult human cardiac fibroblasts in vitro offering a new, safer way of delivering a reprogramming cocktail [49]. Alternatively, reprogramming without transgenes could offer a work-around to many of the delivery barriers. Extracellular vesicles were isolated from embryonic stem cells that were undergoing cardiac differentiation and showed their ability to convert mouse embryonic fibroblasts to cardiomyocytes with 60% efficiency [50]. Many of these techniques unfortunately require intra-myocardial injection that in itself can cause secondary injury, but alternatively, in 2021, systemic administration became possible through the use of nanoparticles [51••]. The nanoparticle is composed of a hybrid membrane of modified lipid membranes fused with neutrophil membrane proteins surrounding a silica nanoparticle core that could carry miRNAs [51••]. This membrane allows the nanoparticle to move more freely and would allow them to enter the injured heart following an MI in a similar manner as immune cells travel throughout the tissue [51••]. The recent advancements in the delivery of reprogramming factors or alternative methods have significantly increased the feasibility of clinical translation and continue to offer new solutions and answers to many of the challenges that face direct cardiac reprogramming.
Conclusion
The ability to revitalize cardiac tissue following injury through direct cardiac reprogramming continues to offer a promising and revolutionary way to treat cardiac disease. Since the initial evidence of successful direct cardiac reprogramming, there has been significant progress in understanding the various contributing factors and mechanisms by which a cardiac fibroblast becomes an iCM. Through further characterization of cardiac fibroblasts, an increased understanding of endogenous environment of the heart, a breakdown of the signaling pathways and the epigenetic landscape, and exploration of alternative delivery methods, reprogramming has continued to improve in its efficiency and translatability. It is imperative that the inner workings of reprogramming continue to be elucidated and the information gathered be integrated together to result in a new, effective therapeutic treatment for cardiovascular disease.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Tsao CW, Aday AW, Almarzooq ZI, Alonso A, Beaton AZ, Bittencourt MS, et al. Heart disease and stroke statistics-2022 update: a report from the American Heart Association. Circulation. 2022;145:e153-639.
Sutton MG, Sharpe N. Left ventricular remodeling after myocardial infarction: pathophysiology and therapy. Circulation. 2000;101:2981–8.
Kikuchi K, Poss KD. Cardiac regenerative capacity and mechanisms. Annu Rev Cell Dev Biol. 2012;28:719–41.
Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76.
Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res. 2009;104:e30-41.
Ieda M, Fu J-D, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–86.
Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–8.
Song K, Nam Y-J, Luo X, Qi X, Tan W, Huang GN, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604.
Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, et al. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110:1465–73.
Jayawardena TM, Finch EA, Zhang L, Zhang H, Hodgkinson CP, Pratt RE, et al. MicroRNA induced cardiac reprogramming in vivo: evidence for mature cardiac myocytes and improved cardiac function. Circ Res. 2015;116:418–24.
Zhou H, Dickson ME, Kim MS, Bassel-Duby R, Olson EN. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc Natl Acad Sci USA. 2015;112:11864–9.
Protze S, Khattak S, Poulet C, Lindemann D, Tanaka EM, Ravens U. A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. J Mol Cell Cardiol. 2012;53:323–32.
Hirai H, Katoku-Kikyo N, Keirstead SA, Kikyo N. Accelerated direct reprogramming of fibroblasts into cardiomyocyte-like cells with the MyoD transactivation domain. Cardiovasc Res. 2013;100:105–13.
Christoforou N, Chellappan M, Adler AF, Kirkton RD, Wu T, Addis RC, et al. Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming. PLoS ONE. 2013;8: e63577.
Addis RC, Ifkovits JL, Pinto F, Kellam LD, Esteso P, Rentschler S, et al. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J Mol Cell Cardiol. 2013;60:97–106.
Muraoka N, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Isomi M, et al. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. 2014;33:1565–81.
Nam Y-J, Song K, Luo X, Daniel E, Lambeth K, West K, et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci USA. 2013;110:5588–93.
Litviňuková M, Talavera-López C, Maatz H, Reichart D, Worth CL, Lindberg EL, et al. Cells of the adult human heart. Nature. 2020;588:466–72.
Petrov VV, Fagard RH, Lijnen PJ. Stimulation of collagen production by transforming growth factor-beta1 during differentiation of cardiac fibroblasts to myofibroblasts. Hypertension. 2002;39:258–63.
Daseke MJ, Tenkorang MAA, Chalise U, Konfrst SR, Lindsey ML. Cardiac fibroblast activation during myocardial infarction wound healing: Fibroblast polarization after MI. Matrix Biol. 2020;91–92:109–16.
Zhang Z, Zhang W, Blakes R, Sundby LJ, Shi Z, Rockey DC, et al. Fibroblast fate determination during cardiac reprogramming by remodeling of actin filaments. Stem Cell Reports. 2022;17:1604–19.
• Wang L, Yang Y, Ma H, Xie Y, Xu J, Near D, et al. Single-cell dual-omics reveals the transcriptomic and epigenomic diversity of cardiac non-myocytes. Cardiovasc Res. 2022;118:1548–63. This study emphasizes the heterogeneity of heart tissues and defines the complexity of various cell types within the heart.
Kurotsu S, Sadahiro T, Fujita R, Tani H, Yamakawa H, Tamura F, et al. Soft matrix promotes cardiac reprogramming via inhibition of YAP/TAZ and suppression of fibroblast signatures. Stem Cell Reports. 2020;15:612–28.
Li Y, Dal-Pra S, Mirotsou M, Jayawardena TM, Hodgkinson CP, Bursac N, et al. Tissue-engineered 3-dimensional (3D) microenvironment enhances the direct reprogramming of fibroblasts into cardiomyocytes by microRNAs. Sci Rep. 2016;6:38815.
Yamakawa H, Muraoka N, Miyamoto K, Sadahiro T, Isomi M, Haginiwa S, et al. Fibroblast growth factors and vascular endothelial growth factor promote cardiac reprogramming under defined conditions. Stem Cell Reports. 2015;5:1128–42.
• Paoletti C, Marcello E, Melis ML, Divieto C, Nurzynska D, Chiono V, et al. Cardiac tissue-like 3D microenvironment enhances route towards human fibroblast direct reprogramming into induced cardiomyocytes by microRNAs. Cells. 2022;11. This study emphasizes the importance of the heart environment to cell fate and integrates information from many of the previous structural studies to improve cardiac reprogramming.
Ifkovits JL, Addis RC, Epstein JA, Gearhart JD. Inhibition of TGFβ signaling increases direct conversion of fibroblasts to induced cardiomyocytes. PLoS ONE. 2014;9: e89678.
Lalit PA, Salick MR, Nelson DO, Squirrell JM, Shafer CM, Patel NG, et al. Lineage reprogramming of fibroblasts into proliferative induced cardiac progenitor cells by defined factors. Cell Stem Cell. 2016;18:354–67.
Jang J, Ku SY, Kim JE, Choi K, Kim YY, Kim HS, et al. Notch inhibition promotes human embryonic stem cell-derived cardiac mesoderm differentiation. Stem Cells. 2008;26:2782–90.
Chen VC, Stull R, Joo D, Cheng X, Keller G. Notch signaling respecifies the hemangioblast to a cardiac fate. Nat Biotechnol. 2008;26:1169–78.
Li H, Yu B, Zhang Y, Pan Z, Xu W, Li H. Jagged1 protein enhances the differentiation of mesenchymal stem cells into cardiomyocytes. Biochem Biophys Res Commun. 2006;341:320–5.
Koyanagi M, Bushoven P, Iwasaki M, Urbich C, Zeiher AM, Dimmeler S. Notch signaling contributes to the expression of cardiac markers in human circulating progenitor cells. Circ Res. 2007;101:1139–45.
Abad M, Hashimoto H, Zhou H, Morales MG, Chen B, Bassel-Duby R, et al. Notch inhibition enhances cardiac reprogramming by increasing MEF2C transcriptional activity. Stem Cell Reports. 2017;8:548–60.
Hodgkinson CP, Pratt RE, Kirste I, Dal-Pra S, Cooke JP, Dzau VJ. Cardiomyocyte maturation requires TLR3 activated nuclear factor kappa B. Stem Cells. 2018;36:1198–209.
Hu J, Hodgkinson CP, Pratt RE, Lee J, Sullenger BA, Dzau VJ. Enhancing cardiac reprogramming via synthetic RNA oligonucleotides. Mol Ther Nucleic Acids. 2021;23:55–62.
Zhao H, Zhang Y, Xu X, Sun Q, Yang C, Wang H, et al. Sall4 and myocd empower direct cardiac reprogramming from adult cardiac fibroblasts after injury. Front Cell Dev Biol. 2021;9: 608367.
Wang L, Ma H, Huang P, Xie Y, Near D, Wang H, et al. Down-regulation of Beclin1 promotes direct cardiac reprogramming. Sci Transl Med. 2020;12.
Zhou Y, Wang L, Liu Z, Alimohamadi S, Yin C, Liu J, et al. Comparative gene expression analyses reveal distinct molecular signatures between differentially reprogrammed cardiomyocytes. Cell Rep. 2017;20:3014–24.
Liu Z, Chen O, Zheng M, Wang L, Zhou Y, Yin C, et al. Re-patterning of H3K27me3, H3K4me3 and DNA methylation during fibroblast conversion into induced cardiomyocytes. Stem Cell Res. 2016;16:507–18.
Zhou Y, Wang L, Vaseghi HR, Liu Z, Lu R, Alimohamadi S, et al. Bmi1 is a key epigenetic barrier to direct cardiac reprogramming. Cell Stem Cell. 2016;18:382–95.
Zhou Y, Alimohamadi S, Wang L, Liu Z, Wall JB, Yin C, et al. A loss of function screen of epigenetic modifiers and splicing factors during early stage of cardiac reprogramming. Stem Cells Int. 2018;2018:3814747.
Dal-Pra S, Hodgkinson CP, Mirotsou M, Kirste I, Dzau VJ. Demethylation of H3K27 is essential for the induction of direct cardiac reprogramming by mir combo. Circ Res. 2017;120:1403–13.
Stone NR, Gifford CA, Thomas R, Pratt KJB, Samse-Knapp K, Mohamed TMA, et al. Context-specific transcription factor functions regulate epigenomic and transcriptional dynamics during cardiac reprogramming. Cell Stem Cell. 2019;25:87-102.e9.
Garry GA, Bezprozvannaya S, Chen K, Zhou H, Hashimoto H, Morales MG, et al. The histone reader PHF7 cooperates with the SWI/SNF complex at cardiac super enhancers to promote direct reprogramming. Nat Cell Biol. 2021;23:467–75.
•• Wang H, Keepers B, Qian Y, Xie Y, Colon M, Liu J, et al. Cross-lineage potential of Ascl1 uncovered by comparing diverse reprogramming regulatomes. Cell Stem Cell. 2022;29:1491-1504.e9. This paper was the first to study the epigenetic regulatomes across three types of lineage reprogramming, with the unexpected discovery of Ascl1’s cross lineage potential.
Sadahiro T, Yamanaka S, Ieda M. Direct cardiac reprogramming: progress and challenges in basic biology and clinical applications. Circ Res. 2015;116:1378–91.
Miyamoto K, Akiyama M, Tamura F, Isomi M, Yamakawa H, Sadahiro T, et al. Direct in vivo reprogramming with sendai virus vectors improves cardiac function after myocardial infarction. Cell Stem Cell. 2018;22:91-103.e5.
Kaur K, Hadas Y, Kurian AA, Żak MM, Yoo J, Mahmood A, et al. Direct reprogramming induces vascular regeneration post muscle ischemic injury. Mol Ther. 2021;29:3042–58.
Nicoletti L, Paoletti C, Tarricone G, Andreana I, Stella B, Arpicco S, et al. Lipoplexes for effective in vitro delivery of microRNAs to adult human cardiac fibroblasts for perspective direct cardiac cell reprogramming. Nanomedicine. 2022;45: 102589.
Kim H, Song B-W, Park S-J, Choi SW, Moon H, Hwang K-C, et al. Ultraefficient extracellular vesicle-guided direct reprogramming of fibroblasts into functional cardiomyocytes. Sci Adv. 2022;8:eabj6621.
•• Wang Q, Song Y, Chen J, Li Q, Gao J, Tan H, et al. Direct in vivo reprogramming with non-viral sequential targeting nanoparticles promotes cardiac regeneration. Biomaterials. 2021;276:121028. This study was the first to identify a direct reprogramming technique that would allow for systemic administration instead of an intramyocardial injection.
Funding
PT is supported by UNC Integrative Vascular Biology Training Grant (T32HL069768). LQ is funded by the American Heart Association (AHA20EIA35310348) and the National Institute of Health (NIH/NHLBI R35HL155656).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
There are no conflicts of interest.
Human and Animal Rights and Informed Consent
All reported studies/experiments with human or animal subjects performed by the authors have been previously published and complied with all applicable ethical standards.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Takasugi, P., Qian, L. Exploring the Inner Workings of Direct Cardiac Reprogramming. Curr Cardiol Rep 25, 467–472 (2023). https://doi.org/10.1007/s11886-023-01868-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11886-023-01868-9