Abstract
Purpose of Review
This review aims to summarize and discuss the function and molecular mechanism of miRNA and lncRNA in the heart, focusing on ischemic and non-ischemic cardiomyopathy.
Recent Findings
Extensive studies in the past decades have identified numerous protein-coding genes that are highly expressed in the heart, playing essential roles in the regulation of cardiac gene expression, heart development, and function. Furthermore, mutations in many of these genes have been identified and are linked to cardiovascular disease. Intriguingly, it is now recognized that majority of our genome is “non-coding,” which produces a large amount of non-coding RNAs (ncRNAs), including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs). Emerging evidence has indicated that these classes of non-coding RNAs participate in most (if not all) aspects of cardiac gene expression, cardiomyocyte proliferation, differentiation, and cardiac remodeling in response to stress.
Summary
Recent findings have demonstrated important functions for non-coding RNA in ischemic and non-ischemic cardiomyopathy. It is expected that non-coding RNAs will become promising therapeutic targets for cardiovascular diseases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The myocardium consists of three main components that are integral to each other: cardiomyocytes, extracellular matrix, and the vascular microcirculation that supply nutrient and oxygen to the heart. The heart can adapt to a wide array of extrinsic or genetic factors. Cardiomyopathies are diseases of the myocardium that manifest structural and functional abnormalities that can lead to heart failure (HF). The pathogenesis of HF, in general, can be placed into two categories, i.e., ischemic cardiomyopathy (ICM) and non-ischemic cardiomyopathy (NICM), which was defined based on the presence or absence of ischemic injury.
ICM is defined as left ventricular (LV) systolic dysfunction followed by myocardial infarction events or has more than 75% stenosis occur in the left coronary artery [1]. ICM is one of the most severe consequences of coronary artery disease (CAD), which cause over seven million deaths globally each year [2]. The development of CAD is a chronic process, starting from endothelial dysfunction promoting plaque development, which results in narrowing of the coronary artery [3]. A plaque rupture event or the formation of thrombus on the plaque surface can both lead to myocardial infarction (MI), which is an acute event that can lead to severe tissue damage in the heart [4]. The ischemia-induced cardiomyocyte necrosis can induce a cascade of cellular and intracellular signaling process to initiate the repair process and compensatory remodelings, such as ventricular dilation, hypertrophy, and the formation of scar tissues. The neutrophils recruited into the infarct zone release and activate serine protease and matrix metalloproteinases which result in the degradation of the extracellular matrix surrounding the infarct zone; this leads to an expansion of the infarct area and thinning of the injured myocardium [5]. To preserve the stroke volume of the heart, the adaptive remodeling of hypertrophy follows in the non-infarcted myocardium to compensate for the increased wall stress due to the loss of contractility in the infarcted area. Unfortunately, the adult heart has limited regenerative potential after injury [6, 7], and the repair processes usually involved the removal of dead cardiomyocyte followed by activation of cardiac fibroblast into myofibroblast, and deposit extracellular matrix protein to promote a pro-fibrotic response [8]. The fibrotic tissue reduces the compliance of the chamber wall, which further reduces cardiac function and accelerates the progression of HF [9, 10].
In contrast, NICM refers to the forms of cardiomyopathy independent of the ischemic event. NICM is sometimes inherited; could be secondary to hypertension, valvular disease, or toxins; and is often associated with mechanical or electrical dysfunction. The pathology of NICM usually displays as hypertrophic cardiomyopathy (HCM) with a thickened myocardium, or dilated cardiomyopathy (DCM) with thinning of ventricular wall, restrictive cardiomyopathy (RCM) (in which the ventricular wall lost diastolic function and becomes rigid), and arrhythmogenic right ventricular dysplasia (ARVD) where the right ventricle myocardium is replaced by adipose or fibrous tissue, which usually associates with arrhythmia [11]. Of these, HCM and DCM are the two most prevalent NICMs, and this cardiac remodeling induced by pathological stress leads to progressive declines in cardiac output, increased cardiomyocyte apoptosis, and increased fibrosis [12, 13], which usually results in heart failure.
The investigation of the causes, prevention, and treatment of ICM and NICM has been an active field of research. Despite the improvement of the outcome and survival of patients with cardiomyopathies through strategies such as pharmacological treatment or mechanical support, such therapies are unable to prevent further progression of the disease [14]. Therefore, a more thorough understanding of the underlying mechanism for ICM and NICM is needed.
The World of Non-coding—MicroRNAs (miRNAs) and Long Non-coding RNA (lncRNAs)
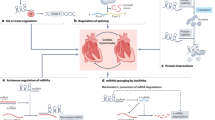
The vast majority of the genome is transcribed, but only a small portion (~ 2%) codes for protein [15], leaving most transcripts non-coding RNAs (ncRNAs). ncRNA can be categorized as transfer RNA (tRNA), ribosomal RNA (rRNA), microRNA (miRNA), small interfering RNA (siRNA), piwi-interacting RNA (piwiRNA), exosomal RNA (exRNA), small nucleolar RNA (snoRNA), small nuclear RNA (snRNA), and long non-coding RNA (lncRNA) [16,17,18,19,20]. The last few decades of studies have demonstrated that ncRNA can perform a variety of biological function, including regulation of gene expression by transcription, RNA processing and translation, and regulation of the epigenome, to form scaffolds and serve as cellular signaling component. Here, we will focus our scope on two categories of ncRNA, i.e., miRNAs and lncRNAs, in the context of heart development, function, and cardiomyopathy (Fig. 1).
Role of miRNAs in Cardiac Function, Remodeling, and Disease
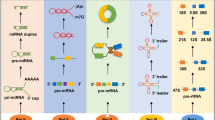
MicroRNAs (miRNAs) are a class of highly conserved non-coding RNAs widely expressed by metazoan eukaryotes [21], which inhibit gene expression through modulating translation, mRNA stability, and mRNA sequestration [22,23,24]. The primary miRNAs (pri-miRNAs) are initially produced in the nucleus (transcription step). The RNA hairpins of pri-miRNAs will then be processed by Drosha and Pasha complex, which cleaved into an imperfect stem–loop-structured miRNA precursors (pre-miRNAs). Pre-miRNAs will then be exported to the cytoplasm and further processed by Dicer and its associated double-stranded RNA-binding proteins TRBP and PACT into miRNA duplex. The unwound miRNA duplex became the mature miRNA, loaded onto argonaute2 (AGO2) proteins and their associated co-factors to form the RNA-induced silencing complex (RISC). The RISC assembly carries the single-stranded miRNA to bind complementarily to the 3′UTR of the transcribed mRNA to facilitate either mRNA cleavage or translational repression [23, 24].
miRNAs can be classified based on their genomic location and gene structure into three categories: intronic, intergenic, and exonic [25]. Over 2000 miRNAs have been found in human. Majority of the known miRNA exist as a single gene or as a cluster of genes located in the intergenic region, and their expression is controlled by their own promoters [26, 27]. Exonic miRNAs are located in regions that overlap with an exon of a gene [28]. Intronic miRNAs are located within the intron of a gene, which is usually transcribed with their host gene or regulated by their own promoter.
The emerging field of miRNA biology has revealed essential roles for these regulatory ncRNAs in a wide variety of biological processes, including heart development and cardiovascular disease [29,30,31]. Several landmark studies have established the crucial role of miRNA in cardiac development and cardiac function in the adult heart. Dicer is a critical component in miRNA biogenesis; cardiac-specific deletion of Dicer using Nkx2.5-Cre leads to a malformed ventricular myocardium and cardiac outflow tract, and septal defects [32]. Deletion of Dicer in the heart in later development with αMHC-Cre results in severe DCM and HF without septation defect and mutant mice die within 4 days after birth [33]. Chen et al. also reported that Dicer expression is decreased in human end-stage DCM and failing heart and its expression is increased after installation of left ventricular assist device (LVAD) to improve heart function. To further investigate the role of miRNA in adult heart, a tamoxifen-inducible αMHC-MerCreMer was utilized to facilitate cardiac-specific deletion of Dicer in adult mice [34]. Deletion of Dicer in 3-week old mice results in premature death within 1 week. Dicer deletion in 8-week-old mice leads to HCM, fibrosis, and reduced contractility. Overall, these studies demonstrated a critical role of miRNAs in both normal and pathological cardiac function.
The Expression and Function of miRNAs During Heart Development and Cardiac Gene Expression
The role of individual miRNA in cardiac development has been actively investigated. For example, two muscle-specific miRNAs, i.e., miR-1 and miR-133, promote mesoderm differentiation while repressing the endoderm and ectoderm differentiation and are critical for skeletal and cardiac muscle lineage specification and development [35,36,37,38]. miR-1 is a direct transcriptional target of myogenic transcription factors SRF, MyoD, and MEF2 while repressing the expression of transcription factors HAND2 and MYOCD; therefore miR-1 acts as a modulator for cardiac lineage commitment and morphogenesis [36, 37]. The expression of miR-133 is also regulated by SRF and MEF2, and the double deletion of miR-133a and miR-133b leads to ventricular septal defect with about 50% penetrance, and the mutant that survived eventually developed DCM. The loss of both miR-133a and miR-133b results in ectopic expression of smooth muscle genes as a result of increased SRF expression, thereby forming a negative-feedback myogenic transcriptional circuit [38]. Loss of both miR-133a and miR-133b also results in an increased cardiomyocyte proliferation, which can also attribute to the miR-133 inhibition of SRF and cell cycle genes such as CCND2 [38]. Together, these studies demonstrated an essential role for the miR-1/133 cluster in myogenic differentiation and cardiac development.
MiR-138 is expressed in the zebrafish heart and is required for the establishment of appropriate chamber-specific genes during development [39]. Morton et al. demonstrated that when the function of miR-138 is disrupted, it led to the expansion of atrial-ventricular canal-specific gene expression into the ventricle, resulting in disruption of ventricular cardiomyocyte morphology and function. Knockdown of miR-138 by antagomirs showed that miR-138 function is required during a specific temporal window of zebrafish development, 24–34-h post-fertilization. miR-138 functions by targeting multiple players in the retinoic acid signaling cascade, including Aldh1a2 and Cspg2, to establish appropriate chamber-specific gene pattern during cardiac morphogenesis [39].
miRNAs Are Important Regulators of Cardiac Remodeling
miR-195 is upregulated in hypertrophic human hearts, and overexpression of miR-195 in mouse heart under the control of α-myosin heavy-chain promoter leads to HCM [40]. Overexpression of miR-195 in the heart increases the size of the cardiomyocyte, and the ratio of heart weight to body weight was also dramatically increased in transgenic mice, indicating that miR-195 expression is sufficient to induce hypertrophic growth of cardiomyocytes. miR-195 transgenic mice eventually progressed to DCM by 6 weeks of age [40]. This suggests that the expression of miR-195 is sufficient to induce hypertrophic signaling and led to HF. It would be interesting to test whether the loss of function of miR-195 can restore cardiac function in DCM or HF models.
In mammalian system, cardiomyocyte contraction function is regulated by two myosin heavy-chain proteins, i.e., Myh6/αMHC and Myh7/βMHC. The fast-twitch Myh6 is expressed predominantly in the adult mouse heart, and the slow-twitch Myh7 is expressed predominantly in the embryonic mouse heart [41]. In the human heart, Myh7 expression continues to adulthood, but the level of Myh7 is similarly induced in hypertrophic human heart compared to hypertrophic mouse heart [42].
The miR-208/miR-499 superfamily has been extensively studied in the heart and found to be positioned in the interphase of regulation of this myosin heavy-chain protein [43, 44]. miR-208a, miR-208b, and miR-499 are respectively encoded within the introns of Myh6, Myh7, and Myh7b [43, 44]. The expression pattern of these intronic microRNAs is closely associated with that of their host genes. miR-208a overexpression specifically in the heart is sufficient to induce HCM and arrhythmia in mice, and the targeted deletion of miR-208a leads to a defective cardiac conduction system and misexpression of connexin 40 [43]. Genetic deletion of miR-208a also results in the expression of fast skeletal muscle genes such as TNNT3 and TNNL2 in the heart [45]. Moreover, deletion of miR-208a abolished the expression of miR-499 and Myh7b in the postnatal heart and reduced the expression of miR-208b under propylthiouracil inhibition of T3 biogenesis. Overexpression of miR-499 in the heart of miR-208a null mice was sufficient to restore the level of fast skeletal muscle genes TNNT3 and TNNL2 to wild-type level, suggesting that miR-208a regulates the expression of Myh7b/miR-499, and their function in regulating the fast skeletal muscle gene switch in the heart is redundant. This regulation is in a spatial-temporal specific manner, as the expression of miR-499 is undisturbed in the neonatal heart or skeletal muscle of miR-208a null mice [44]. This evidence suggests that miR-208a functions upstream of miR-208b and miR-499 to fine-tune the expression of fast and slow muscle gene expressions in the heart. A recent study demonstrated that miR-208a directly inhibits SOX6, which subsequently inhibits the slow-twitch muscle gene expression in the heart [46•], providing a better explanation of the molecular mechanisms of miR-208a in the heart. Modulation of miR-208a level tightly regulates this cardiac muscle program by Trbp, an RNA-binding protein that is required for maintenance of normal cardiac contraction and function. Cardiac-specific deletion of Trbp abolishes the expression of miR-208a, and overexpression of miR-208a in Trbp knockout heart rescued the lethality and defective cardiac function [46•]. This further demonstrates a linear genetic pathway in modulating this important cardiac muscle program via the miR-208/miR-499 superfamily.
miRNAs Play a Key Role in Cardiomyocyte Proliferation and Cardiac Regeneration
miRNAs are important in regulating cardiomyocyte proliferation. In zebrafish, cardiac regeneration was found to be dependent on the reduced level of miR-99/100 and Let-7a/Let-7c [47]. Aguirre et al. identified Smarca5 and Fntb as the downstream protein-coding targets of these microRNAs in regulating the dedifferentiation and proliferation of cardiomyocytes. When reducing miR-99/10 and Let7a/Let7c following myocardial infarction (MI) injury, cardiomyocyte proliferation was induced, and the scar size at 3 months post-surgery was reduced. As a result, cardiac function was partially rescued. This suggests the microRNA program of miR-99/100 and Let7a/Let7c in regulating proliferation and regeneration is conserved [47, 48].
The miR-17-92 cluster has also been implicated in regulating cardiomyocyte proliferation. Cardiac-specific overexpression of miR-17-92 cluster in a transgenic mouse model induced cardiomyocyte proliferation in both neonatal and adult hearts, and the overexpression of this miRNA cluster has a protective effect in MI model [49]. Mechanistically, the miR-19-92 cluster is found to reduce the expression of PTEN, which is a repressor for proliferation [49].
miR-195 was identified to be upregulated in mouse heart on postnatal day 10 (P10) compared to P1, when cardiomyocytes exit cell cycle. Overexpression of miR-195 in the embryonic heart under βMHC promoter leads to ventricular hyperplasia and ventricular septal defect, indicating premature cell cycle exit [50]. Gene profiling reveals miR-195 regulates a large number of the cell cycle genes, including Chek1, a conserved direct target of miR-195 [50]. miR-195 belongs to the miR-15 family, and the knockdown of the miR-15 family of microRNAs induced cardiomyocyte proliferation in neonatal and early postnatal age [51]. Porrello et al. further demonstrated that inhibition of the miR-15 family of miRNAs in early postnatal age can improve LV systolic function following MI, and that the miR-15 family is responsible for the loss of postnatal regenerative capacity in the heart [51]. Using high-throughput screening method to identify human miRNAs that can promote neonatal cardiomyocyte proliferation, Enlaliao et al. identified some candidates including hsa-miR-590 and hsa-miR-199a [52]. Expression of these two miRNAs in ex vivo culture of adult cardiomyocytes can induce cell cycle re-entry. Moreover, administration of these two miRNAs after MI injury in mice stimulated cardiac regeneration, and the cardiac function was recovered. Taken together, these studies demonstrated the critical role for miRNAs in regulating cardiomyocyte proliferation and regeneration and showcase the potential to manipulate the activity of specific miRNAs to induce cardiomyocyte proliferation for regenerative therapy.
Long Non-coding RNA Expression in Normal and Diseased Hearts
lncRNAs are defined as RNA genes larger than 200 bp that do not have coding potential. This size cutoff distinguished lncRNA from small non-coding RNAs. Over the last decade, it has been increasingly recognized that the vast majority of the genome is pervasively transcribed; many of these result in the production of lncRNAs. As the importance of ncRNAs is gaining recognition in the cardiac development and pathology, many groups had screened for lncRNAs that have implications in ICM or NICM. Huang et al. utilized an RNA-seq approach to identify differentially expressed lncRNA between human hearts with ICM and healthy individuals [53]. They found that the expression of lncRNAs is highly correlated to their neighboring coding genes, suggesting a cis-regulatory circuit of lncRNAs. A similar observation was reported in cardiac development comparing the human fetal heart and adult heart [54]. Using an unsupervised clustering approach, Huang et al. found that the differentially expressed lncRNAs in ICM samples correlate with extracellular matrix coding genes. This was validated using loss and gain of function on a subset of lncRNAs identified and revealed a potential role for these lncRNAs in participating the TGF-β pathway [53]. Greco et al. have also performed similar lncRNA screening in heart tissue from ICM patients [55]. They performed RNA-seq on heart samples from non-end-stage dilated ICM and validated their result in an independent cohort of end-stage HF patient’s heart samples. Fourteen lncRNAs were identified in non-end-stage HF patient, nine of these were validated in end-stage HF patients, and three of these (H19, RMRP, and HOTAIR) were also induced in a mouse transverse-aortic constriction (TAC) model. Similar modulation of a subset of lncRNAs (ANRIL, HOTAIR, and TUSC7) has also been observed in peripheral blood mononuclear cell and heart tissue [55, 56]. These studies provide convincing evidence that lncRNAs are expressed in normal and diseased hearts. Interestingly, an increase of lncRNA H19 in the plasma has been found to associate with CAD in a Chinese population [57], suggesting that circulating lncRNA can be a potential biomarker for ICM (Fig. 1; Table 1).
Yang et al. performed a systematic screening of mRNA, miRNA, and lncRNA in the heart samples of NICM patients by RNA-seq, before and after mechanical circulatory support by left ventricular assist device (LVAD) [92]. Interestingly, using unsupervised hierarchical clustering of expression profiles, the authors found that the expression signature of lncRNAs, but not mRNA or miRNAs, distinguishes ICM and NICM in human failing hearts. Moreover, the expression signature of lncRNAs also differentiates failing hearts before and after mechanical circulatory support by LVAD [92], suggesting that lncRNA expression level in the heart is regulated by the pathological state of HF. Li et al. performed similar screening and validation in the heart samples of DCM-induced congestive HF patients; they identified TAF10 as a potential regulator for a subset of differentially expressed lncRNA in DCM [93]. Taken together, this large-scale screening identified lncRNAs as potential players in the pathogenesis of ICM and NICM (Table 1).
The Molecular Mechanisms of lncRNA Function in the Heart
lncRNAs play critical regulatory roles in diverse cellular processes, including chromatin remodeling, transcription, post-translational regulation, and intracellular processes [94, 95]. Increasing evidence has also implicated that lncRNAs are involved in cardiac development, contractile function, and cardiomyopathies.
The polycomb repressive complex (PRC1/PRC2) and the trithorax/MLL (TrxG/MLL) complex are critical regulators for the chromatin structures to repress or activate gene expression [96]. In particular, the PRC2 promotes the H3K27me3 to repress gene expression [97], while TrxG/MLL promotes the H3K4me3 which is an activating mark for gene expression [98]. Interestingly, several lncRNAs are involved in epigenetic regulation of cardiac gene expression by interacting with these epigenetic modifiers [62, 67•, 74, 75, 99]. The lncRNA Braveheart (Bvht) was identified to interact with PRC2 complex during cardiac lineage commitment [62, 63]. Klattenhoff et al. performed RNA-seq in Bvht-depleted embryonic stem cells (ESC) and found that the expression of more than 548 genes was changed significantly when comparing to the control ESC. Among many dysregulated genes, the authors identified Mesp1, a potent regulator for cardiovascular commitment, is regulated by Bvht, to drive cardiac differentiation [62]. Using an in vitro cardiomyocyte differentiation model, the authors found that Bvht directly interacts with SUZ12, a core component of the PRC2 complex, suggesting the Bvht might modulate the epigenetic state of the chromatin to regulate the gene expression program during cardiac differentiation. Through a more thorough analysis of the secondary RNA structure, Xue et al. identified a 5′ asymmetric G-rich internal loop (AGIL) of Bvht to be necessary for mesoderm to cardiomyocyte progenitor differentiation [63]. This AGIL region interacts with and functionally antagonizes a zinc-finger transcription factor CNBP, to promote the cardiac commitment [63]. Together, this evidence suggests that Bvht can regulate both the epigenetic state and transcription of gene expression during cardiac differentiation. While these reports clearly pointed the function of Bvht to the regulation of cardiac gene expression, questions remain how the specificity of such regulation is generated. The initial screen identified Bvht is a heart-enriched lncRNA, but it is also expresses in several other mouse tissues, questioning whether it plays a role in cells/tissues outside the heart. Given the established role of Bvht in cardiac commitment in vitro, it would be particularly important to further investigate its in vivo function in cardiovascular development and disease. Finally, it remains mysterious whether the human homolog of mouse Bvht can be identified.
Chaer is another example of lncRNA that interacts with epigenetic modifiers. Chaer was identified as a cardiac-enriched lncRNA, that its expression increased in hypertrophic heart [67•]. Chaer negatively regulates H3K27 methylation state via its interaction with the catalytic core of PRC2 complex (SUZ12 and EZH2), but not TrxG/MLL complex. Interestingly, overexpression of Chaer in mouse embryonic fibroblasts reduced the interaction of PRC2 with other lncRNAs (Hotair or Fendrr), suggesting that Chaer binding to PRC2 reduces the PRC2 binding to other lncRNAs. This observation indicates that Chaer may modulate PRC2 function or targeting location [67•]. Conversely, Chaer deletion increases H3K27me2 and H3K27me3 in the heart. H3K27me3 repressive mark on a subset of hypertrophic genes (Anf, Myh7, and Acta1) is significantly reduced after TAC operation. However, Chaer deletion restores the level of H3K27me3 upon TAC operation. To investigate the timing significance of the Chaer-PRC2 interaction in the contact of cardiac hypertrophy, the authors utilized siRNAs to knockdown Chaer before TAC operation or 1 day after TAC operation. Interestingly, only knocking down Chaer before TAC operation could the cardiac function be preserved and fibrotic remodeling be decreased, associating with the attenuation of the H3K27me3 level, while silencing Chaer 1 day after TAC operation was unable to affect the progression of hypertrophy. Taken together, these data suggest that Chaer functions at a crucial time window immediately following hypertrophic stress to repress the PRC2-mediated chromatin repression on a subset of hypertrophic genes [67•] (Fig. 2). It will be important to determine whether Chaer is involved in human cardiomyopathy.
One of the well-characterized examples for divergently transcribed lncRNA that regulates its neighboring protein-coding gene is the Hand2-Upperhand (Uph) locus. The bHLH transcription factor Hand2 gene is a critical regulator of heart development [100,101,102], and its expression is tightly regulated by upstream enhancers [103, 104]. A conserved lncRNA, Uph, was identified to transcribed in the opposite direction upstream of the Hand2 locus, and the Uph expression has a similar pattern as Hand2 in mouse embryos [82••]. In order to define the in vivo function of Uph, the investigators generated mutant mice lacking functional Uph. Since Uph encompasses two of the upstream enhancers for Hand2, to avoid potential effect of disturbing the upstream regulatory element, an exon 2 insertion of polyadenylation sequence was introduced to terminate the transcription of Uph (Uph KO) without affecting the upstream enhancers of Hand2 gene. The Uph KO mouse is embryonically lethal and displays pericardial effusion by E10.5. The Uph KO embryos failed to develop a right ventricular chamber, resembling the phenotype of Hand2 KO embryos. Hand2 expression is absent in the Uph KO heart, but remains undisturbed in branchial arches and limb buds, suggesting the specificity for the regulation of Hand2 expression in the developing heart by the transcription of Uph [82••]. The authors also demonstrated that knockdown of mature transcript of Uph does not affect Hand2 expression, and the transcription of Uph is required for maintaining the active enhancer marks H3K4me1 and H3K27ac, and recruitment of GATA4 to the regulatory region [82••]. Moreover, double heterozygous of Uph and Hand2 (Uph+/−; Hand2+/−) is embryonically lethal and phenocopy the heart phenotype of Hand2 KO, suggesting that Uph transcription regulates the expression of Hand2 in-cis [82••] (Fig. 3). Given that there are many genetic loci of lncRNAs and protein-coding genes share similar bidirectional promoter structure [105], the Uph-Hand2 regulation circuit provides an interesting and important example for the divergent transcription of lncRNA regulates the expression of neighboring protein-coding genes in vivo. It is predictable that additional neighboring lncRNA/mRNA regulatory circuits will be identified and it will be extremely interesting to learn how they participate in the regulation of divergent biological function and disease status.
H19 is one of the cardiomyopathy-associated lncRNAs identified to be upregulated in the left ventricle biopsies of post-ischemic dilated cardiomyopathy from a cohort of 13 heart failure patients [55]. The increased plasma level of H19 had also been associated with increased risk of coronary artery disease in a cohort of 480 in a Chinese population [57]. The H19 gene encodes a 2.3-kb lncRNA and also contains a miRNA miR-675 in the first exon [106]. H19 is located in a conserved imprinted cluster on chromosome 7 in mouse and 11 in human. H19 is expressed from the maternal allele, while its neighbor gene Igf2, which is 90 kb away, is paternally express. Their expression is controlled by an intergenic differentially methylated region, where it is highly methylated in the paternal allele, which allows the downstream enhancer to promoter Igf2 expression. In the maternal allele, this intergenic region is unmethylated, which recruits insulator CTCF [107, 108], and allows the downstream enhancer to promote expression of H19 lncRNA. The expressed H19 lncRNA can act in-cis to recruit Mbd1 and Setdb1 to the promoter region of Igf2 and several other genes to promote H3K9me2 [109], which in turn silence their maternal allelic expression. Targeted deletion of H19 has led to an overgrowth phenotype, and Igf2 becomes biallelically expressed, which can be explained by the cis-effect of H19 [76, 110]. To test whether H19 can act in-trans, Gabory et al. utilized a transgenic allele which expresses H19 in the Ndn locus which was not imprinted. The expression of this transgene is sufficient to rescue the overgrowth phenotype displayed in the H19 knockout [77]. Moreover, the expression of the H19 transgene can rescue the biallelic expression of the Igf2 caused by H19-targeted deletion. Interestingly, the authors observed that many imprinted genes outside of this locus which were mis-expressed in the H19 knockout are rescued to normal level in the presence of this H19 transgene, suggesting that H19 can also act in-trans [77]. H19 lncRNA has also been shown capable of changing the DNA methylation genome-wide [111, 112]. Zhou et al. showed that H19 can bind and inhibit S-adenosylhomocysteine hydrolase and subsequently influences Dnmt3b-dependent DNA methylation. It was further demonstrated that H19 suppresses methylation on DNA methylation regions of many gene targets outside of the H19 locus in a myotube [111]. Taken together, this suggests that H19 can act in-cis or in-trans to modify the epigenetic marks in the targeted locus (Fig. 4). Although H19 is associated with HF and CAD, mechanistic studies in vivo are needed to elucidate its function in the context of cardiomyopathy further.
A methylation-sensitive enhancer region (imprinting control region, ICR) between lncRNA H19 and protein-coding gene Igf2 controls the allele-specific expression of these two genes. In maternal allele, ICR is hypomethylated and allows CTCF interaction; this direct the distal enhancers to drive H19 expression from the maternal allele, and the lncRNA H19 product recruits transcriptional repressors (such as MBD1) to repress Igf2 in-cis. On the other hand, the ICR in the paternal allele is methylated, and the expression of H19 is silenced; this leads to Igf2 expression from this locus of in an allelic specific manner
CAIF is a lncRNA that directly interacts with p53 protein and interferes with its binding to the myocardin (Myocd) promoter, thus blocking the p53-mediated transcription of Myocd, which subsequently attenuates cardiac autophagy [64]. Knockdown of p53 or overexpression of CAIF in the heart can reduce cardiac injury upon ischemia and reperfusion. Although it is shown that CAIF can bind p53 and functions as a transcriptional repressor for Myocd transcription, it will be interesting to determine whether such CAIF/p53 binding can influence transcription of other p53-dependent genes as well. This study suggests that lncRNA CAIF participates in the regulation of the p53-Myocd axis and is important in modulating the pathological progression of ICM [64].
LncRNA MIAT can function as a competing endogenous RNA (ceRNA). MIAT is short for myocardial infarction-associated transcript and was identified as a risk allele for MI in a large-scale study in the Japanese population [113]. Several MIAT variants were identified to have higher susceptibility to MI. MIAT was found to be upregulated in the myocardium of diabetic rats, and the knockdown of MIAT could improve the LV dysfunction induced by hyperglycemia [87]. Mechanistically, it was shown that MIAT interacts with miR-22-3p and Ago2 and effectively sponges the miR-22-3p, preventing it from the inhibition of its target Dapk2. This demonstrated a role for lncRNA to function as a ceRNA or miRNA sponge to regulate pathways involved in the pathogenesis of diabetic cardiomyopathy.
Emerging Role of Circular RNA in Cardiomyopathy
Circular RNA (circRNA) is a newly identified class of non-coding RNA that is formed during 5′ to 3′ back-splicing event. Many circRNAs are highly conserved and widely expressed [114], possessing many recently discovered functions, including as transcription regulators [115] or as miRNA sponges [116] (Table 1, Fig. 1). A recent study by Tan et al. had surveyed and validated circRNA expression in the heart, and also during cardiomyocyte differentiation [73]. They found that the cardiac-expressed circRNAs are derived from highly express cardiac genes [73]. For example, one circRNA abundantly expressed with a large number of isoforms is derived from the Titin gene [71, 73]. Intriguingly, some circTitin isoforms are differentially expressed in HCM and DCM hearts, suggesting they might be involved in the regulation of cardiomyopathy. RBM20 is an RNA-binding protein that is previously linked to human DCM and can regulate splicing [117,118,119]. Kahn et al. demonstrated that RBM20 could also regulate the biogenesis of a subset of circTitin isoforms [71], which provides an important insight of regulation for generating circRNA isoforms. However, the function of the diverse circTitin isoforms is largely unknown and future investigation is warranted. Most importantly, it is essential to determine whether genetic mutations of the Titin gene that have been linked to cardiomyopathy and heart failure are caused by altered expression and function of circTitin isoforms.
Two other cardiomyopathy-related circRNAs, i.e., circFoxo3 [70] and HRCR [81], were reported recently. circFoxo3 is highly expressed in the heart of aged mice and human and its expression is closely correlated with cellular senescence [70]. Ectopic expression of circFoxo3 promotes cellular senescence and aggravates doxorubicin-induced cardiomyopathy. Inversely, silencing of circFoxo3 can partially rescue the pathology of doxorubicin-induced cardiomyopathy. circFoxo3 is predominantly localized in the cytoplasm, and it interacts with ID1, E2F1, HIF1α, and FAK to prevent their nuclear (ID1, E2F1, and HIF1α) or mitochondria (FAK) localization under stress conditions. This presented a role for circRNA to interact with cellular proteins to modulate their function to induce senescence. The expression of HRCR decreases in failing mouse heart; Wang et al. demonstrated that HRCR interacts with miR-233, acting as a ceRNA or miRNA sponge. They found that the miR-233 transgenic mice developed HCM and HF, while the miR-233 deficient mice were protected from isoproterenol-induced hypertrophy [81]. Ectopic expression of HRCR in the heart can inhibit cardiac hypertrophy upon isoproterenol treatment. Taken together, these studies suggest an novel role for circRNAs in modulating the regulatory pathways leading to cardiac hypertrophy and HF.
Conclusions and Future Directions
Since the realization that the vast majority of the human genome is transcribed but does not code for protein, the role of ncRNAs has been actively investigated in many biological systems and a variety of biological functions has been discovered for these RNA species. miRNAs have clearly been established as critical modulators for cardiovascular development and pathogenesis. Numerous current efforts are focused on building miRNAs as useful diagnostic biomarkers and powerful therapeutic targets for human diseases. Although lncRNA research is still at its infancy, emerging evidence has already pointed to their important roles for a variety of processes. While many putative ncRNAs have been identified to associate with cardiomyopathy, only relatively few have been studied in detail. We anticipate that there will be many more mechanistic studies and discoveries of novel functions of miRNAs and lncRNAs in regulating the pathways involved in cardiomyopathy in the coming years. Given the versatility for the modes of regulation by lncRNAs, further mechanistic studies will provide a better understanding of the biological machinery and provide an attractive area for pharmacological targeting for cardiomyopathy and regenerative therapy.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Felker GM, Shaw LK, O'Connor CM. A standardized definition of ischemic cardiomyopathy for use in clinical research. J Am Coll Cardiol. 2002;39(2):210–8.
Benjamin EJ, Virani SS, Callaway CW, Chamberlain AM, Chang AR, Cheng S, et al. Heart Disease and Stroke Statistics-2018 update: a report from the American Heart Association. Circulation. 2018;137(12):e67–e492.
da Luz PL et al. Endothelium in atherosclerosis: plaque formation and its complications. In: Endothelium and Cardiovascular Diseases. 2018. Elsevier. p. 493–512.
Arroyo LH, Lee RT. Mechanisms of plaque rupture: mechanical and biologic interactions. Cardiovasc Res. 1999;41(2):369–75.
Phatharajaree W, Phrommintikul A, Chattipakorn N. Matrix metalloproteinases and myocardial infarction. Can J Cardiol. 2007;23(9):727–33.
Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331(6020):1078–80.
van Berlo JH, et al. c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature. 2014;509(7500):337–41.
Talman V, Ruskoaho H. Cardiac fibrosis in myocardial infarction—from repair and remodeling to regeneration. Cell Tissue Res. 2016;365(3):563–81.
Cleutjens JP, et al. Collagen remodeling after myocardial infarction in the rat heart. Am J Pathol. 1995;147(2):325–38.
Cohn JN, Ferrari R, Sharpe N. Cardiac remodeling—concepts and clinical implications: a consensus paper from an international forum on cardiac remodeling. J Am Coll Cardiol. 2000;35(3):569–82.
Braunwald E. Cardiomyopathies: an overview. Circ Res. 2017;121(7):711–21.
Hein S, et al. Progression from compensated hypertrophy to failure in the pressure-overloaded human heart: structural deterioration and compensatory mechanisms. Circulation. 2003;107(7):984–91.
Gaasch WH, Zile MR. Left ventricular structural remodeling in health and disease. J Am Coll Cardiol. 2011;58(17):1733–40.
Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3(11):e442.
Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921.
Dahlberg AE. The functional role of ribosomal RNA in protein synthesis. Cell. 1989;57(4):525–9.
Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat Rev Mol Cell Biol. 2017;19:45–58.
Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–55.
Dieci G, Preti M, Montanini B. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics. 2009;94(2):83–8.
Ishizu H, Siomi H, Siomi MC. Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev. 2012;26(21):2361–73.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97.
Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11(12):1753–61.
Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–5.
Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions--beyond repression of gene expression. Nat Rev Genet. 2014;15(9):599–612.
Olena AF, Patton JG. Genomic organization of microRNAs. J Cell Physiol. 2010;222(3):540–5.
Rodriguez A, et al. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14(10A):1902–10.
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–60.
Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–39.
Hata A. Functions of microRNAs in cardiovascular biology and disease. Annu Rev Physiol. 2013;75:69–93.
Novák J, et al. Mechanistic role of MicroRNAs in coupling lipid metabolism and atherosclerosis. In: Santulli G, editor. microRNA: basic science: from molecular biology to clinical practice. Cham: Springer International Publishing; 2015. p. 79–100.
Callis TE, Wang D-Z. Taking microRNAs to heart. Trends Mol Med. 2008;14(6):254–60.
Saxena A, Tabin CJ. miRNA-processing enzyme Dicer is necessary for cardiac outflow tract alignment and chamber septation. Proc Natl Acad Sci U S A. 2010;107(1):87–91.
Chen J-F, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, et al. Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A. 2008;105(6):2111–6.
da Costa Martins PA, Bourajjaj M, Gladka M, Kortland M, van Oort RJ, Pinto YM, et al. Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation. 2008;118(15):1567–76.
Chen J-F, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38(2):228–33.
Wystub K, et al. miR-1/133a clusters cooperatively specify the cardiomyogenic lineage by adjustment of myocardin levels during embryonic heart development. PLoS Genet. 2013;9(9):e1003793.
Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436(7048):214–20.
Liu N, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22(23):3242–54.
Morton SU, et al. microRNA-138 modulates cardiac patterning during embryonic development. Proc Natl Acad Sci U S A. 2008;105(46):17830–5.
van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103(48):18255–60.
Lompre AM, Nadal-Ginard B, Mahdavi V. Expression of the cardiac ventricular alpha-and beta-myosin heavy chain genes is developmentally and hormonally regulated. J Biol Chem. 1984.
Chien KR. Genomic circuits and the integrative biology of cardiac diseases. Nature. 2000;407(6801):227–32.
Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119(9):2772–86.
van Rooij E, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17(5):662–73.
van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316(5824):575–9.
• Ding J, et al. Trbp regulates heart function through microRNA-mediated Sox6 repression. Nat Genet. 2015;47(7):776–83. This study thoroughly demonstrated a unique Trbp/miR-208a/Sox6 pathway that controls fast- and slow-twitch myofiber balance in the heart, and cardiomyopathy.
Aguirre A, Montserrat N, Zacchigna S, Nivet E, Hishida T, Krause MN, et al. In vivo activation of a conserved microRNA program induces mammalian heart regeneration. Cell Stem Cell. 2014;15(5):589–604.
Hodgkinson CP, Dzau VJ. Conserved microRNA program as key to mammalian cardiac regeneration: insights from zebrafish. Circ Res. 2015;116(7):1109–11.
Chen J, et al. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ Res. 2013;112(12):1557–66. https://doi.org/10.1161/CIRCRESAHA.112.300658.
Porrello ER, et al. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ Res. 2011;109(6):670–9.
Porrello ER, Mahmoud AI, Simpson E, Johnson BA, Grinsfelder D, Canseco D, et al. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc Natl Acad Sci U S A. 2013;110(1):187–92.
Eulalio A, Mano M, Ferro MD, Zentilin L, Sinagra G, Zacchigna S, et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492(7429):376–81.
Huang Z-P, et al. Long non-coding RNAs link extracellular matrix gene expression to ischemic cardiomyopathy. Cardiovasc Res. 2016;112(2):543–54.
He C, et al. Systematic characterization of long noncoding RNAs reveals the contrasting coordination of cis- and trans-molecular regulation in human fetal and adult hearts. Circ Cardiovasc Genet. 2016;9(2):110–8.
Greco S, Zaccagnini G, Perfetti A, Fuschi P, Valaperta R, Voellenkle C, et al. Long noncoding RNA dysregulation in ischemic heart failure. J Transl Med. 2016;14(1):183.
Gao L, et al. Circulating long noncoding RNA HOTAIR is an essential mediator of acute myocardial infarction. Cell Physiol Biochemist. 2017;44(4):1497–508.
Zhang Z, et al. Increased plasma levels of lncRNA H19 and LIPCAR are associated with increased risk of coronary artery disease in a Chinese population. Sci Rep. 2017;7(1):7491.
Pasmant E, Sabbagh A, Vidaud M, Bièche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25(2):444–8.
Vausort M, Wagner DR, Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ Res. 2014;115(7):668–77.
Arslan S, Berkan Ö, Lalem T, Özbilüm N, Göksel S, Korkmaz Ö, et al. Long non-coding RNAs in the atherosclerotic plaque. Atherosclerosis. 2017;266:176–81.
Burd CE, et al. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6(12):e1001233.
Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152(3):570–83.
Xue Z, et al. A G-rich motif in the lncRNA Braveheart interacts with a zinc-finger transcription factor to specify the cardiovascular lineage. Mol Cell. 2016;64(1):37–50.
Liu C-Y, et al. LncRNA CAIF inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nat Commun. 2018;9(1):29.
Yu L, et al. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One. 2016;11(7):e0158347.
Zhang Y, Sun L, Xuan L, Pan Z, Li K, Liu S, et al. Reciprocal changes of circulating long non-coding RNAs ZFAS1 and CDR1AS predict acute myocardial infarction. Sci Rep. 2016;6:22384.
• Wang Z, et al. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat Med. 2016;22(10):1131–9. This study provided convincing evidence for a novel lncRNA Chaer in regulating cardiac hypertrophy through interaction with the PRC2 complex to modulate epigenetic state of cardiac hypertrophic gene expression.
Wang K, et al. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res. 2014;114(9):1377–88.
Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429.
Du WW, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38(18):1402–12.
Khan MAF, et al. RBM20 regulates circular RNA production from the Titin gene. Circ Res. 2016;119(9):996–1003.
Werfel S, Nothjunge S, Schwarzmayr T, Strom TM, Meitinger T, Engelhardt S. Characterization of circular RNAs in human, mouse and rat hearts. J Mol Cell Cardiol. 2016;98:103–7.
Tan WLW, et al. A landscape of circular RNA expression in the human heart. Cardiovasc Res. 2017;113(3):298–309.
Grote P, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24(2):206–14.
Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749.
Leighton PA, et al. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature. 1995;375(6526):34–9.
Gabory A, Ripoche MA, le Digarcher A, Watrin F, Ziyyat A, Forne T, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136(20):3413–21.
Kallen AN, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52(1):101–12.
Li X, Wang H, Yao B, Xu W, Chen J, Zhou X. lncRNA H19/miR-675 axis regulates cardiomyocyte apoptosis by targeting VDAC1 in diabetic cardiomyopathy. Sci Rep. 2016;6:36340.
Liu L, et al. The H19 long noncoding RNA is a novel negative regulator of cardiomyocyte hypertrophy. Cardiovasc Res. 2016;111(1):56–65.
Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37(33):2602–11.
•• Anderson KM, et al. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature. 2016;539(7629):433–6. This study demonstrated elegantly that the divergent transcription of lncRNA upperhand (Uph), but not the RNA product from the lncRNA locus, regulates neighboring gene Hand2 in cardiac development. This is an important example for the divergently transcribed lncRNA and coding RNA pairs present in the genome.
Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen C, et al. LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation. 2014;130(17):1452–65.
Groff AF, Sanchez-Gomez DB, Soruco MML, Gerhardinger C, Barutcu AR, Li E, et al. In vivo characterization of Linc-p21 reveals functional cis-regulatory DNA elements. Cell Rep. 2016;16(8):2178–86.
Dimitrova N, et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. 2014;54(5):777–90.
Wang K, Gan TY, Li N, Liu CY, Zhou LY, Gao JN, et al. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24(6):1111–20.
Zhou X, et al. lncRNA MIAT functions as a competing endogenous RNA to upregulate DAPK2 by sponging miR-22-3p in diabetic cardiomyopathy. Cell Death Dis. 2017;8(7):e2929.
Yan B, et al. LncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res. 2015; https://doi.org/10.1161/CIRCRESAHA.114.305510.
Qu X, du Y, Shu Y, Gao M, Sun F, Luo S, et al. MIAT is a pro-fibrotic long non-coding RNA governing cardiac fibrosis in post-infarct myocardium. Sci Rep. 2017;7:42657.
Jiang Y, et al. Downregulation of long non-coding RNA Kcnq1ot1: an important mechanism of arsenic trioxide-induced long qt syndrome. Cell Physiol Biochemist. 2018;45(1):192–202.
Korostowski L, Sedlak N, Engel N. The Kcnq1ot1 long non-coding RNA affects chromatin conformation and expression of Kcnq1, but does not regulate its imprinting in the developing heart. PLoS Genet. 2012;8(9):e1002956.
Yang K-C, Yamada KA, Patel AY, Topkara VK, George I, Cheema FH, et al. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation. 2014;129(9):1009–21.
Li H, et al. Identification of cardiac long non-coding RNA profile in human dilated cardiomyopathy. Cardiovasc Res. 2018;114(5):747–58.
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–41.
Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–14.
Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128(4):735–45.
Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–9.
Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol. 2011;12(12):799–814.
Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–23.
Srivastava D, et al. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet. 1997;16(2):154–60.
Yamagishi H, Olson EN, Srivastava D. The basic helix-loop-helix transcription factor, dHAND, is required for vascular development. J Clin Invest. 2000;105(3):261–70.
Han Z, et al. Hand, an evolutionarily conserved bHLH transcription factor required for Drosophila cardiogenesis and hematopoiesis. Development. 2006;133(6):1175–82.
Charité J, et al. Role of Dlx6 in regulation of an endothelin-1-dependent, dHAND branchial arch enhancer. Genes Dev. 2001;15(22):3039–49.
McFadden DG, et al. A GATA-dependent right ventricular enhancer controls dHAND transcription in the developing heart. Development. 2000;127(24):5331–41.
Lepoivre C, Belhocine M, Bergon A, Griffon A, Yammine M, Vanhille L, et al. Divergent transcription is associated with promoters of transcriptional regulators. BMC Genomics. 2013;14:914.
Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13(3):313–6.
Pant V, et al. The nucleotides responsible for the direct physical contact between the chromatin insulator protein CTCF and the H19 imprinting control region manifest parent of origin-specific long-distance insulation and methylation-free domains. Genes Dev. 2003;17(5):586–90.
Fedoriw AM, et al. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science. 2004;303(5655):238–40.
Monnier P, Martinet C, Pontis J, Stancheva I, Ait-Si-Ali S, Dandolo L. H19 lncRNA controls gene expression of the imprinted gene network by recruiting MBD1. Proc Natl Acad Sci U S A. 2013;110(51):20693–8.
Ripoche MA, Kress C, Poirier F, Dandolo L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 1997;11(12):1596–604.
Zhou J, Yang L, Zhong T, Mueller M, Men Y, Zhang N, et al. H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat Commun. 2015;6:10221.
Hadji F, Boulanger MC, Guay SP, Gaudreault N, Amellah S, Mkannez G, et al. Altered DNA methylation of long noncoding RNA H19 in calcific aortic valve disease promotes mineralization by silencing NOTCH1. Circulation. 2016;134(23):1848–62.
Ishii N, Ozaki K, Sato H, Mizuno H, Susumu Saito, Takahashi A, et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet. 2006;51(12):1087–99.
Rybak-Wolf A, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58(5):870–85.
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–64.
Piwecka M, Glažar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357(6357):eaam8526.
Brauch KM, Karst ML, Herron KJ, de Andrade M, Pellikka PA, Rodeheffer RJ, et al. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J Am Coll Cardiol. 2009;54(10):930–41.
Li D, et al. Identification of novel mutations in RBM20 in patients with dilated cardiomyopathy. Clin Transl Sci. 2010;3(3):90–7.
Guo W, et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18(5):766–73.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Yao Wei Lu and Da-Zhi Wang declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Myocardial Disease
Rights and permissions
About this article
Cite this article
Lu, Y.W., Wang, DZ. Non-coding RNA in Ischemic and Non-ischemic Cardiomyopathy. Curr Cardiol Rep 20, 115 (2018). https://doi.org/10.1007/s11886-018-1055-y
Published:
DOI: https://doi.org/10.1007/s11886-018-1055-y