Abstract
Purpose of Review
This review describes the recent progress in nuclease-based therapeutic applications for inherited heart diseases in vitro, highlights the development of the most recent genome editing technologies and discusses the associated challenges for clinical translation.
Recent Findings
Inherited cardiovascular disorders are passed from generation to generation. Over the past decade, considerable progress has been made in understanding the genetic basis of inherited heart diseases. The timely emergence of genome editing technologies using engineered programmable nucleases has revolutionized the basic research of inherited cardiovascular diseases and holds great promise for the development of targeted therapies.
Summary
The genome editing toolbox is rapidly expanding, and new tools have been recently added that significantly expand the capabilities of engineered nucleases. Newer classes of versatile engineered nucleases, such as the “base editors,” have been recently developed, offering the potential for efficient and precise therapeutic manipulation of the human genome.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cardiovascular diseases are the leading cause of death worldwide [1, 2]. According to World Heart Federation, an estimated 17.3 million people die of cardiovascular diseases every year. According to the Center of Disease Control, about 610,000 people die of heart disease in the USA every year—that is one in every four deaths—and are responsible for $300 billion in direct and indirect costs. Many different types of heart diseases are usually associated with a mutation in a single gene and are passed from generation to generation with autosomal dominant mode of inheritance. Such monogenic inherited heart diseases include cardiomyopathies, such as dilated and hypertrophic cardiomyopathies as well as conditions that affect the electric system of the heart, causing arrhythmias. Clinical therapies for these inherited diseases focus on disease management without addressing the underlying genetic defects and remain largely insufficient. The identification of disease-causing genes underlying the pathogenesis of inherited heart diseases has opened up new therapeutic opportunities for gene therapy approaches.
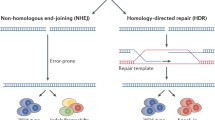
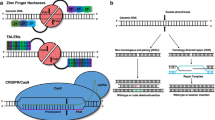
Classical gene therapy approaches comprise the delivering of a functional copy of the affected gene using a variety of viral vectors. However, most of the developing therapies are focused on acquired forms of heart disease. Recently, the advent of nuclease-based genome editing has facilitated development of a broad range of therapeutic strategies in vitro and has raised the possibility that this technology could be used to treat diseases that are refractory to traditional therapies. Genetic modification of mammalian cells has been greatly facilitated by the development of customized zinc-finger nucleases (ZFNs) [3], transcription activator-like effector nucleases (TALENs) [4], and clustered regularly interspaced short palindromic repeat/Cas9 (CRISPR/Cas9) [5]. A common feature of all these nuclease-based systems is that they can generate targeted double-stranded breaks (DSBs) at specific loci in the genome. The nuclease-induced DSBs are repaired by two different endogenous mechanisms, the non-homologous end joining (NHEJ) or the homology-directed repair (HDR) pathway [6, 7]. While NHEJ creates random insertions/deletions at the point of DSB, the HDR pathway can create precise genetic alterations by employing the homologous strands from the sister chromatids, homologous chromosomes, or exogenous DNA templates. Current genome editing approaches are harnessing both mechanisms to introduce targeted modifications in the genome, allowing the development of new model systems, including animal models and stem cell-derived in vitro models for studying inherited heart diseases [8]. Genome editing-based therapies can be achieved through several approaches including correction or inactivation of pathogenic mutations, or addition of therapeutic transgenes. Although the nuclease-based gene therapies have the potential to permanently modify the genome and correct the underlying genetic defects, there are numerous hurdles, such as efficacy, safety, and delivery, that must first be overcome before these tools can be used as therapeutics.
Over the past few years, the emergence of more precise and efficient genome editing approaches using engineered nuclease systems, especially the CRISPR-Cas9 system, has advanced both basic and translational cardiovascular research. Particularly, combinatorial approaches using engineered nucleases and induced pluripotent stem cells (iPSCs) have revolutionized disease modeling under normal and pathological conditions in vitro. The combination of the patient-derived iPSCs and the accesibility of CRISPR/Cas9-mediated genome editing has become a very powerful experimental tool. Together these two technologies have revolutionized stem cell research, disease modeling and human genetics.
In this review, we will summarize the recent studies that combined the nuclease-based genome editing and iPSC technologies in vitro, discuss the major challenges that will need to be addressed to realize their clinical translation, and finally highlight the most recent genome editing technologies and their potential therapeutic uses.
Genome Editing in Human iPSCs
Genome editing in human iPSCs is a novel experimental platform that has now become a versatile tool in stem cell research and human disease modeling. Isogenic iPSC-based disease models have been generated for several inherited cardiomyopathies and channelopathies, including dilated cardiomyopathy, Barth syndrome, and long QT and Brugada syndromes that have either corrected a disease-relevant mutation in iPSCs or introduced a disease-relevant allele in wild-type iPSCs (extensively reviewed elsewhere [8]). For example, the genetic correction of a pathogenic mutation in the phospholamban gene (PLN, p. Arg14del) in patient-specific iPSCs was sufficient to rescue the calcium defects that are associated with the dilated cardiomyopathy pathology, exemplifying how this approach can be successfully implemented in vitro [9•].
More recently, Ang and colleagues harness the power of iPSCs and CRISPR-Cas9 technologies to generate a congenital heart disease (CHD) model associated with a mutation in the GATA4 gene in vitro [10]. Using transcriptomics analysis by RNA-Seq on isogenic human iPSC-derived cardiomyocytes (hiPSC-CMs), it was found that the heterozygous GATA4 G296S mutation impaired expression of the cardiac gene program and sonic hedgehog signaling while upregulating genes of alternative fates, particularly the endothelial lineage and those related to cardiac septation. Conversely, the GATA4-G296S mutation led to failure of GATA4 and TBX5-mediated repression at non-cardiac genes and enhanced open chromatin states at endothelial/endocardial promoters. These results provide a better understanding of how disease-causing missense mutations can disrupt transcriptional cooperativity, leading to aberrant chromatin states and cellular dysfunction, including those related to morphogenetic defects. This study reveals that a single missense mutation in a key cardiac transcription factor leads to CHD by regulating the recruitment of transcription factor complexes to enhancers in a dose-dependent manner, and points to potential nodes for therapeutic intervention.
In another study, Liang and colleagues [11] generated hiPSC-CMs from a patient affected by Brugada syndrome, carrying a deletion mutation in the SCN5A gene encoding for the sodium (Na+) channel Nav1.5. Upon demonstrating a strong genotype-phenotype correlation, the authors employed CRISPR/Cas9-mediated genome editing technology to correct the SCN5A deletion mutation in the patients’ iPSCs. The genetic correction of the SCN5A ameliorated the Brugada syndrome-associated phenotype at the single-cell level, including the restoration of electrical properties and Ca2+ handling kinetics.
Lastly, in a recent study, the NHEJ-mediated repair of nuclease-generated breaks was harnessed to generate gene knockouts in iPSCs [12]. We developed a library of TALENs to disrupt the expression of a comprehensive set of genes associated with cardiomyopathies and CHDs in human iPSCs. When introduced in iPSCs, each TALEN pair generated a DSB located around the start codon of each targeted gene that was repaired by the error-prone NHEJ mechanism. The formation of deletions (indels) on the gene of interest often results in a frameshift mutations that generates a premature stop codon resulting in efficient gene knockout. We demonstrated that TALEN-mediated gene knockout strategies in iPSCs could be used to better understand the biological function of genes and the pathological significance of genetic variants as well as to model complex diseases in vitro. This study suggests that nuclease-based approaches for efficient disruption of gene expression provide an unprecedented opportunity to better understand the molecular basis of cardiomyopathies and decipher the relationships between genotype and phenotype of human cardiovascular diseases.
Challenges for Therapeutic Translation
These recent studies have elegantly illustrated the power of genome editing and iPSC technologies in providing mechanistic insight into patient-specific phenotypes. Despite the obvious advances, several challenges remain to translate genome editing technologies to the clinic.
The introduction of precise genetic modifications in the post-mitotic cardiomyocytes is critical for therapeutic strategies for many cardiovascular disorders that require targeted insertion or in situ correction of mutated DNA sequence. Current approaches for point mutation correction are inefficient and mainly induce an abundance of random insertions and indels at the targeted locus [13]. Precise genome editing for efficient gene insertion and correction requires the HDR-mediated incorporation of exogenous DNA fragments-a process that inherently occurs at very low frequencies-especially in post-mitotic cells. HDR primarily occurs in the S and G2 phases of the cell cycle [14], whereas NHEJ occurs throughout the cell cycle and is the major repair pathway in mammalian cells. It is expected that DSBs in cardiomyocytes would likely be repaired predominantly by the mutagenic NHEJ and rarely, if at all, by the HDR mechanism. Therefore, the cardiomyocytes are unlikely to be amenable to HDR-based genome editing, limiting the overall applicability of this strategy. Enabling HDR with higher efficacy in post-mitotic cells would therefore be beneficial and will open the door to therapeutic applications of genome editing in genetic heart disorders. In this respect, strategies to suppress NHEJ and enhance the HDR efficiency such as small molecule inhibitors of ligase IV, an essential enzyme in the NHEJ pathway, have been shown to increase HDR efficiency [15]. However, this strategy is challenging in post-mitotic cells, and its therapeutic value is limited by the potential risk of inhibiting ligase IV in non-target cells.
New Genome Editing Technologies
Cpf1
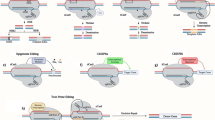
Scientists are continuously trying to overcome the shortcomings of existing genome editing systems by discovering new technologies. The recent introduction of Cpf1, a class 2 CRISPR-Cas system that includes an effective single RNA-guided endonuclease with distinct properties from Cas9, has the potential to substantially advance our ability to manipulate eukaryotic genomes [16]. The Cpf1 system uses the same basic principles as Cas9, but cleaves DNA and generates a staggered DNA double-stranded break with a 4- or 5-nt overhangs, in contrast to the blunt ends generated by Cas9. This cleavage pattern offers a directional gene transfer, analogous to traditional restriction enzyme cloning that could be particularly advantageous for targeting non-dividing cells. Eventually, the newly discovered Cpf1 system could provide an effective way to precisely introduce DNA into the genome via non-HDR mechanisms in cardiomyocytes.
Base Editors
Perhaps, the most exciting advance in the genome editing field in the past year is the development of a novel class of nuclease-derived “base editors,” a new approach to genome editing that enables the direct and irreversible conversion of a DNA base into another at a target genomic locus in a HDR-independent manner without DNA cleavage and exogenous donor DNA template delivery. Recently, programmable deaminases by fusing activation-induced deaminase (AID) and apolipoprotein B mRNA editing enzyme catalytic polypeptide-like family proteins (APOBECs) with catalytically dead Cas9, ZFN, or TALEN-DNA-binding modules have been reported [17••, 18]. Deaminases are naturally occurring proteins that operate in various important cellular processes. These enzymes convert cytidines (C) to uracils (U) in DNA. If DNA replication occurs before uracil repair, the replication machinery will treat the uracil as thymine (T), leading to a C:G to T:A base pair conversion. Although APOBEC- and AID-based systems use different cytidine deaminases and protein fusion arrangements, both have been reported to have high on target C > T base-editing efficiency and relatively low indel rates. Combining the precise targeting of programmable nucleases and the ability to manipulate the damage and repair pathways utilizing the cytidine deaminases provide a powerful strategy to correct or introduce specific C > T point mutations (or G > A on the opposite strand) independent of the HDR processes. Komor et al. [17••] estimated that 3000 genetic variants listed in ClinVar would be correctable with a C > T substitution. However, given the current limitations of the system, such as the narrow base-editing window and the potential of editing multiple cytidines in the editing window, it was estimated that only 300–900 variants would be targetable using this approach.
To overcome this limitation, additional engineering may increase the versatility of the base-editors technology. Specifically, the ability to convert A•T base pairs to G•C base pairs at target loci in the genomic DNA could make it possible to correct approximately half of known disease-associated point mutations in humans. Recently, Gaudelli and colleagues used directed evolution with several rounds of selection to create a adenosine deaminase base editor (ABE) that would convert adenine to inosine, resulting in an A > G change [19]. When this novel enzyme was tethered to an inactive RNA-guided Cas9 complex, it transformed A•T into G•C base pairs in the genome of living cells with surprising efficiency across multiple target sites with a very low mutagenic background.
Conclusion
With the advent of programmable genome editing technologies, the precise modification of the human genome has proceeded with remarkable speed and breadth, bringing patient-specific therapies for inherited cardiomyopathies within reach. In addition, the versatility of the nuclease-mediated genome editing tools together with the iPSC technology have become a very powerful experimental approach to study the effects of an individual’s genetic mutation within the context of his or her own genetic background. The recently discovered RNA-guided CRISPR-Cas9 system has been widely adopted in research and is currently considered the state-of-the-art tool for genome editing in vitro. This technology has the potential to dramatically accelerate our understanding of the underlying pathogenic mechanisms of inherited cardiomyopathies, paving the way for precision medicine.
The recent introduction of the sophisticated base-editing systems offers a simple and effective genome editing tool that circumvents the limitations associated with HDR-mediated approaches. Theoretically, using these programmable base editors, all the so-called transition mutations—C to T, T to C, A to G, or G to A—that collectively account for two thirds of all disease-causing point mutations could be therapeutically targeted. These novel technologies hold great promise as new therapeutic agents. However, numerous challenges still lie ahead, and additional research is required to develop these enzymes as safe and efficient therapeutics for genetic cardiovascular diseases. For example, a key limitation to the clinical use of CRISPR-Cas9 nucleases for therapeutic genome editing is the non-specific activity that can induce unwanted off-target mutations. In a search for better Cas9-mediated editing tools, several Cas9 variants have been recently generated by engineering or evolution, including the high fidelity (SpCas9-HF1), the enhanced specificity (eSpCas9(1.1)), the evoCas9, the expanded PAM SpCas9 (xCas9), and hyper-accurate Cas9 (HypaCas9), which exhibit substantially reduced off-target cleavage in human cells without compromising on-target activity [20,21,22,23,24]. Nevertheless, off-target cleavage remains a problem for some sites.
Furthermore, the translation of these new technologies will depend on the development of appropriate and efficacious delivery methods. For example, CRISPR/Cas9-mediated therapies will require significant advances in methods to deliver these relatively large transgenes to the heart. The AAV vector system provides major advantages for gene therapeutic applications and gene transfer in the cardiovascular system [25], and can be utilized to deliver CRISPR/Cas9 constructs in the heart for therapeutic in vivo genome editing [26]. As ongoing human clinical trials are currently utilizing the AAV system to deliver ZFNs for the treatment of hemophilia B and mucopolysaccharidosis (NCT03041324, NCT02702115, and NCT02695160), it is likely that the AAV system will be also used to develop CRISPR/Cas9-based therapeutic approaches for genetic cardiovascular diseases. Recently, several proof-of-principle studies have demonstrated the promise of AAV-mediated CRISPR/Cas9-based genome editing as a potential new therapeutic platform in restoring the dystrophic cardiomyopathy phenotype of a small animal model of Duchenne muscular dystrophy in vivo [27, 28, 29•]. Nevertheless, future studies that consider the optimization of vector design, including evaluation of various AAV capsids, tissue-specific promoters and choice of Cas9 variants, will be important to bring the CRISPR-Cas9 technology into the clinic.
Although in its infancy, the astounding pace of technological advances of genome editing approaches suggest that their clinical application for inherited cardiovascular diseases will undoubtedly evolve beyond a conceptual framework in the next decade. Genome editing has already revolutionized cardiovascular research, and this is just the beginning.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, et al. Heart disease and stroke statistics—2015 update: a report from the American Heart Association. Circulation. 2015;131(4):e29–322.
Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–128.
Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci U S A. 1996;93(3):1156–60.
Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326(5959):1509–12.
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–21.
Jasin M, Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol. 2013;5(11):a012740.
Chiruvella KK, Liang Z, Wilson TE. Repair of double-strand breaks by end joining. Cold Spring Harb Perspect Biol. 2013;5(5):a012757.
Termglinchan V, Karakikes I, Seeger T et al. Current status of genome editing in cardiovascular medicine. In: Turksen K, editor. Genome editing. Cham: Springer International Publishing; 2016. p. 107–26.
• Karakikes I, et al. Correction of human phospholamban R14del mutation associated with cardiomyopathy using targeted nucleases and combination therapy. Nat Commun. 2015;6:6955. This proof-of-principle study describes how engineered nucleases can be used to correct a pathogenic mutation associated with dilated cardiomyopathy in vitro.
Ang YS, et al. Disease model of GATA4 mutation reveals transcription factor cooperativity in human cardiogenesis. Cell. 2016;167(7):1734–1749 e22.
Liang P, Sallam K, Wu H, Li Y, Itzhaki I, Garg P, et al. Patient-specific and genome-edited induced pluripotent stem cell-derived cardiomyocytes elucidate single-cell phenotype of Brugada syndrome. J Am Coll Cardiol. 2016;68(19):2086–96.
Karakikes I, Termglinchan V, Cepeda DA, Lee J, Diecke S, Hendel A, et al. A comprehensive TALEN-based knockout library for generating human-induced pluripotent stem cell-based models for cardiovascular diseases. Circ Res. 2017;120(10):1561–71.
Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21(2):121–31.
Pardo B, Gomez-Gonzalez B, Aguilera A. DNA repair in mammalian cells: DNA double-strand break repair: how to fix a broken relationship. Cell Mol Life Sci. 2009;66(6):1039–56.
Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33(5):538–42.
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–71.
•• Komor AC, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature, 2016;533(7603):420–4. This study demonstrates that genome editing can be used to make precise changes in DNA without causing double-strand breaks.
Yang L, Briggs AW, Chew WL, Mali P, Guell M, Aach J, et al. Engineering and optimising deaminase fusions for genome editing. Nat Commun. 2016;7:13330.
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017;551(7681):464–71.
Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–5.
Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351(6268):84–8.
Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550(7676):407–10.
Casini A, Olivieri M, Petris G, Montagna C, Reginato G, Maule G, et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat Biotechnol. 2018;36(3):265–71.
Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. In: Nature, vol. 556; 2018. p. 57–63.
Zacchigna S, Zentilin L, Giacca M. Adeno-associated virus vectors as therapeutic and investigational tools in the cardiovascular system. Circ Res. 2014;114(11):1827–46.
Lau CH, Suh Y. In vivo genome editing in animals using AAV-CRISPR system: applications to translational research of human disease. F1000Res. 2017;6:2153.
El Refaey M, et al. In vivo genome editing restores dystrophin expression and cardiac function in dystrophic mice. Circ Res. 2017;121(8):923–9.
Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351(6271):400–3.
• Nelson CE, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351(6271):403–7. In this study, the gene defect responsible for Duchenne muscular dystrophy was corrected in a mouse model by AAV-mediated genome editing in vivo.
Author information
Authors and Affiliations
Ethics declarations
Conflict of Interest
Balpreet Kaur, Isaac Perea Gil, and Ioannis Karakikes declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Regenerative Medicine
Rights and permissions
About this article
Cite this article
Kaur, B., Perea-Gil, I. & Karakikes, I. Recent Progress in Genome Editing Approaches for Inherited Cardiovascular Diseases. Curr Cardiol Rep 20, 58 (2018). https://doi.org/10.1007/s11886-018-0998-3
Published:
DOI: https://doi.org/10.1007/s11886-018-0998-3