Abstract
Purpose of Review
Eosinophilic esophagitis (EoE) is an allergic inflammatory esophageal disorder with a complex underlying genetic and molecular etiology. The interest of the scientific community in EoE has grown considerably over the past three decades, and the understanding of the genetic and molecular mechanisms involved in this disease has greatly increased.
Recent Findings
In this article, we aim to provide both historic aspects and updates on the recent genetic and molecular advances in the understanding of EoE. Although EoE is a relatively newly described disorder, much progress has been made toward identifying the genetic and molecular factors contributing to the disease pathogenesis by a variety of approaches with next-generation sequencing technologies, including genome-wide association study, whole exome sequencing, and bulk and single-cell RNA sequencing.
Summary
This review highlights the multifaceted impacts of various findings that have shaped the current molecular and genetic landscape of EoE, providing insights that facilitate further understanding of the disease process.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Eosinophilic esophagitis (EoE) is a chronic inflammatory disorder of the esophagus characterized by eosinophilic infiltration and esophageal symptoms associated with esophageal dysfunction [1]. Over the past three decades, the incidence of EoE has been increasing dramatically, with a current overall prevalence estimated at 56.3 cases per 100,000 persons [2]. EoE is costly, with annual health care estimates ranging from 500 million to 1.4 billion dollars [3, 4]. Epidemiologic studies have revealed a consistent gender discrepancy, with males affected three to four times more commonly than females. EoE is also more frequently reported in individuals who self-identify as white compared with other races, indicating a genetic component in this disease [1, 5].
EoE is a type 2 immune disease that often co-occurs with atopic diseases [1]. As such, EoE is characterized by increased levels of the type 2 cytokines, including IL-5 and IL-13, in the esophagus [6]. Furthermore, there is marked overexpression of approximately 1% of the human genome in the esophagus of patients with active EoE. This EoE transcriptome is highly conserved among patients regardless of sex, age, or history of atopy.
The pathology of EoE has been uncovered in the last three decades; cumulative evidence indicates that the risk of EoE involves the complex interplay of genetic and immunologic components [7]. Herein, we review the available evidence concerning both historic aspects and updates on the recent genetic and molecular advances in the understanding of EoE, including recent knowledge from genome-wide association studies (GWAS), whole exome sequencing (WES), and bulk and single-cell RNA sequencing (Fig. 1).
Schematic summary for the timeline of relevant events for the progress of genetic and molecular progress in EoE. Created with BioRender.com. EDP, EoE Diagnostic Panel; EGID, eosinophilic gastrointestinal disease; EoG, eosinophilic gastritis; EoD, eosinophilic duodenitis; EoC, eosinophilic colitis; GWAS, genome-wide association study; WES, whole exome sequencing
Genetic Contributors in EoE
Familial Components and Twin Study
The high prevalence of EoE among individuals of European descent and males that was noted across multiple epidemiologic studies implicates a genetic contribution to the etiology of disease. EoE occurs predominantly in individuals self-identifying as white, and males constitute approximately three quarters of all cases [2, 8]. In a 2009 study of 620 patients with EoE enrolled over a period of 14 years, 90% reported white ancestry and 75% were males [9]. Additionally, EoE often occurs in multiple family members in a non-Mendelian pattern, indicating that the heritable component of EoE is likely complex in nature [10, 11]. Indeed, a family history of EoE in first-degree relatives has been reported in at least 6.8% of patients with EoE [12]. In a 2014 study of 914 patients with EoE and family members, the relative risk ratios (RRR) for EoE in family members ranged from 10 to 64, with highest values for brothers (RRR = 64.0) and fathers (RRR = 42.9) [13]. Overall, nearly 2% of patients with EoE had a relative with EoE [13]. This is a substantial rate of heritability, particularly given that the RRR in related diseases, such as asthma and food allergy, is about doubled [13]. Furthermore, analyses of EoE concordance in twins have provided important information about the genetic basis of EoE. Alexander et al. reported an approximately 41% concordance between monozygotic twins and 22% concordance between dizygotic twins [13]. In contrast, concordance in siblings is 2.4% [13]. Collectively, these findings suggest that EoE clusters in families and can be attributed to both common family environment and additive genetic heritability [13]. Thus, early life exposures may prime genetically susceptible individuals to develop EoE.
Genetic, Mendelian, and Autosomal Disorders
Despite the frequent observation of non-Mendelian inheritance patterns in multiple family members with EoE, a subset of patients develop EoE in conjunction with another genetic condition, particularly connective tissue disorders with hypermobility syndromes, including Loeys-Dietz syndrome and Ehlers-Danlos syndrome, hypermobility type [14, 15]. A common denominator between these 2 conditions is the increased production and/or signaling of TGF-β, which may lead to increased smooth muscle contractility, tissue remodeling, and type 2 immune responses [16, 17]. Additionally, a loss-of-function mutation in ERBB2-interacting protein (ERBIN), which negatively regulates TGF-β signaling, was identified as a pathway associated with increased TGF-β production [18].
EoE is also associated with several Mendelian diseases, including PTEN hamartoma tumor syndrome (PHTS), hyper-IgE syndromes, and SAM syndrome [19,20,21,22]. In PHTS, the phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) signaling pathway is inhibited due to the mutation of PTEN, which also disrupts the esophageal immune system and is related to EoE susceptibility [19]. In autosomal dominant hyper-IgE syndromes, due to the mutations of STAT3, dysregulated response to IL-6 and possibly IL-5 contribute to EoE; in autosomal recessive hyper-IgE syndrome, loss-of-function in DOCK8 is considered to cause loss of T cell homeostasis and/or lack of durable secondary antibody response against specific antigens [21, 22]. Homozygous mutations in desmoglein 1 (DSG1) or desmoplakin (DSP), which disrupt the esophageal barrier, are observed in SAM syndrome [22]. Furthermore, EoE was associated with Netherton’s syndrome, a disorder caused by autosomal dominant mutations in the protease inhibitor SPINK5, which are typically expressed in the skin [23]. Notably, most of the genes known to be involved in these conditions were associated with EoE by recently reported transcriptome analysis (Table 1).
Finally, EoE has been associated with a variety of autoimmune conditions, although the mode of inheritance is not always straightforward. These associated conditions include Hashimoto’s thyroiditis, rheumatoid arthritis, celiac disease, inflammatory bowel disease, combined variable immunodeficiency, multiple sclerosis, and Sjögren’s syndrome [24].
Common Genetic Variants
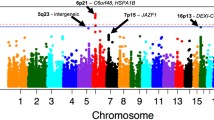
According to the genome-wide association studies (GWAS) reported from 2010 to 2019, genetic variants at 4 loci have consistently been found at genome-wide significance (5q22 [TSLP/WDR36], 2p23 [CAPN14], 11q13 [LRRC32/C11orf30], and 12q13 [STAT6]) (Table 2) [25,26,27]. Rothenberg et al. first conducted a GWAS for EoE and identified a genetic variant at 5q22 [TSLP/WDR36] [25]. They described how activated epithelial cells in EoE release TSLP, which regulates dendritic cell–mediated Th2 responses following IL-13 stimulation. In 2014, Kottyan et al. newly identified association at 2p23, which encodes CAPN14 [26]. Their experiment further demonstrated that CAPN14 is specifically expressed in esophageal epithelium, dynamically upregulated by IL-13, and involved in epithelial homeostasis and repair. Subsequently, Sleiman et al. also reported additional significant genome-wide associations at 4 loci [27], representing 4 genes (c11orf30, STAT6, ANKRD27, CAPN14). c11orf30 and STAT6 are associated with both atopic and autoimmune diseases although conditional analyses of sensitization status at the STAT6 locus indicate that the observed association with EoE is independent of sensitization. STAT6 is a key player in the IL-4 and IL-13 pathways and controls the Th2 response. In contrast to c11orf30 and STAT6, CAPN14 and ANKRD27 are at EoE-specific loci. CAPN14 encodes a calpain whose expression is highly enriched in the esophagus, and ANKRD27 regulates the trafficking of melanogenic enzymes to epidermal melanocytes. In a recent study, significant genome-wide associations with EoE were also found at 16p13 (CLEC16A) [28].
Given the limited sample size of prior GWAS reported from 2010 to 2019 [25,26,27,28], meta-analyses were recently undertaken to boost the statistical power to detect genetic variants underlying EoE. Kottyan et al. conducted a meta-analysis to replicate known and suggestive EoE genetic loci, which showed another 7 loci with suggestive significance (P < 10−6): 1p31, 5q23, 6q15, 6q21, 8p21, 17q12, and 22q13 [29]. From these risk loci, including ones identified by replication of known and suggestive EoE genetic risk loci previously reported, 13 protein-coding EoE candidate risk genes were expressed in a genotype-dependent manner. Expression quantitative trait loci (eQTL)–associated genes near identified EoE genetic risk variants were expressed in cell types relevant in EoE pathogenesis, including esophageal epithelial cells and immune cells [29]. Chang et al. recently conducted the most extensive GWAS to date, comprising 1930 affected European subjects and 13,634 controls. Their study identified 11 novel, genome-wide significant loci, including 5q31.1 (rs2106984, RAD50), 15q22.2 (rs2279293, RORA), and 15q23 (rs56062135, SMAD3), which have been previously associated with allergic conditions [30]. Furthermore, considering observed sex differences in EoE, they undertook meta-analyses of sex-stratified GWAS to identify sex-specific loci. Meta-analysis of males identified 7 genome-wide significant loci, of which the low-frequency variant loci at 3q22.1 (rs554318837, CPNE4) and 7p13 (rs188483654, URGCP) were male specific. Meta-analysis of females found 2 low-frequency variant loci at 7q22.3 (rs147307036, NAMPT) and 10p11.21 (rs191051238, CCNY) and a common variant locus at 9p24.1 (rs62541556, JAK2), all reaching genome-wide significance [31]. As described above, multiple genetic risk factors linked to EoE have been identified; however, these studies have focused primarily on populations of European ancestry. Recent GWAS and admixture mapping for EoE in Black or African American populations identified an African ancestry–specific genetic susceptibility locus at 1p22.3, 9p13.3, and 12q24.23, providing evidence of ancestral specific inheritance of EoE [32]. The GWAS findings showed that only three specific regions, 15q22.2 (RORA), 9p24.1 (JAK2), and 15q13.3 (LINC02352-KLF13), were first discovered in European ancestry population studies and subsequently observed in the African American population, underscoring the importance of using population-specific genomic resources in genetic research on EoE. Reported risk loci for EoE are summarized in Table 2.
The interplay of gene–gene interaction, particularly between IL4 and TSLP, and gene-environment interactions was demonstrated to contribute to the genetic susceptibility of EoE [33]. This susceptibility is mediated by EoE-specific genes, including but not limited to TSLP and CAPN14, and general atopic disease loci (encoding IL4/KIF3A), which may exert synergistic effects. This interaction may help explain the high rate of atopy in patients with EoE. A separate study identified genetic epistasis between TSLP variants and the uPA-encoding gene, PLAU [34]. Notably, uPA mediates eosinophil activation downstream from the loss of SPINK7, and the pro-inflammatory responses by excessive cytokine production, including TSLP, are triggered by lack of esophageal SPINK7. Moreover, the genetic association between 2p23 (CAPN14) and EoE was largely driven by patients with early disease onset and presentation [35].
Although responses to drugs are diverse and complex, genetic factors have been associated with variable responsiveness to individual drugs. The genetic variant STAT6 rs324011 was shown to synergize with CYP2C19*17 to predict the responsiveness to proton pump inhibitor (PPI) in pediatric patients with EoE [36]. Pediatric patients who initially respond to PPI therapy and carry STAT6 variants rs324011, rs167769, or rs12368672 were at increased risk of poor long-term response to PPI maintenance therapy [37]. Thus, individual patient genetic variations have been shown to influence response to PPIs in EoE. It was suggested that in the future, precision medicine may enable appropriate patient and dose selection of PPI to optimize efficacy and minimize toxicity [38].
The combined genetic burden of identified EoE risk variants can be utilized to create a patient-based assessment of genetic risk to help identify patients at the highest risk of developing EoE. According to Kottyan et al.’s findings, individuals with EoE exhibited a greater genetic risk burden from EoE-associated genetic variants than did the control group [29].
Collectively, there has been a growing understanding of the common genetic variants associated with EoE pathogenesis. Genetic variants associated with EoE pathogenesis have received increasing clarity in the last two decades. Genetic risk scoring will provide an invaluable tool for the future of preventive medicine, and identifying EoE-associated genetic variants advances the potential for targeted precision medicine.
Rare Genetic Variants
Although the number of common genetic variants associated with EoE continues to grow, a recent emphasis has been placed on identifying causal variants that alter the expression or protein function of the associated genes. For GWAS on low-prevalence diseases such as EoE, rare variants are excluded due to the lack of statistical power [11]. Accordingly, protein-coding variants of EoE have been analyzed in recent whole exome sequencing (WES) studies. Rochman et al. utilized WES to analyze 33 unrelated patients with EoE and identified 39 rare mutations in 18 esophagus-specific genes that were transcriptionally altered in the disease. Functional enrichment analysis identified proteolytic activity and tissue differentiation as the most significant pathways associated with these genes [39]. Sherrill et al. conducted a WES study of 37 unrelated families containing 63 patients with EoE (25 trios and 12 multiplex families) and unaffected family members. They identified an enrichment of rare, damaging variation in dehydrogenase E1 and transketolase domain-containing I (DHTKDI). Genetic insult of DHTKD1 and oxoglutarate dehydrogenase L (OGDHL) drives mitochondrial dysfunction and allergic inflammation involving viperin that has been shown to promote Th2 cytokine production in T cells [40]. Subsequently, we analyzed WES of 62 unrelated families (a multi-generation EoE pedigree and 61 additional multiplex families with EoE) and identified a series of rare variants in the genes encoding desmosome-associated proteins (DSP and PPL) in 21% of the multiplex families [41]. These variants affected barrier integrity, cell motility, and RhoGTPase activity in esophageal epithelial cells and had increased susceptibility to calpain-14–mediated degradation, providing a deeper mechanistic understanding of tissue-specific allergic responses.
There are some case reports that identified suggestive rare variants associated with EoE; (1) cases of STAT6 variants with early-life onset of profound allergic immune dysregulation, treatment-resistant allergic disorders, including EoE [42]; (2) STAT1 gain-of-function as a primary immune dysregulatory disorder, and (3) members of a multi-generation family with treatment-refractory EoE [43].
Taken together, WES studies identified several protein-coding rare variants of EoE, although most variants identified by GWAS are in noncoding regions. The aforementioned evidence supports the independent action of these genetic variants on the esophageal epithelium in EoE. These findings also indicate that the disease risk of EoE is multifactorial and involves the complex interaction of genetic and environmental factors.
Challenges to EoE Genetic Studies
Even with considerable progress in investigating the genetic basis of EoE, there are still areas to explore and improve, such as population structure, epistasis and gene-environment interactions, data-related issues (e.g., ancestry diversity and rare genetic variants), and specific challenges related to heritability estimates and polygenic risk scores [11, 44, 45]. In addition to statistical robustness, investigating biological function is also important [46]. Future studies are warranted to overcome these barriers with further advances in technology (e.g., high-throughput sequencing and data processing, artificial intelligence) and resources (e.g., biobanks, data sharing).
Molecular Contributors in EoE
Identification of EoE Transcriptome
For the molecular profiling, advances in the technology have facilitated the identification of a distinct transcriptional signature in EoE. At the onset of EoE research, the utilization of microarray chip technology, which arrays a large set of genes in a compact and regular manner, had been instrumental in this endeavor. Gene expression profiling of esophageal biopsy specimens by microarray technology revealed the EoE transcriptome, comprising approximately 574 genes that can distinguish patients with EoE from healthy control subjects and patients with chronic esophagitis. Gene expression profiles are stable regardless of patients’ gender, age, and/or history of allergies and are also strongly correlated with esophageal eosinophil levels [47]. It is notable that other research groups replicated similar gene expression profiles in patients with EoE [48, 49]. Of particular interest, the eosinophil chemoattractant eotaxin-3 (CCL26) was identified as the most highly expressed gene in the esophagus of patients with EoE (i.e., 53-fold increase in EoE compared to controls). Additionally, a genomic hotspot on chromosome 1q21, which encodes the epidermal differentiation complex, was identified as having the strongest transcriptional changes in EoE [50]. This region contains genes involved in squamous cell differentiation, including filaggrin (FLG). Notably, the expression of these genes is markedly decreased in EoE, suggesting a disruption of barrier function associated with loss of epithelial cell differentiation. Furthermore, a subset of the EoE transcriptome (approximately 2%) was found to remain unchanged after clinical remission with glucocorticoids, including the genes cadherin-like 26 (CDH26), uroplakin 1B (UPK1B), periostin (POSTN), and desmoglein-1 (DSG1), which are involved in the regulation of homeostatic and pathogenic responses in the epithelium [51, 52]. More recently, RNA sequencing has emerged as an attractive alternative to traditional microarray platforms for conducting transcriptional profiling and has identified 1607 transcripts dysregulated in EoE (1096 upregulated and 511 downregulated) [53], further expanding our understanding of the EoE transcriptome. In patients with EoE, it has been observed that approximately 39% of esophageal enriched/specific genes were altered, with a majority of these alterations resulting in downregulation (~90%). EoE pathophysiology, with its significant loss of esophageal tissue differentiation, is considered essential and specific [39]. Collectively, transcriptome analysis technologies have established EoE as a distinct disease with unique molecular profiles and have provided valuable insights into key molecules that contribute to various changes observed in EoE.
EoE Diagnostic Panel
The discovery of the EoE transcriptome has made substantial strides in the diagnosis of the disease. Given the cost and technical barrier of a genome-wide expression approach, the real-time PCR–-based array (e.g., TaqMan-qPCR–based, low-density array system) has been utilized in EoE, representing the EoE Diagnostic Panel (EDP), which is a set of informative esophageal transcripts with major gene categories (i.e., “cell adhesion,” “epithelial related,” “inflammatory process,” “remodeling,” “eosinophil/mast cell,” “chemokine and cytokine”). Currently, the EDP has been widely used for EoE diagnosis and monitoring, particularly in demonstrating significant reversibility in nearly all EoE-related genes, including decreased expression of eosinophil chemoattractants (e.g., CCL26) and mast cell signature genes (e.g., CPA3) and increased expression of barrier genes (e.g., DSG1) [54]. Notably, the EDP has high sensitivity and specificity (92–100% and 96–100%, respectively), enabling it to effectively differentiate EoE from controls, including those with gastroesophageal reflux disease (GERD) and other digestive diseases [54]. Furthermore, the EDP has been established as a minimally invasive and accurate diagnostic tool. Despite the patchy distribution of tissue eosinophils, the EDP can predict inflammation along its spatial length with a single distal biopsy [55]. Another advantage of the EDP is that it can utilize RNA from formalin-fixed or paraffin-embedded tissue for diagnosis [54], eliminating the need for additional biopsies for RNA extraction. Moreover, the EDP has been found to possess predictive ability for subclinical histology patients, underscoring the importance of closely monitoring these patients from an early stage [54]. The EDP has also revealed that the transcriptome of EoE and proton pump inhibitor–responsive esophageal eosinophilia (PPI-REE) are nearly overlapping, and the same holds true for PPI’s effect on the epithelium in inhibiting transcriptomic processes involved in cell proliferation and IL-13–mediated responses [56]. At present, PPI-REE is an entity of EoE distinct from GERD, and PPI is a treatment option for EoE, not a diagnostic tool to differentiate EoE from PPI-REE [1].
Recently, several studies have been conducted to correlate clinical information with the EDP: (1) the expression of TRPV1 and mast cells, as well as associated genes (e.g., CPA3 and HPGDS), was proposed to be linked to chest pain [57]; (2) unbiased clustering of the molecular signature classified EoE into three endotypes (referred to as EoEe1-3, respectively) comprising a relatively mild phenotype, an inflammatory and steroid-refractory phenotype due to type 2 immune response, and a severe fibrostenotic phenotype [58], and (3) endothelial TSPAN12 contributed to fibrostenotic EoE, and this loss modulated endothelial dysfunction and gene expression, leading to remodeling [59]. Dysregulation of TSPAN12 was also replicated in a recent RNA sequencing study in fibrostenotic EoE [60]. Taken together, with the simplification of EoE diagnosis by the EDP, our understanding of EoE in terms of pathophysiology, subtypes, and gene expression has advanced one after another. However, further investigation of gene expression beyond the current EDP set will be necessary for continued advancements in understanding and practice in the future.
Recent Advance of EoE Transcriptome by Single-Cell RNA Sequencing
Single-cell transcriptome analysis techniques have enabled the elucidation of heterogeneous cell subtypes and rare cells, reconstructing cell developmental trajectories and modeling transcriptional dynamics that were previously masked by bulk measurements [61]. Single-cell RNA sequencing has allowed us to learn more about disease-specific microenvironments, heterogeneity, and immune interactions in various conditions, such as tumors. In the context of EoE, T cell heterogeneity was reported in 2019 and was demonstrated to closely associate with allergic diseases [62]. A total of 8 heterogeneous T cell subclasses were identified in the esophagus, with increased concentrations of disease-associated CD4 + populations, such as T7 (i.e., putative T regulatory cells [FOXP3 +]) and T8 (i.e., effector Th2-like cells [GATA3 +]), found in active EoE. A significant proportion (~30%) of CD4 + T effector cells in the active state showed Th2 cytokine production. IL-13 was highly expressed in most Th2 cells, whereas IL-4 and IL-5 were expressed to a lesser extent. In addition, esophageal eosinophils were present only during active disease, whereas some esophageal mast cells were a prominent cellular source of inflammatory mediators during active disease and remission [63]. In a recent study, single-cell analysis of human esophageal epithelial cells in homeostasis and EoE provided a global view of the cellular and molecular mechanisms of the esophageal epithelium in homeostatic and allergic inflammatory conditions at the single-cell level [64]. Previous studies based on single-cell RNA sequencing have shown that eosinophils and other granulocytes have low RNA content and high levels of ribonucleases, which limits their detection ability [65]. However, another technique (i.e., Seq-Well) was recently used to profile key allergic mediators, such as tissue-resident eosinophils and pathogenic effector Th2 (peTh2) cells present in the esophageal and duodenal biopsies of patients with EoE [65].
Recent Advances of Non-esophageal EGID Transcriptomes
Transcriptome analysis has dramatically advanced our understanding of the pathogenesis in EoE. In recent years, RNA sequencing has also facilitated the identification of tissue-specific transcriptomes in other eosinophilic gastrointestinal disorders (EGIDs), such as eosinophilic gastritis (EoG) [66,67,68], eosinophilic colitis (EoC) [69, 70], and, more recently, eosinophilic duodenitis (EoD) [71]. These tissue-specific transcriptomes have substantiated the validation of disease concepts and diagnostic criteria. Although the EoG transcriptome revealed the same underlying type 2 allergic inflammation as EoE, the respective EoE and EoG transcriptomes overlapped by only a few percent, suggesting that EoG has a unique pathophysiology [66, 68]. Molecular validation of histologic diagnostic criteria (≥ 30 eosinophils/HPF and ≥ 5 HPF) was also demonstrated, suggesting the possibility of a molecular diagnosis in 63% of cases, even in more ambiguous cases (≥ 30 eosinophils/HPF and 1–4 HPF) [68]. The EoC transcriptome showed that EoC is an independent disease with unique molecular profiles compared to those of healthy subjects and patients with inflammatory bowel disease (IBD) [70]. A higher proportion of the EoC transcriptome correlates with local eosinophil counts in patients with EoC than IBD, and molecularly, EoC is strongly associated with eosinophil infiltration. Notably, the EoE and EoG transcriptomes only overlap in 9 genes (1%) with the EoC transcriptome, suggesting differing pathogenesis in EGID of the lower gastrointestinal tract [70]. There was no increase in type 2 cytokines (IL13, IL4, IL5) in EoC, and thus little evidence supporting strong type 2 allergic inflammation in EoC [70]. Further elucidation of the EGID transcriptomes promises the discovery of disease biomarkers and therapeutic targets, which will contribute to clinical applications, such as precision medicine and personalized diagnosis and treatment in the future.
Challenges to EoE Molecular Studies
Despite the recent advancements of molecular findings in EoE and other EGID, knowledge about spatial transcriptomics and omics is insufficient, and there are unmet needs regarding molecular data for clinical application. With the progress of next-generation sequencing technology, single-cell RNA sequencing has gradually replaced conventional bulk RNA sequencing to provide new and important findings. Although single-cell RNA sequencing is unable to analyze positional information, the advent of spatial transcriptomics [72, 73], an alternate approach synthesizing single-cell RNA sequencing and spatial genomics, has emerged as a potential solution; spatial transcriptomics enables visualization of RNA transcripts at the resolution of a single cell and can assign cell types (identified by the mRNA readouts) to their locations in histologic sections or determine subcellular localization of mRNA molecules. Similar to this synthesis of approaches in spatial transcriptomics, the further development of molecular biological research necessitates integrating not only molecular analyses, but also other omics, such as genome, proteome, and metabolome analyses. In terms of clinical application, previous molecular data revealed several endotypes of EoE [58, 74]. Recent anti–IL-13 antibody therapies and other treatments have demonstrated significant effects, yet therapeutic success has not been achieved in all patients [75,76,77]. Therefore, further studies using genome-wide approaches could advance a more accurate and precise stratification of patient population in EoE, which is pivotal for appropriate entry into therapeutic trials, especially in the context of new biologics targeting specific pathways. Furthermore, the recent remarkable progress in artificial intelligence may provide a unique opportunity for creating an efficient analytic platform in combination with RNA sequencing technology in the near future [78, 79].
Conclusion
EoE is an increasingly prevalent disease with the potential for significant implications for quality of life of patients. We have reviewed the known genetic and molecular mechanisms involved in this disease.
Early work with population-based studies has helped to establish the combined genetic and environmental predispositions at play in the development of EoE. Further, recent GWAS and WES studies have identified significant genome-wide associations at particular genetic loci and genetic risk variants that may be beneficial in identifying individuals at highest risk for disease. These advances aid in the understanding of genetic risk factors, their interactions with environmental factors, and targeted treatment of disease. Advances in technology have also aided in the identification of the distinct transcriptional signature in EoE. Currently, the EDP has been particularly useful in diagnosing EoE, demonstrating significant reversibility in nearly all EoE-related genes, and correlating clinical information to define distinct EoE endotypes. RNA sequencing has also enabled identification of tissue-specific transcriptomes of other EGIDs and their overlap, or lack thereof, with EoE.
Looking forward, recent and ongoing technological advances will enable further discovery of the genetic and molecular mechanisms involved in EoE, increasing the potential for targeted approaches to diagnosis and treatment.
References
Dellon ES, Liacouras CA, Molina-Infante J, Furuta GT, Spergel JM, Zevit N, et al. Updated international consensus diagnostic criteria for eosinophilic esophagitis: proceedings of the AGREE conference. Gastroenterology. 2018;155:1022–1033 e10.
Dellon ES, Jensen ET, Martin CF, Shaheen NJ, Kappelman MD. Prevalence of eosinophilic esophagitis in the United States. Clin Gastroenterol Hepatol. 2014;12:589-96.e1.
Jensen ET, Kappelman MD, Martin CF, Dellon ES. Health-care utilization, costs, and the burden of disease related to eosinophilic esophagitis in the United States. Am J Gastroenterol. 2015;110:626–32.
Eke R, Dellon ES. Hospitalization trends and determinants of inpatient costs for eosinophilic esophagitis patients in the United States: results from the Nationwide Inpatient Sample analysis. Ann Gastroenterol Hepatol. 2021;34:643–50.
Sperry SL, Woosley JT, Shaheen NJ, Dellon ES. Influence of race and gender on the presentation of eosinophilic esophagitis. Am J Gastroenterol. 2012;107:215–21.
Rothenberg ME. Molecular, genetic, and cellular bases for treating eosinophilic esophagitis. Gastroenterology. 2015;148:1143–57.
Lyles J, Rothenberg M. Role of genetics, environment, and their interactions in the pathogenesis of eosinophilic esophagitis. Curr Opin Immunol. 2019;60:46–53.
Molina-Infante J, Gonzalez-Cordero PL, Ferreira-Nossa HC, Mata-Romero P, Lucendo AJ, Arias A. Rising incidence and prevalence of adult eosinophilic esophagitis in midwestern Spain (2007–2016). United Euro Gastroenterol J. 2018;6:29–37.
Spergel JM, Brown-Whitehorn TF, Beausoleil JL, Franciosi J, Shuker M, Verma R, et al. 14 years of eosinophilic esophagitis: clinical features and prognosis. J Pediatr Gastroenterol Nutr. 2009;48:30–6.
Sherrill JD, Blanchard C. Genetics of eosinophilic esophagitis. Dig Dis. 2014;32:22–9.
Kottyan LC, Parameswaran S, Weirauch MT, Rothenberg ME, Martin LJ. The genetic etiology of eosinophilic esophagitis. J Allergy Clin Immunol. 2020;145:9–15.
Noel RJ, Putnam PE, Rothenberg ME. Eosinophilic esophagitis. N Engl J Med. 2004;351:940–1.
Alexander ES, Martin LJ, Collins MH, Kottyan LC, Sucharew H, He H, et al. Twin and family studies reveal strong environmental and weaker genetic cues explaining heritability of eosinophilic esophagitis. J Allergy Clin Immunol. 2014;134:1084-1092.e1.
Abonia JP, Wen T, Stucke EM, Grotjan T, Griffith MS, Kemme KA, et al. High prevalence of eosinophilic esophagitis in patients with inherited connective tissue disorders. J Allergy Clin Immunol. 2013;132:378–86.
Frischmeyer-Guerrerio PA, Guerrerio AL, Oswald G, Chichester K, Myers L, Halushka MK, et al. TGFβ receptor mutations impose a strong predisposition for human allergic disease. Sci Transl Med. 2013;5:195ra94.
Aceves SS, Chen D, Newbury RO, Dohil R, Bastian JF, Broide DH. Mast cells infiltrate the esophageal smooth muscle in patients with eosinophilic esophagitis, express TGF-β1, and increase esophageal smooth muscle contraction. J Allergy Clin Immunol. 2010;126:1198-204.e4.
Beppu LY, Anilkumar AA, Newbury RO, Dohil R, Broide DH, Aceves SS. TGF-β1-induced phospholamban expression alters esophageal smooth muscle cell contraction in patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2014;134:1100-1107.e4.
Lyons JJ, Liu Y, Ma CA, Yu X, O’Connell MP, Lawrence MG, et al. ERBIN deficiency links STAT3 and TGF-β pathway defects with atopy in humans. J Exp Med. 2017;214:669–80.
Henderson CJ, Ngeow J, Collins MH, Martin LJ, Putnam PE, Abonia JP, et al. Increased prevalence of eosinophilic gastrointestinal disorders in pediatric PTEN hamartoma tumor syndromes. J Pediatr Gastroenterol Nutr. 2014;58:553–60.
Crosby K, Swender D, Chernin L, Hafez-Khayyata S, Ochs H, Tcheurekdjian H, et al. Signal transducer and activator of transcription 3 mutation with invasive eosinophilic disease. Allergy Rhinol. 2012;3:e94–7.
Lyons JJ, Sun G, Stone KD, Nelson C, Wisch L, O’Brien M, et al. Mendelian inheritance of elevated serum tryptase associated with atopy and connective tissue abnormalities. J Allergy Clin Immunol. 2014;133:1471–4.
Samuelov L, Sarig O, Harmon RM, Rapaport D, Ishida-Yamamoto A, Isakov O, et al. Desmoglein 1 deficiency results in severe dermatitis, multiple allergies and metabolic wasting. Nat Genet. 2013;45:1244–8.
Paluel-Marmont C, Bellon N, Barbet P, Leclerc-Mercier S, Hadj-Rabia S, Dupont C, et al. Eosinophilic esophagitis and colonic mucosal eosinophilia in Netherton syndrome. J Allergy Clin Immunol. 2017;139:2003-2005.e1.
Xue Z, Miller TL, Abramson L, Thakkar KP, Ketchem CJ, Reddy S, et al. Association of eosinophilic esophagitis with autoimmune and connective tissue disorders, and the impact on treatment response. Dis Esophagus. 2022;36:doac043.
Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet. 2010;42:289–91.
Kottyan LC, Davis BP, Sherrill JD, Liu K, Rochman M, Kaufman K, et al. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet. 2014;46:895–900.
Sleiman PMA, Wang M-L, Cianferoni A, Aceves S, Gonsalves N, Nadeau K, et al. GWAS identifies four novel eosinophilic esophagitis loci. Nat Commun. 2014;5:5593.
Kottyan LC, Maddox A, Braxton JR, Stucke EM, Mukkada V, Putnam PE, et al. Genetic variants at the 16p13 locus confer risk for eosinophilic esophagitis. Genes Immun. 2019;20:281–92.
Kottyan LC, Trimarchi MP, Lu X, Caldwell JM, Maddox A, Parameswaran S, et al. Replication and meta-analyses nominate numerous eosinophilic esophagitis risk genes. J Allergy Clin Immunol. 2021;147:255–66.
Ferreira MA, Vonk JM, Baurecht H, Marenholz I, Tian C, Hoffman JD, et al. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat Genet. 2017;49:1752–7.
Chang X, March M, Mentch F, Nguyen K, Glessner J, Qu H, et al. A genome-wide association meta-analysis identifies new eosinophilic esophagitis loci. J Allergy Clin Immunol. 2022;149:988–98.
Gautam Y, Caldwell J, Kottyan L, Chehade M, Dellon ES, Rothenberg ME, et al. Genome-wide admixture and association analysis identifies African ancestry-specific risk loci of eosinophilic esophagitis in African Americans. J Allergy Clin Immunol. 2022;S0091–6749:01474–9.
Martin LJ, He H, Collins MH, Abonia JP, Biagini Myers JM, Eby M, et al. Eosinophilic esophagitis (EoE) genetic susceptibility is mediated by synergistic interactions between EoE-specific and general atopic disease loci. J Allergy Clin Immunol. 2018;141:1690–8.
Azouz NP, Ynga-Durand MA, Caldwell JM, Jain A, Rochman M, Fischesser DM, et al. The antiprotease SPINK7 serves as an inhibitory checkpoint for esophageal epithelial inflammatory responses. Sci Transl Med. 2018;10:eaap9736.
Lyles JL, Martin LJ, Shoda T, Collins MH, Trimarchi MP, He H, et al. Very early onset eosinophilic esophagitis is common, responds to standard therapy, and demonstrates enrichment for CAPN14 genetic variants. J Allergy Clin Immunol. 2021;147:244-254.e6.
Mougey EB, Williams A, Coyne AJK, Gutiérrez-Junquera C, Fernández-Fernández S, Cilleruelo ML, et al. CYP2C19 and STAT6 variants influence the outcome of proton pump inhibitor therapy in pediatric eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2019;69:581–7.
Mougey EB, Nguyen V, Gutiérrez-Junquera C, Fernández-Fernández S, Cilleruelo ML, Rayo A, et al. STAT6 variants associate with relapse of eosinophilic esophagitis in patients receiving long-term proton pump inhibitor therapy. Clin Gastroenterol Hepatol. 2021;19:2046-2053.e2.
Franciosi JP, Mougey EB, Dellon ES, Gutierrez-Junquera C, Fernandez-Fernandez S, Venkatesh RD, et al. Proton pump inhibitor therapy for eosinophilic esophagitis: history, mechanisms, efficacy, and future directions. J Asthma Allergy. 2022;15:281–302.
Rochman M, Travers J, Miracle CE, Bedard MC, Wen T, Azouz NP, et al. Profound loss of esophageal tissue differentiation in patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2017;140:738-749.e3.
Sherrill JD, Kc K, Wang X, Wen T, Chamberlin A, Stucke EM, et al. Whole-exome sequencing uncovers oxidoreductases DHTKD1 and OGDHL as linkers between mitochondrial dysfunction and eosinophilic esophagitis. JCI Insight. 2018;3: e99922.
Shoda T, Kaufman KM, Wen T, Caldwell JM, Osswald GA, Purnima P, et al. Desmoplakin and periplakin genetically and functionally contribute to eosinophilic esophagitis. Nat Commun. 2021;12:6795.
Sharma M, Lu HY, Vaseghi-Shanjani M, Del Bel KL, Fornes O, van der Lee R, et al. Human germline heterozygous gain-of-function STAT6 variants cause severe allergic disease [Internet]. bioRxiv. 2022. Available from: https://www.medrxiv.org/content/10.1101/2022.04.25.22274265.abstract.
Scott O, Sharfe N, Dadi H, Vong L, Garkaby J, Abrego Fuentes L, et al. Case report: Eosinophilic esophagitis in a patient with a novel STAT1 gain-of-function pathogenic variant. Front Immunol. 2022;13: 801832.
Chang JW, Jensen ET, Dellon ES. Nature with nurture: the role of intrinsic genetic and extrinsic environmental factors on eosinophilic esophagitis. Curr Allergy Asthma Rep. 2022;22:163–70.
Brandes N, Weissbrod O, Linial M. Open problems in human trait genetics. Genome Biol. 2022;23:131.
MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–76.
Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–47.
Matoso A, Mukkada VA, Lu S, Monahan R, Cleveland K, Noble L, et al. Expression microarray analysis identifies novel epithelial-derived protein markers in eosinophilic esophagitis. Mod Pathol. 2013;26:665–76.
Shoda T, Morita H, Nomura I, Ishimura N, Ishihara S, Matsuda A, et al. Comparison of gene expression profiles in eosinophilic esophagitis (EoE) between Japan and Western countries. Allergol Int. 2015;64:260–5.
Blanchard C, Stucke EM, Burwinkel K, Caldwell JM, Collins MH, Ahrens A, et al. Coordinate interaction between IL-13 and epithelial differentiation cluster genes in eosinophilic esophagitis. J Immunol. 2010;184:4033–41.
Blanchard C, Mingler MK, Vicario M, Abonia JP, Wu YY, Lu TX, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120:1292–300.
Caldwell JM, Blanchard C, Collins MH, Putnam PE, Kaul A, Aceves SS, et al. Glucocorticoid-regulated genes in eosinophilic esophagitis: a role for FKBP51. J Allergy Clin Immunol. 2010;125:879-888.e8.
Sherrill JD, Kiran KC, Blanchard C, Stucke EM, Kemme KA, Collins MH, et al. Analysis and expansion of the eosinophilic esophagitis transcriptome by RNA sequencing. Genes Immun. 2014;15:361–9.
Wen T, Stucke EM, Grotjan TM, Kemme KA, Abonia JP, Putnam PE, et al. Molecular diagnosis of eosinophilic esophagitis by gene expression profiling. Gastroenterology. 2013;145:1289–99.
Min S, Shoda T, Wen T, Rothenberg ME. Diagnostic merits of the Eosinophilic Esophagitis Diagnostic Panel from a single esophageal biopsy. J Allergy Clin Immunol. 2022;149:782-787.e1.
Wen T, Dellon ES, Moawad FJ, Furuta GT, Aceves SS, Rothenberg ME. Transcriptome analysis of proton pump inhibitor–responsive esophageal eosinophilia reveals proton pump inhibitor–reversible allergic inflammation. J Allergy Clin Immunol. 2015;135:187-197.e4.
Zhang S, Shoda T, Aceves SS, Arva NC, Chehade M, Collins MH, et al. Mast cell-pain connection in eosinophilic esophagitis. Allergy. 2022;77:1895–9.
Shoda T, Wen T, Aceves SS, Abonia JP, Atkins D, Bonis PA, et al. Eosinophilic oesophagitis endotype classification by molecular, clinical, and histopathological analyses: a cross-sectional study. Lancet Gastroenterol Hepatol. 2018;3:477–88.
Shoda T, Wen T, Caldwell JM, Ben-Baruch Morgenstern N, Osswald GA, Rochman M, et al. Loss of endothelial TSPAN12 promotes fibrostenotic eosinophilic esophagitis via endothelial cell-fibroblast crosstalk. Gastroenterology. 2022;162:439–53.
Menard-Katcher C, Liu C, Galbraith MD, Benson T, Burger C, Dobias D, et al. Fibrostenotic eosinophilic esophagitis phenotype is defined by a proliferative gene signature. Allergy. 2023;78:579–83.
Liu S, Trapnell C. Single-cell transcriptome sequencing: recent advances and remaining challenges. F1000Res. 2016;5:F1000.
Wen T, Aronow BJ, Rochman Y, Rochman M, Kc K, Dexheimer PJ, et al. Single-cell RNA sequencing identifies inflammatory tissue T cells in eosinophilic esophagitis. J Clin Invest. 2019;129:2014–28.
Ben-Baruch Morgenstern N, Ballaban AY, Wen T, Shoda T, Caldwell JM, Kliewer K, et al. Single-cell RNA sequencing of mast cells in eosinophilic esophagitis reveals heterogeneity, local proliferation, and activation that persists in remission. J Allergy Clin Immunol. 2022;149:2062–77.
Rochman M, Wen T, Kotliar M, Dexheimer PJ, Ben-Baruch Morgenstern N, Caldwell JM, et al. Single-cell RNA-Seq of human esophageal epithelium in homeostasis and allergic inflammation. JCI Insight. 2022;7: e159093.
Morgan DM, Ruiter B, Smith NP, Tu AA, Monian B, Stone BE, et al. Clonally expanded, GPR15-expressing pathogenic effector TH2 cells are associated with eosinophilic esophagitis. Sci Immunol. 2021;6:eabi5586.
Caldwell JM, Collins MH, Stucke EM, Putnam PE, Franciosi JP, Kushner JP, et al. Histologic eosinophilic gastritis is a systemic disorder associated with blood and extragastric eosinophilia, TH2 immunity, and a unique gastric transcriptome. J Allergy Clin Immunol. 2014;134:1114–24.
Sato M, Shoda T, Shimizu H, Orihara K, Futamura K, Matsuda A, et al. Gene expression patterns in distinct endoscopic findings for eosinophilic gastritis in children. J Allergy Clin Immunol Pract. 2017;5:1639-1649.e2.
Shoda T, Wen T, Caldwell JM, Collins MH, Besse JA, Osswald GA, et al. Molecular, endoscopic, histologic, and circulating biomarker-based diagnosis of eosinophilic gastritis: Multi-site study. J Allergy Clin Immunol. 2020;145:255–69.
Shoda T, Matsuda A, Arai K, Shimizu H, Morita H, Orihara K, et al. Sera of patients with infantile eosinophilic gastroenteritis showed a specific increase in both thymic stromal lymphopoietin and IL-33 levels. J Allergy Clin Immunol. 2016;138:299–303.
Shoda T, Collins MH, Rochman M, Wen T, Caldwell JM, Mack LE, et al. Evaluating eosinophilic colitis as a unique disease using colonic molecular profiles: a multi-site study. Gastroenterology. 2022;162:1635–49.
Shoda T, Rochman M, Collins MH, Caldwell JM, Mack LE, Osswald GA, et al. Molecular analysis of duodenal eosinophilia. J Allergy Clin Immunol. In press.
Williams CG, Lee HJ, Asatsuma T, Vento-Tormo R, Haque A. An introduction to spatial transcriptomics for biomedical research. Genome Med. 2022;14:68.
Ahmed R, Zaman T, Chowdhury F, Mraiche F, Tariq M, Ahmad IS, et al. Single-cell RNA sequencing with spatial transcriptomics of cancer tissues. Int J Mol Sci. 2022;23:3042.
Sallis BF, Erkert L, Moñino-Romero S, Acar U, Wu R, Konnikova L, et al. An algorithm for the classification of mRNA patterns in eosinophilic esophagitis: Integration of machine learning. J Allergy Clin Immunol. 2018;141:1354-1364.e9.
Rothenberg ME, Wen T, Greenberg A, Alpan O, Enav B, Hirano I, et al. Intravenous anti-IL-13 mAb QAX576 for the treatment of eosinophilic esophagitis. J Allergy Clin Immunol. 2015;135:500–7.
Dellon ES, Rothenberg ME, Collins MH, Hirano I, Chehade M, Bredenoord AJ, et al. Dupilumab in adults and adolescents with eosinophilic esophagitis. N Engl J Med. 2022;387:2317–30.
Dellon ES, Spergel JM. Biologics in eosinophilic gastrointestinal diseases. Ann Allergy Asthma Immunol. 2023;130:21–7.
Larey A, Aknin E, Daniel N, Osswald GA, Caldwell JM, Rochman M, et al. Harnessing artificial intelligence to infer novel spatial biomarkers for the diagnosis of eosinophilic esophagitis. Front Med. 2022;9: 950728.
Römmele C, Mendel R, Barrett C, Kiesl H, Rauber D, Rückert T, et al. An artificial intelligence algorithm is highly accurate for detecting endoscopic features of eosinophilic esophagitis. Sci Rep. 2022;12:11115.
Funding
This study was supported by NIH grant K99/R00 AI158660 (to T.S.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hiroki Sato and Kasumi Osonoi are co-first authors.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sato, H., Osonoi, K., Sharlin, C.S. et al. Genetic and Molecular Contributors in Eosinophilic Esophagitis. Curr Allergy Asthma Rep 23, 255–266 (2023). https://doi.org/10.1007/s11882-023-01075-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11882-023-01075-0