Abstract
Atopic dermatitis (AD) is a common skin disease that affects a large proportion of the population worldwide. The incidence of AD has increased over the last several decades along with AD’s burden on the physical and psychological health of the patient and family. However, current advances in understanding the mechanisms behind the pathophysiology of AD are leading to a hopeful outlook for the future. Staphylococcus aureus (S. aureus) colonization on AD skin has been directly correlated to disease severity but the functions of other members of the skin bacterial community may be equally important. Applying knowledge gained from understanding the role of the skin microbiome in maintaining normal skin immune function, and addressing the detrimental consequences of microbial dysbiosis in driving inflammation, is a promising direction for development of new treatments. This review discusses current preclinical and clinical research focused on determining how the skin microbiome may influence the development of AD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Atopic dermatitis (AD) is a chronic allergic skin disease the includes frequent flares, and manifests with characteristic findings of dry, red, and pruritic skin in a typical distribution [1]. AD is one of the most common skin disorders and affects 5–20 % of infants worldwide with lesser prevalence in adulthood [2]. Although AD is rarely lethal, several studies have shown that AD frequently leads to a severely compromised way of life for patients and their families and thus is a major health care burden [3, 4]. Furthermore, AD often associates with other allergic diseases including asthma and allergic rhinitis [5], thus further compounding the importance of this disease. Finally, the number of infants and adults affected by AD is higher in industrialized nations and has also grown over the past several decades. Many suspect that a link to excessive hygiene in these industrialized regions leads to a lack of beneficial host immune education provided by microbes on or within the body [6, 7]. Understanding the mechanisms behind AD and ways to treat it are therefore extremely important and highly relevant topics in the fields of allergy and dermatology.
Both genetic and environmental factors play a substantial role in the onset of AD. Genome-wide scans have revealed that there are several shared chromosome loci among AD patients where gene expression has been altered. These studies, however, clearly demonstrate that AD is a complex disease with many potential genetic factors at play [1]. Specific genetic mutations that have been linked to AD Include genes involved in formation of the epidermal skin barrier such as filaggrin (FLG), genes involved in tight junctions (TJs), and the serine protease inhibitor Kazal-type 5 (SPINK5) [8–12]. Mutations in T helper cell 2 (Th2) cytokines IL-4 and IL-13 have also been correlated in AD pathogenesis [13, 14]. In most cases, data support that environmental factors including food and aeroallergens, as well as physical stressors including hydration and scratching, can play a substantial role in disease severity as well [15–17].
A complete and in-depth analysis of the hypothesized mechanisms that drive AD has been discussed previously [1, 18, 19]. The purpose of this review is to focus on the rapidly advancing information that has shown a clear role for the skin microbiome in controlling the clinical manifestations of this disease (Fig. 1). Determining how microorganisms that inhabit the skin regulate AD can help us to better understand both the mechanisms behind AD and provide new ways to apply therapeutic strategies to combat the disease.
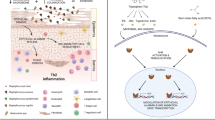
Dysbiosis of the skin microbiome in AD. Genetic defects in both physical (e.g., FLG and SPINK5) and immune (e.g., IL-4 and IL-13) skin barrier genes lead to increased skin barrier disruption. A decreased skin barrier response allows for increased susceptibility of the skin to allergens. Second, the skin becomes dry and itchy leading to more physical stress to the already damaged skin barrier. Overall, the decreased skin barrier leads to an increase in skin pH, altered keratinocyte adhesion properties, and both increased serine protease activity and inflammation. An acute Th2 response is thought to dampen certain antimicrobial peptide (AMP) responses as well. These events all play a role in dysbiosis of the skin microbiome during AD leading to increased colonization by S. aureus along with decreased overall microbial diversity. S. aureus colonization is speculated to increase AD severity through secreting a series of virulence factors that damage the skin barrier and increase inflammation. Other mechanisms pertaining to the role of dysbiosis of the skin microbiome in altering AD pathogenesis have yet to be determined
The Skin Microbiome and Changes in Atopic Dermatitis
Methods of Determining the Skin Microbiome Composition
The skin is colonized by numerous species of bacteria, fungi, and viruses that together are known as the skin microbiome. The composition of this community varies between individuals but varies even more depending on skin topography [20]. The use of next-generation DNA sequencing methods have allowed us to better understand both the composition and potential roles that these microorganisms, in particular bacteria, play on our skin. Importantly, as the technology has improved, so has our understanding of what specific bacteria are associated with disease.
16S ribosomal RNA (16S rRNA) partial DNA sequencing was initially used to determine the microbial composition on the skin at the genus level and has been followed by the development of full-length 16S rRNA DNA sequencing techniques to allow for more specific identification of the organism at the species level [20, 21]. These initial sequencing methods showed that our normal skin microbial flora consist of four common phyla of bacteria—Firmicutes, Proteobacteria, Bacteroidetes, and Actinobacteria (Table 1). Species of bacteria detected by 16S rRNA DNA sequencing show that hundreds of bacteria frequently colonize the skin, while culture techniques have found that the most commonly detected organisms include both Propionibacterium acnes and Staphylococcus epidermidis (S. epidermidis) among others [24–27]. DNA sequencing has also provided a useful way to track temporal shifts in the skin microbial flora during normal versus disease states, and this will be discussed later in the review. Beyond 16S rRNA DNA sequencing, recent efforts have begun to apply metagenomics [28••]. Metagenomics is a method that has been used for studying the composition of microbial communities in both the gut and oral cavities. It uses DNA sequencing of full-length microbial genomes allowing for identification of microbes at the strain level. This degree of specificity provides a tool for determining which bacterial strains are present on the skin. Knowing the specific strains, and not just the species of bacteria, permits one to explore the molecular mechanisms by which individual bacteria influence skin immune function.
Normal Skin Microbial Flora
The colonization of bacteria on specific regions on the skin depends upon the local skin environment including moisture levels, pH, and keratinocyte cell surface adhesion proteins. For instance, Staphylococcus and Corynebacterium species thrive in specific environments on the skin such as the sole of the foot and the popliteal fossa (back of the knee). Dry environments such as the volar forearm (inside forearm) and hypothenar palm (palm of hand) are more prone to harbor a mixed population of bacteria including both β-Proteobacteria and Flavobacteriales [20]. Environmental factors such as diet, age, and gender also play a role in the makeup of the skin microbial flora [29]. Infants are essentially born microbe free until exposure to the external environment allows for immediate colonization of their skin [30]. Throughout their lifespan, the microbial diversity can change. This environmental influence on our skin microbial flora means that each individual has a unique skin microbiome based in part upon life experiences and in part upon factors genetically predetermined by the host.
The skin microbiome has been investigated extensively for its role in communicating with the immune response. S. epidermidis, a highly abundant commensal bacteria, has been shown to modify innate inflammatory responses through a Toll-like receptor (TLR) cross talk mechanism. In this study, it was revealed that S. epidermidis could suppress TLR3-mediated keratinocyte inflammation due to skin injury by stimulating TLR2 with lipoteichoic acid (LTA) [31]. S. epidermidis can also increase T cell recruitment to the skin in germ-free mouse models and influence T cell maturation [32•]. Similarly, the host skin immune response can act on the microbiome. Keratinocytes in the epidermis detect microbial flora on the skin surface via pattern recognition receptors (PRRs). Such recognition by TLR2 leads to production of host antimicrobial peptides (AMPs) including the beta-defensins 2 and 3 (DEFΒ-2/DEFΒ-3) that defend the skin from microbial evasion. TLR2 activation in the skin by the microbial flora can also influence mast cell recruitment and increase tight barrier junctions leading to a stronger skin barrier [33–35]. Interestingly, commensal bacteria still survive on the skin surface despite TLR activation and AMP production by keratinocytes. These findings have led to the speculation that there must be a permissive relationship with common skin microbial flora. This prevents both a constitutively active immune response as well as allowing for commensal bacteria to help protect our skin from pathogens.
Recently, it was discovered that bacteria are normally present within the different layers of the skin and not just on the skin surface [36••]. This demonstrated that bacteria could penetrate the outer skin barrier and interact normally with many different cell types that exist in the deeper dermis. Based upon this finding, the capacity for the skin microbiome to influence immune homeostasis in normal and disease states becomes more apparent. In the future, it will be important to determine how microbes that have penetrated the skin barrier and that reside in either the epidermis, dermis, or subcutaneous layers communicate with the host and vice versa.
Commensal skin microbes seeking to retain their niche on specific skin sites have also developed defense mechanisms against other bacteria that may seek to colonize the skin surface. For example, some strains of S. epidermidis produce bacteriocins that are toxic to other bacterial species such as Staphylococcus aureus [37]. S. epidermidis can also target S. aureus through production of both the serine protease Esp and phenol-soluble modulins (PSMs) that prevent S. aureus biofilm formation and growth, respectively [38, 39]. Another common commensal bacteria on the skin, P. acnes, inhibits S. aureus growth as well [40]. Thus, commensal skin microbes can communicate with our skin in order to promote a stronger skin barrier and immune response, and have methods of their own to prevent invasion by pathogenic bacteria. Taken together, it is apparent that the normal skin microbiome has multiple essential actions to maintain immune homeostasis, and some commensal bacteria might even be considered as another immunocyte functioning in coordination with host-derived immune cells.
Dysbiosis of the Skin Microbiome in AD
The skin microbial flora can enter a state of dysbiosis, defined as a change in relative composition of the different microbes compared to normal, during a disease state. This is most well described in AD. It was first observed in the 1970s that there was increased S. aureus colonization on AD lesions [22]. Since then, multiple studies have observed this phenomenon, although the sheer complexity of how microbial composition changes in AD was not established until the use of 16S rRNA DNA sequencing [23•]. Sequencing revealed that AD patients have an overall increase of colonization by Staphylococcae species along with an overall decrease in the number of different types of bacteria (microbial diversity) on involved skin. It particular S. aureus colonization of the skin is increased, as previously shown, while S. epidermidis is increased to a lesser extent. These increases result in dysbiosis since the expansion of these populations is not accompanied by a proportional increase in the other bacteria found on normal skin.
The prevalence of S. aureus colonization on AD lesional skin varies, with some reporting the capacity to culture S. aureus from AD to be approximately 80–100 % while it is detectable with much lower frequency (5–20 %) in healthy individuals [41]. Increased colonization of S. aureus on AD skin is strongly linked to increased severity of the disease [41, 42]. The role of S. aureus colonization in increasing AD severity will be discussed below, and this association highlights why it is important to understand the mechanism of S. aureus virulence on AD skin in order to come up with more appropriate therapeutic solutions. However, the association is not clearly one of cause and effect as elimination of S. aureus is not a solution for this disease.
Dysbiosis of the Skin Microbiome in Other Allergic Skin Diseases
Besides AD, other rare skin diseases associated with allergy also have been reported to experience shifts in the skin microbiome. Netherton syndrome (NS) is a rare skin disease caused by a loss-of-function mutation in SPINK5, a key serine protease inhibitor in the epidermis [43]. Decreased SPINK5 expression leads to a hyperactive serine protease response and increased inflammation and desquamation, or stratum corneum shedding. This disease has been associated with both increased IgE levels in the blood as well as increased colonization by S. aureus although in depth 16S rRNA sequencing of NS patients has not yet been studied to reveal the total microbial composition [44, 45].
Hyper IgE syndrome (HIES), a primary immunodeficiency disease, represents another allergic skin disease where STAT3, a crucial signaling kinase, is mutated. HIES patients experience eczema as well as increased blood IgE levels. 16S rRNA sequencing has revealed that the skin microbiome of HIES patients has increased colonization of S. aureus along with, to a lesser extent, Corynebacterium [46•]. Thus, several allergic skin diseases, which display dry and flaky skin and a compromised skin barrier, display dysbiosis of the skin microbiome and in particular increased S. aureus colonization.
Microbial Mechanisms for Increased AD Severity
Physical Changes to the Skin Barrier and the Role of Antimicrobial Peptides in Skin Microbiome Dysbiosis
AD skin harbors a very different environment for bacterial growth than that of normal skin, and this may be the fundamental explanation for the dysbiosis observed in AD. A dysfunctional physical skin barrier leads to an increase in pH on the skin surface that favors S. aureus growth [47, 48]. Differentiated keratinocytes in the epidermis that are directly exposed to microbes have altered cell surface marker expression as well. In particular, it has been found that both fibronectin and fibrinogen expressions are increased in keratinocytes from AD patients and these markers directly bind to S. aureus in vitro [49, 50].
Immunological forces at play in the skin also influence changes to the skin microbial composition during AD. Endogenous AMPs are important for preventing pathogenic microbes from infecting the skin. Two main classes of AMPs in the skin are cathelicidin and DEFΒs [51, 52]. Both of these have the ability to kill S. aureus in vitro. However, in AD skin, cathelicidin as well as DEFΒ-2 and DEFΒ-3 expression is decreased in comparison to similarly inflamed psoriatic skin [53]. Second, keratinocyte models show that the AD-associated Th2 cytokines IL-4, IL-5, and IL-13 can decrease expression of host AMPs in vitro [54]. Besides Th2-specific cytokines, IL-10 has also been linked to decreasing AMP production [55]. Overall, the lack of host AMPs represents another reason for decreased microbial diversity and enhanced S. aureus growth on AD skin.
S. aureus Virulence Factors in AD Severity
It is proposed that the association of dysbiosis with AD disease severity is more complex than simply AD leading to changes in bacterial growth due to the factors described above. It is apparent that, in some cases, the bacterial community can further drive the disease. A well-known example of this is the hypothesis that S. aureus can increase the severity of AD by secreting a variety of virulence factors (Table 2). The most well studied of these are the superantigens (SAgs). SAgs function mechanistically by binding to the non-peptide groove of major histocompatibility complex class II (MHC-II) on antigen-presenting cells (APCs), including skin keratinocytes, as well as to the T cell receptor (TCR) β-chains. This leads to non-specific activation of approximately 5–20 % of all naïve T cells and systemic inflammation through production of pro-inflammatory cytokines including TNF-α and IL-1β [70–73]. SAgs produced by S. aureus include the staphylococcal enterotoxins (SEs) and toxic shock syndrome toxin 1 (TSST-1) [56–58]. Several studies have shown direct correlations between SAg-producing S. aureus strains on AD patients and AD severity. It was observed in one study that 57 % of AD patient S. aureus skin isolates produced SAgs as opposed to 33 % of control patient isolates. The SAg-positive S. aureus–colonized AD patients also displayed increased AD severity based upon Scoring Atopic Dermatitis (SCORAD) with 58 ± 17 as opposed to 41 ± 7 in control patients [57]. A second study revealed that elevated levels of SEA- and SEB-specific IgE antibodies in the blood correlated with increased AD severity [59].
Another factor that is proposed to increase AD severity is that of hemolysin-α or α-toxin. α-Toxin monomers form a heterodimer complex on the cell membrane that creates a porous channel leading to cell lysis. This has been shown in in vitro studies where α-toxin is severely toxic to keratinocytes [60, 61]. It has been proposed that Th2 cytokines can also increase α-toxin-induced keratinocyte toxicity [62]. Murine models with subcutaneous injections of α-toxin display increased inflammation at the site of injection [63]. Although high concentrations of α-toxin are toxic to host cells, low concentration can also stimulate keratinocyte cytokine production leading to increased inflammation as well [64].
Other S. aureus virulence factors also can possibly influence AD severity. δ-Toxin can target skin mast cells leading to degranulation and increased inflammation [65]. Protein A can induce TNF-α production in keratinocytes in a non-toxic manner [64]. Finally, S. aureus-secreted PSMs are also known to increase inflammation in keratinocytes [69]. Although much is known of the virulence factors discussed above, more are still being discovered. Understanding all of these virulence factors and how they affect the skin barrier during AD is crucial to our ability to combat S. aureus-mediated AD severity.
Role of Proteases in Microbial Dysbiosis and AD Severity
Maintaining the balance of activity between proteases and their inhibitors is essential to upholding a functional skin barrier. During AD, patients display an increase in serine protease activity. Specifically, the serine protease family known as the kallikreins (KLKs) is observed to have increased activity [74, 75]. Hyperactive KLK responses have been studied in NS patients and appear to be responsible for increased desquamation of the skin, altered cathelicidin and filaggrin processing, and increased PAR-2 activity and inflammation [76–79]. Thus, the increased KLK activity in AD skin helps create a compromised skin barrier and possibly aids in increasing S. aureus colonization of the skin.
Exogenous serine proteases released from S. aureus have also been studied for their role in damaging the skin barrier. The V8 serine protease increases desquamation of the epidermis in vitro as well as being able to alter skin barrier integrity in murine models [66, 67]. The serine-like proteases, exfoliative toxins A and B (ETA/B), have mostly been described for their role in inducing staphyloccocal scalded skin syndrome (SSSS) [68]. In SSSS, ETA/B cleave desmoglein-1, a corneodesmosomal adhesion protein that plays a crucial role in regulating desquamation. Excess shedding of the stratum corneum, or the uppermost epidermal layer, leads to a disrupted skin barrier and increased bacterial invasion. Although S. aureus serine proteases have not been directly linked to AD, their virulence can easily suggest another mechanism for enhanced AD severity.
Effect of Other Bacterial Strains on AD Pathogenesis
Besides increased S. aureus colonization of the skin in AD, 16S rRNA DNA sequencing has revealed increased S. epidermidis colonization. However, the function of increased S. epidermidis colonization on AD skin remains unclear. The presence of S. epidermidis on germ-free mouse skin can lead to an increase in T cell effector function in an IL-1-dependent manner. This seems to have a protective role against infections [32•]. S. epidermidis has also been shown in both murine and keratinocyte models to have a protective role through the amplification of endogenous AMPs [80, 81]. Finally, there is evidence that S. epidermidis can prevent S. aureus biofilm formation in the nasal cavities as well as produce its own AMPs to prevent other pathogens from colonizing the skin [37, 82]. Therefore, increased S. epidermidis colonization in AD skin may represent a way for the skin to naturally prevent increased S. aureus colonization in AD. However, the increased abundance of S. epidermidis in AD may not correlate with the protective and beneficial effects discussed above since these effects are strain-dependent, and it is not clear if these beneficial strains are active in AD. The use of metagenomics and species-specific bacterial sequencing should provide useful insight into the role of S. epidermidis on AD skin.
Recently, it has been reported that Corynebacterium bovis colonization is increased in an ADAM17 knockout model of AD in mice [83]. Increased Corynebacterium bovis colonization led to a robust Th2 response in the skin, a key characteristic of acute AD. 16S rRNA DNA sequencing has only revealed increased Corynebacterium in HIES though, and not in AD skin. Thus, this might serve as a way that dysbiosis of the skin microbiome can increase HIES severity by altering the immune response versus that seen in AD.
Therapeutic Strategies to Combat S. aureus on AD Skin
Since S. aureus colonization has been linked to increased AD severity, multiple therapeutic strategies have attempted to target specifically S. aureus colonization. The most common used treatments for S. aureus on AD skin include both topical/oral antibiotics and mild bleach baths. Studies conducted so far, however, have produced conflicting results. Many topical/oral antibiotic treatments can kill S. aureus in the short term on AD patients including muripocin, flucloxacillin, retapamulin, and cephalexin [84–86]. However, few of these patients see decreased AD severity. S. aureus colonization also typically relapses in AD patients after 4 to 8 weeks of antibiotic treatment [87]. A study in 2002 showed that a combination therapy of multiple antibiotics including muripocin, chlohexidine, and cephalexin improved clinical AD scores in 9 out of 10 patients with S. aureus colonization [88], but this is not the typical clinical experience. Corticosteriods, antiinflammatory reagents used as a staple for treatment of AD, have also been used in combination with antibiotics to improve AD severity [41]. This suggests that perhaps removal of S. aureus alone is not sufficient for AD treatment but antiinflammatory reagents are also necessary. Furthermore, there also are other important issues that deter using antibiotics as a treatment method for AD. The recent rise of methicillin-resistant S. aureus (MRSA) strains and their ability to colonize AD skin is a clear demonstration of one major issue [89]. Antibiotic therapy for AD patients is also relatively non-specific, targeting mostly all Gram-positive bacteria. Thus, treating S. aureus colonization on AD skin with antibiotics could affect the beneficial microbes such as S. epidermidis on the skin as well.
Bleach baths in combination with antibiotics can also decrease the severity of AD [90, 91]. In 2013, a study in Malaysia revealed that dilute bleach baths performed twice a week for 2 months decreased both AD severity and S. aureus colonization [92]. Although bleach baths have potential to successfully treat AD through clearance of S. aureus, more studies need to be conducted to confirm the current results with or without antibiotic activity. In particular, it is unlikely that the highly dilute bleach bath solution is directly antimicrobial for the skin surface. Beneficial actions of the bleach bath solution may relate to other hydrating or immunological effects [93]. In the few studies where S. aureus has decreased with bleach bath treatments, it is important to understand if this effect occurred as a consequence of improved function of the skin rather than the action of the bath directly on the bacteria. It is also important to understand how bleach baths affect the total skin microbiome. Overall, it is clear that the current treatment of S. aureus in AD patients represents at best a limited solution for some patients suffering from frank super-infection and other more targeted therapies are needed to correct the dysbiosis seen on AD skin.
Conclusions
We have known for over 30 years that the community of bacteria changes on the skin during AD, and recent DNA sequencing approaches have further described in greater detail the magnitude of dysbiosis seen in this disease. S. aureus colonization has been studied in greatest depth and appears to be a significant factor that exacerbates the disorder. However, other species such as S. epidermidis also show increased colonization and may modulate AD pathogenesis. Today, it appears that understanding how the full community of different bacteria interact and communicate with each other is necessary to best treat this disease.
Since S. aureus colonization increases the most during AD flares and is correlated with AD severity, much work has focused on the mechanisms of S. aureus virulence on the skin. As discussed earlier, S. aureus produces a series of virulence factors that can directly disrupt the skin barrier and potentially act as the reason for S. aureus-mediated increased AD severity. It is important, however, to consider mechanisms that still have yet to be studied. One possible future direction is to study how S. aureus colonization can lead to changes in gene expression by the host. If S. aureus virulence factors were to target the expression of skin barrier proteins (e.g., filaggrin, keratins, and skin-specific proteases and inhibitors) that are vital for maintaining an intact skin barrier against pathogens, then AD severity would likely be increased. Another interesting event to study is that of increased bacterial penetration into the skin. It is now known that the skin microbiome extends into the epidermis, dermis, and subcutaneous layers. Thus, an altered skin barrier in AD can result in increased S. aureus penetration into deeper layers of skin. Perhaps this could lead to increased disease severity as well by mechanisms yet to be investigated. Recently, it has been published that skin adipocytes respond as immune cells to S. aureus infection [94]. Perhaps S. aureus colonization and penetration can target adipocytes to mount an immune response and increase AD severity. Thus, there remain various hypotheses to be studied in terms of how S. aureus or other microbes may modulate AD severity.
Current methods of treatment for AD do not address how resident microbes on the skin such as S. epidermidis are altered. Some novel small-scale studies have attempted to remove S. aureus colonization to improve the clinical symptoms of AD. Such approaches have even included adding antimicrobial factors to clothing [95, 96]. These methods, however, face similar problems as previous studies in that they lack an approach to selectively protect the normal microbiome. Some of the most promising therapeutic strategies to combat S. aureus-mediated AD severity may be linked to methods already being used in the gut. Fecal microbiota therapy (FMT) is a method for replacing a diseased gut microbiome in a state of dysbiosis with bacteria from normal patients. FMT has been shown to successfully revert gut microbial dysbiosis in patients with recurrent Clostridium difficile infection (CDI) as well as provided promising results for other gastrointestinal diseases [97]. Similarly, the use of microbial skin transplant therapy could be useful in treating AD. It is worth exploring if the skin microbiome of a healthy donor can be transplanted onto a patient’s skin with AD and evaluate if this can repair the skin microbiome. Overall, the most promising future therapeutic strategies will seek to prevent microbial dysbiosis.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of very importance
Beiber T. Mechanisms of disease atopic dermatitis. NEJM. 2008;358:12.
Carroll CL, Balkrishnan R, Feldman SR, Fleischer AB, Manuel JC. The burden of atopic dermatitis: impact on the patient, family, and society. Pediatr Dermatol. 2005;22:8.
Beikert FC, Langenbruch AK, Radtke MA, Kornek T, Purwins S, Augustin M. Willingness to pay and quality of life in patients with atopic dermatitis. Arch Dermatol Res. 2014;306:279–86.
Su JC, Kemp AS, Varigos GA, Nolan TM. Atopic eczema: its impact on the family and financial cost. Arch Dis Child. 1997;76:4.
Kapoor R, Menon C, Hoffstad O, Bilker W, Leclerc P, Margolis DJ. The prevalence of atopic triad in children with physician-confirmed atopic dermatitis. J Am Acad Dermatol. 2008;58:68–73.
Ring J, Kramer U, Schafer T, Behrendt H. Why are allergies increasing? Curr Opin Immunol. 2001;13:8.
Williams H, Stewart A, von Mutius E, Cookson W, Anderson HR, International Study of, A., Allergies in Childhood Phase, O., and Three Study, G. Is eczema really on the increase worldwide? J Allergy Clin Immunol. 2008;121:947–54. e915.
Kato A, Fukai K, Oiso N, Hosomi N, Murakami T, Ishii M. Association of SPINK5 gene polymorphisms with atopic dermatitis in the Japanese population. Br J Dermatol. 2003;148:665.
Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–6.
Weidinger S, Illig T, Baurecht H, Irvine AD, Rodriguez E, Diaz-Lacava A, et al. Loss-of-function variations within the filaggrin gene predispose for atopic dermatitis with allergic sensitizations. J Allergy Clin Immunol. 2006;118:214–9.
Zhao LP, Di Z, Zhang L, Wang L, Ma L, Lv Y, et al. Association of SPINK5 gene polymorphisms with atopic dermatitis in Northeast China. J Eur Acad Dermatol Venereol. 2012;26:572–7.
De Benedetto A, Rafaels NM, McGirt LY, Ivanov AI, Georas SN, Cheadle C, et al. Tight junction defects in patients with atopic dermatitis. J Allergy Clin Immunol. 2011;127(773–786):e771–7.
Kawashima T, Noguchi E, Arinami T, Yamakawa-Kobayashi K, Nakagawa H, Otsuka F, et al. Linkage and association of an interleukin 4 gene polymorphism with atopic dermatitis in Japanese families. J Med Genet. 1998;35:502–4.
Tsunemi Y, Saeki H, Nakamura K, Sekiya T, Hirai K, Kakinuma T, et al. Interleukin-13 gene polymorphism G4257A is associated with atopic dermatitis in Japanese patients. J Dermatol Sci. 2002;30:100–7.
Eigenmann PA, Sicherer SH, Borkowski TA, Cohen BA, Sampson HA. Prevelance of IgE-mediated food allergy among children with atopic dermatitis. Pediatrics. 1998;101:6.
Fuiano N, Fusilli S, Incorvaia C. House dust mite-related allergic diseases: role of skin prick test, atopy patch test, and RAST in the diagnosis of different manifestations of allergy. Eur J Pediatr. 2010;169:819–24.
Hong J, Buddenkotte J, Berger TG, Steinhoff M. Management of itch in atopic dermatitis. Semin Cutan Med Surg. 2011;30:71–86.
Leung DYM, Bieber T. Atopic dermatitis. Lancet. 2003;361:151–60.
Oyoshi MK, He R, Kumar L, Yoon J, Geha RS. Chapter 3 cellular and molecular mechanisms in atopic. Dermatitis. 2009;102:135–226.
Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–2.
Grice EA, Segre JA. The skin microbiome. Nature reviews. Microbiology. 2011;9:244–53.
Leyden JJ, Marples RR, Kligman AM. Staphylococcus aureus in the lesions of atopic dermatitis. Br J Dermatol. 1974;90:525–30.
Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22:850–9. First study to use 16S rRNA sequencing to analyze the skin microbiome of normal versus atopic dermatitis patients.
Fitz-Gibbon S, Tomida S, Chiu BH, Nguyen L, Du C, Liu M, et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J Invest Dermatol. 2013;133:2152–60.
Tomida S, Nguyen L, Chiu BH, Liu J, Sodergren E, Weinstock GM, et al. Pan-genome and comparative genome analyses of propionibacterium acnes reveal its genomic diversity in the healthy and diseased human skin microbiome. mBio. 2013;4:e00003–13.
Wang Y, Kuo S, Shu M, Yu J, Huang S, Dai A, et al. Staphylococcus epidermidis in the human skin microbiome mediates fermentation to inhibit the growth of Propionibacterium acnes: implications of probiotics in acne vulgaris. Appl Microbiol Biotechnol. 2014;98:411–24.
Kloos WE, Musselwhite MS. Distribution and persistence of Staphylococcus and Micrococcus species and other aerobic bacteria on human skin. Appl Microbiol. 1975;30:381–5.
Oh J, Byrd AL, Deming C, Conlan S, Program NCS, Kong HH, et al. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514:59–64. Major study involving the use of metagenomics to define the skin microbiome.
Belkaid Y, Segre JA. Dialogue between skin microbiota and immunity. Science. 2014;346:7.
Capone KA, Dowd SE, Stamatas GN, Nikolovski J. Diversity of the human skin microbiome early in life. J Invest Dermatol. 2011;131:2026–32.
Lai Y, Di Nardo A, Nakatsuji T, Leichtle A, Yang Y, Cogen AL, et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat Med. 2009;15:1377–82.
Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, et al. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–9. Demonstrated direct communication between skin commensal bacteria and host immune response in murine models.
Lai Y, Cogen AL, Radek KA, Park HJ, Macleod DT, Leichtle A, et al. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J Invest Dermatol. 2010;130:2211–21.
Wang Z, MacLeod DT, Di Nardo A. Commensal bacteria lipoteichoic acid increases skin mast cell antimicrobial activity against vaccinia viruses. J Immunol. 2012;189:1551–8.
Yuki T, Yoshida H, Akazawa Y, Komiya A, Sugiyama Y, Inoue S. Activation of TLR2 enhances tight junction barrier in epidermal keratinocytes. J Immunol. 2011;187:3230–7.
Nakatsuji T, Chiang HI, Jiang SB, Nagarajan H, Zengler K, Gallo RL. The microbiome extends to subepidermal compartments of normal skin. Nat Commun. 2013;4:1431. Revolutionary study indentifying that microbial communities can penetrate and reside in deeper layers of the skin.
Otto M. Staphylococcus colonization of the skin and antimicrobial peptides. Expert Rev Dermatol. 2010;5:183–95.
Sugimoto S, Iwamoto T, Takada K, Okuda K, Tajima A, Iwase T, et al. Staphylococcus epidermidis Esp degrades specific proteins associated with Staphylococcus aureus biofilm formation and host-pathogen interaction. J Bacteriol. 2013;195:1645–55.
Cogen AL, Yamasaki K, Sanchez KM, Dorschner RA, Lai Y, MacLeod DT, et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J Invest Dermatol. 2010;130:192–200.
Shu M, Wang Y, Yu J, Kuo S, Coda A, Jiang Y, et al. Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PLoS One. 2013;8, e55380.
Gong JQ, Lin L, Lin T, Hao F, Zeng FQ, Bi ZG, et al. Skin colonization by Staphylococcus aureus in patients with eczema and atopic dermatitis and relevant combined topical therapy: a double-blind multicentre randomized controlled trial. Br J Dermatol. 2006;155:680–7.
Pascolini C, Sinagra J, Pecetta S, Bordignon V, De Santis A, Cilli L, et al. Molecular and immunological characterization of Staphylococcus aureus in pediatric atopic dermatitis: implications for prophylaxis and clinical management. Clin Dev Immunol. 2011;2011:718708.
Chavanas S, Bodemer C, Rochat A, Hamel-Teillac D, Ali M, Irvine AD, et al. Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nat Genet. 2000;25:2.
Hannula-Jouppi K, Laasanen SL, Heikkila H, Tuomiranta M, Tuomi ML, Hilvo S, et al. IgE allergen component-based profiling and atopic manifestations in patients with Netherton syndrome. J Allergy Clin Immunol. 2014;134:985–8.
Renner ED, Hartl D, Rylaarsdam S, Young ML, Monaco-Shawver L, Kleiner G, et al. Comel-Netherton syndrome defined as primary immunodeficiency. J Allergy Clin Immunol. 2009;124:536–43.
Oh J, Freeman AF, Program NCS, Park M, Sokolic R, Candotti F, et al. The altered landscape of the human skin microbiome in patients with primary immunodeficiencies. Genome Res. 2013;23:2103–14. Second major study analyzing how the skin microbiome composition is effected during disease states by 16S rRNA sequencing.
Knor T, Meholjic-Fetahovic A, Mehmedagic A. Stratum corneum hydration and skin surface pH in patients with atopic dermatitis. Acta Dermatovenerol Croat ADC. 2011;19:242–7.
Rippke F, Schreiner V, Doering T, Maibach HI. Stratum corneum pH in atopic dermatitis: impact on skin barrier function and colonization with Staphylococcus Aureus. Am J Clin Dermatol. 2004;5:217–23.
Bhanu S, Francois PP, NuBe O, Foti M, Hartford OM, Vaudaux P, et al. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin α5β1. Cell Microbiol. 1999;1:17.
Cho SH, Strickland I, Boguniewicz M, Leung DY. Fibronectin and fibrinogen contribute to the enhanced binding of Staphylococcus aureus to atopic skin. J Allergy Clin Immunol. 2001;108:269–74.
Dean SN, Bishop BM, van Hoek ML. Natural and synthetic cathelicidin peptides with anti-microbial and anti-biofilm activity against Staphylococcus aureus. BMC Microbiol. 2011;11:114.
Harder J, Bartels J, Christophers E, Schroder JM. Isolation and characterization of human beta -defensin-3, a novel human inducible peptide antibiotic. J Biol Chem. 2001;276:5707–13.
Ong PY, Ohtake T, Brandt C, Strickland I, Boguniewicz M, Ganz T, et al. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N Engl J Med. 2002;347:10.
Nomura I, Goleva E, Howell MD, Hamid QA, Ong PY, Hall CF, et al. Cytokine milieu of atopic dermatitis, as compared to psoriasis, skin prevents induction of innate immune response genes. J Immunol. 2003;171:3262–9.
Howell MD, Novak N, Bieber T, Pastore S, Girolomoni G, Boguniewicz M, et al. Interleukin-10 downregulates anti-microbial peptide expression in atopic dermatitis. J Invest Dermatol. 2005;125:738–45.
McFadden JP, Noble WC, Camp RD. Superantigenic exotoxin-secreting potential of staphylococci isolated from atopic eczematous skin. Br J Dermatol. 1993;128:631–2.
Zollner TM, Wichelhaus TA, Hartung A, Von Mallinckrodt C, Wagner TO, Brade V, et al. Colonization with superantigen-producing Staphylococcus aureus is associated with increased severity of atopic dermatitis. Clin Exp Allergy. 2000;30:7.
Johnson WM, Tyler SD, Ewan EP, Ashton FE, Pollard DR, Rozee KR. Detection of genes for enterotoxins, exfoliative toxins, and toxic shock syndrome toxin 1 in Staphylococcus aureus by the polymerase chain reaction. J Clin Microbiol. 1991;29:426–30.
Bunikowski R, Mielke M, Skarabis H, Herz U, Bergmann RL, Wahn U, et al. Prevelance and role of serum IgE antibodies to the Staphylococcus aureus-derived superantigens SEA and SEB in children with atopic dermatitis. J Allergy Clin Immunol. 1999;103:6.
Bantel H, Sinha B, Domschke W, Peters G, Schulze-Osthoff K, Janicke RU. alpha-Toxin is a mediator of Staphylococcus aureus-induced cell death and activates caspases via the intrinsic death pathway independently of death receptor signaling. J Cell Biol. 2001;155:637–48.
Walev I, Martin E, Jonas D, Mohamadzadeh M, Muller-Klieser W, Kunz L, et al. Staphylococcal alpha-toxin kills human keratinocytes by permeabilizing the plasma membrane for monovalent ions. Infect Immun. 1993;61:8.
Brauweiler AM, Goleva E, Leung DY. Th2 cytokines increase Staphylococcus aureus alpha toxin-induced keratinocyte death through the signal transducer and activator of transcription 6 (STAT6). J Invest Dermatol. 2014;134:2114–21.
Hong SW, Choi EB, Min TK, Kim JH, Kim MH, Jeon SG, et al. An important role of alpha-hemolysin in extracellular vesicles on the development of atopic dermatitis induced by Staphylococcus aureus. PLoS One. 2014;9, e100499.
Ezepchuk YV, Leung DY, Middleton MH, Bina P, Reiser R, Norris DA. Staphylococcal toxins and protein A differentially induce cytotoxicity and release of tumor necrosis factor-alpha from human keratinocytes. J Invest Dermatol. 1996;107:603–9.
Nakamura Y, Oscherwitz J, Cease KB, Chan SM, Munoz-Planillo R, Hasegawa M, et al. Staphylococcus delta-toxin induces allergic skin disease by activating mast cells. Nature. 2013;503:397–401.
Hanakawa Y, Selwood T, Woo D, Lin C, Schechter NM, Stanley JR. Calcium-dependent conformation of desmoglein 1 is required for its cleavage by exfoliative toxin. J Invest Dermatol. 2003;121:383–9.
Hirasawa Y, Takai T, Nakamura T, Mitsuishi K, Gunawan H, Suto H, et al. Staphylococcus aureus extracellular protease causes epidermal barrier dysfunction. J Invest Dermatol. 2010;130:614–7.
Ladhani S. Understanding the mechanism of action of the exfoliative toxins ofStaphylococcus aureus. FEMS Immunol Med Microbiol. 2003;39:181–9.
Syed AK, Reed TJ, Clark KL, Boles BR, Kahlenberg JM. Staphylococcus aureus phenol soluble modulins stimulate the release of pro-inflammatory cytokines from keratinocytes and are required for induction of skin inflammation. Infect Immun. 2015.
Proft T, Fraser JD. Bacterial superantigens. Clin Exp Immunol. 2003;133:8.
Hirose A, Ikejima T, Gill DM. Established macrophagelike cell lines synthesize interleukin-1 in response to toxic shock syndrome toxin. Infect Immun. 1985;50:6.
Kim KH, Han JH, CHung JH, Cho KH, Eun HC. Role of staphylococcal superantigen in atopic dermatitis: influence on keratinocytes. J Korean Med Sci. 2006;21:9.
Bhardwaj N, Friedman SM, Cole BC, Nisanian AJ. Dendritic cells are potent antigen-presenting cells for microbial superantigens. J Exp Med. 1992;175:7.
Komatsu N, Saijoh K, Kuk C, Liu AC, Khan S, Shirasaki F, et al. Human tissue kallikrein expression in the stratum corneum and serum of atopic dermatitis patients. Exp Dermatol. 2007;16:513–9.
Voegeli R, Rawlings AV, Breternitz M, Doppler S, Schreier T, Fluhr JW. Increased stratum corneum serine protease activity in acute eczematous atopic skin. Br J Dermatol. 2009;161:70–7.
Borgono CA, Michael IP, Komatsu N, Jayakumar A, Kapadia R, Clayman GL, et al. A potential role for multiple tissue kallikrein serine proteases in epidermal desquamation. J Biol Chem. 2007;282:3640–52.
Sakabe J, Yamamoto M, Hirakawa S, Motoyama A, Ohta I, Tatsuno K, et al. Kallikrein-related peptidase 5 functions in proteolytic processing of profilaggrin in cultured human keratinocytes. J Biol Chem. 2013;288:17179–89.
Stefansson K, Brattsand M, Roosterman D, Kempkes C, Bocheva G, Steinhoff M, et al. Activation of proteinase-activated receptor-2 by human kallikrein-related peptidases. J Invest Dermatol. 2008;128:18–25.
Yamasaki K, Schauber J, Coda A, Lin H, Dorschner RA, Schechter NM, et al. Kallikrein-mediated proteolysis regulates the antimicrobial effects of cathelicidins in skin. FASEB J Off Publ Fed Am Soc Exp Biol. 2006;20:2068–80.
Cogen AL, Yamasaki K, Sanchez KM, Dorschner RA, Lai Y, Macleod DT, et al. Selective antimicrobial action is prodived by phenol-soluble modulins derived from staphylococcus epidermidis, a normal resident of the skin. J Investig Dermatol. 2010;130:9.
Wanke I, Steffen H, Christ C, Krismer B, Gotz F, Peschel A, et al. Skin commensals amplify the innate immune response to pathogens by activation of distinct signaling pathways. J Invest Dermatol. 2011;131:382–90.
Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465:346–9.
Kobayashi T, Glatz M, Horiuchi K, Kawasaki H, Akiyama H, Kaplan DH, et al. Dysbiosis and Staphyloccus aureus colonization drives inflammation in atopic dermatitis. Immunity. 2015;42:756–66.
Ewing CI, Ashcroft C, Gibbs AC, Jones GA, Connor PJ, David TJ. Flucloxacillin in the treatment of atopic dermatitis. Br J Dermatol. 1998;138:1022–9.
Lever R, Hadley K, Downey D, Mackie R. Staphylococcal colonization in atopic dermatitis and the effect of topical mupirocin therapy. Br J Dermatol. 1988;119:189–98.
Parish LC, Jorizzo JL, Breton JJ, Hirman JW, Scangarella NE, Shawar RM, et al. Topical retapamulin ointment (1%, wt/wt) twice daily for 5 days versus oral cephalexin twice daily for 10 days in the treatment of secondarily infected dermatitis: results of a randomized controlled trial. J Am Acad Dermatol. 2006;55:1003–13.
Gilani SJ, Gonzalez M, Hussain I, Finlay AY, Patel GK. Staphylococcus aureus re-colonization in atopic dermatitis: beyond the skin. Clin Exp Dermatol. 2005;30:10–3.
Breuer K, Haussler S, Kapp A, Werfel T. Clinical and Laboratory Investigations Staphylococcus aureus: colonizing features and influence of an antibacterial treatment in adults with atopic dermatitis. Br J Dermatol. 2002;147:7.
Herold BC, Immergluck LC, Maranan MC, Lauderdale DS, Gaskin RE, VBoyle-Vavra S, et al. Community-acquired methicilin-resistant staphylococcus aureus in children with no identified presdisposing risk. JAMA. 1998;279:6.
Barnes TM, Greive KA. Use of bleach baths for the treatment of infected atopic eczema. Aust J Dermatol. 2013;54:251–8.
Huang JT, Abrams M, Tlougan B, Rademaker A, Paller AS. Treatment of Staphylococcus aureus colonization in atopic dermatitis decreases disease severity. Pediatrics. 2009;123:e808–14.
Wong SM, Ng TG, Baba R. Efficacy and safety of sodium hypochlorite (bleach) baths in patients with moderate to severe atopic dermatitis in Malaysia. J Dermatol. 2013;40:874–80.
Leung TH, Zhang LF, Wang J, Ning S, Knox SJ, Kim SK. Topical hypochlorite ameliorates NF-kappaB-mediated skin diseases in mice. J Clin Invest. 2013;123:5361–70.
Zhang LJ, Guerrero-Juarez CF, Hata T, Bapat SP, Ramos R, Plikus MV, et al. Innate immunity. Dermal adipocytes protect against invasive Staphylococcus aureus skin infection. Science. 2015;347:67–71.
Koller DY, Halmerbauer G, Bock A, Engstler G. Action of a silk fabric treated with AEGIS in children with atopic dermatitis: a 3-month trial. Pediatr Allergy Immunol Off Publ Eur Soc Pediatr Allergy Immunol. 2007;18:335–8.
Stinco G, Piccirillo F, Valent F. A randomized double-blind study to investigate the clinical efficacy of adding a non-migrating antimicrobial to a special silk fabric in the treatment of atopic dermatitis. Dermatology. 2008;217:191–5.
Aroniadis OC, Brandt LJ. Intestinal microbiota and the efficacy of fecal microbiota transplantation in gastrointestinal disease. Gastroenterol Hepatol. 2014;10:230–7.
Compliance with Ethics Guidelines
Conflict of Interest
The authors declare no conflicts of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection on Allergens
Rights and permissions
About this article
Cite this article
Williams, M.R., Gallo, R.L. The Role of the Skin Microbiome in Atopic Dermatitis. Curr Allergy Asthma Rep 15, 65 (2015). https://doi.org/10.1007/s11882-015-0567-4
Published:
DOI: https://doi.org/10.1007/s11882-015-0567-4