Abstract
Host plant relationships of Australian native and invasive whitefly species in the Bemisia tabaci species complex, namely AUSI and AUSII and Bemisia argentifolii (also called B. tabaci Middle East-Asia Minor 1 (MEAM1), were investigated with three approaches: ecologically in the field with surveys, experimentally in the laboratory, and using population genetics to assess any host-associated differentiation within whitefly species. AUSII and B. argentifolii were collected from various host plant species to test for gene flow using microsatellite genotyping. Neither species showed evidence of population structuring associated with host plant species. Host plant testing in the laboratory showed that only some host plants are reproductive hosts for these three whitefly species. Most individuals of all three species settled on tomato over the other host plant species in a cage with several host species presented simultaneously. Nevertheless, tomato was not a reproductive host for AUSI, and cassava did not support adult survival or nymphal production in any species. AUSI reproduced successfully on cotton, chia, and golden crownbeard. AUSII reproduced best on chia, followed by golden crownbeard, cotton, and tomato. Bemisia argentifolii reproduced well on tomato, followed by cotton, chia, and golden crownbeard. In summary, host plant testing supported the hypothesis that AUSI, AUSII, and B. argentifolii have different host plant relationships from one another and confirmed that the invasive B. argentifolii can use more host plant species for reproduction than the indigenous Australian species. Discrete host associations across cryptic species complexes are likely to be common amongst herbivorous insects.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The name Bemisia tabaci represents a cryptic species complex, with at least 44 distinct genetic groups (Kanakala and Ghanim 2019), although the number of species remains unclear because only some of the designated genotypes have been tested appropriately for their species status (Wongnikong et al. 2020). Bemisia tabaci sensu lato is usually written about as an extreme host plant generalist but it may be only some of the known species in this complex that have a broad host range (Oliveira et al. 2001; Simmons et al. 2008; Abd-Rabou and Simmons 2010; Malka et al. 2018). Amongst the latter, in particular, is Bemisia argentifolii, whose species status has been demonstrated using crossing experiments, assessments of intra- and interspecific mating behaviour, and population genetic studies (Perring et al. 1993; Bellows et al. 1994; Delatte et al. 2006; Simón et al. 2007; Elbaz et al. 2010; Sun et al. 2011; McKenzie et al. 2012; Tahiri et al. 2013), but which is still commonly referred to as B. tabaci B biotype or B. tabaci Middle East-Asia Minor 1 (MEAM1). The host range of some populations (or genotypes) of B. tabaci sensu lato is known to be narrow, for example that of the monophagous Jatropha population in Puerto Rico (Bird 1957). Nevertheless, few studies have quantified the relative host plant use of most species (or designated genotypes) within the complex, either in field studies or in laboratory tests (Sseruwagi et al. 2006; Xu et al. 2011; Malka et al. 2018; Vyskočilová et al. 2019).
Those species and genotypes in the B. tabaci complex that have been investigated with respect to their host species range have generally shown they differ from one another in their performance across different host plant species in terms of adult lifespan, oviposition rate, and development time from egg to adult. Most studies have focussed on the two highly invasive cryptic species B. argentifolii and B. tabaci Mediterranean (MED), either alone (Nava-Camberos et al. 2001; Simmons et al. 2008; Han et al. 2013) or in direct comparison with one another (Muñiz 2000; Kakimoto et al. 2007; Iida et al. 2009; Tsueda and Tsuchida 2011; Jiao et al. 2012, 2013, 2014). For example, low rates of oviposition, nymphal survival, and adult emergence (or none at all) have been reported for B. argentifolii on Capsicum annuum (Solanaceae, sweet pepper), relative to B. tabaci MED (Iida et al. 2009; Tsueda and Tsuchida 2011; Jiao et al. 2014; Watanabe et al. 2019). By contrast, B. argentifolii and B. tabaci MED did not differ significantly from one another in development time, emergence rate, and life span on Solanum lycopersicum (Solanaceae, tomato), and Cucumis sativus (Cucurbitaceae, cucumber) (Tsueda and Tsuchida 2011). Furthermore, B. argentifolii and Asia II 1 (indigenous to South Asia and a pest of cotton) were both shown to develop to the adult stage on tomato, cucumber, Brassica oleracea (Brassicaceae, cabbage) and Gossypium hirsutum (Malvaceae, cotton), but at different rates from one another, and Asia II 1 developed more slowly on vegetables than on cotton (Ahmed et al. 2014). Moreover, in terms of survival, adult lifespan and fecundity, B. argentifolii performed best on tomato, whereas Asia II 1 performed best on cotton (Ahmed et al. 2014).
In Australia, at least three cryptic species in the B. tabaci complex are present. The two species AUSI and AUSII are considered to be native to Australia (De Barro and Hart 2000; Wongnikong et al. 2020), whereas B. argentifolii is invasive and is the major whitefly pest in agricultural crops (Sequeira and Reid 2019; Hopkinson et al. 2020). AUSI and AUSII have been recorded on several host plant species including agricultural crops and weeds. Specifically, AUSI has been recorded on cotton, Helianthus annuus (Asteraceae, sunflower), Glycine max (Fabaceae, soybean), Sonchus oleraceus (Asteraceae, common sowthistle), Euphorbia cyathophora (Euphorbiaceae, painted spurge), and Verbesina encelioides (Asteraceae, golden crownbeard) across Queensland and New South Wales, whereas AUSII has been found on tomato, Cucumis melo (rockmelon), Salvia hispanica (Lamiaceae, chia), and Emilia sonchifolia (Asteraceae, lilac tasselflower) (van Brunschot, unpublished data, and see results). These two whitefly species, however, have never been reported to impact agriculture and consequently remain little-known ecologically.
We postulated that the host plant species used in the field will differ across the three whitefly species (AUSI, AUSII, and B. argentifolii), even where they occur in sympatry. To test this, whiteflies were hand-collected in the field and evaluated with mtCOI sequencing to associate each of the species definitively with particular host plant species in nature. We then used microsatellite markers to test for evidence of genetic differentiation associated with host plant species within AUSII and B. argentifolii (with too few samples of AUSI available for population genetics analysis). In laboratory experiments, we tested our expectation that each whitefly species would respond differentially to the same set of host plant species. This was done through behavioural tests on their landing and settling rates across the plant species, and with tests on various measures of nymphal and adult performance on those host species. We also predicted that preimaginal development of each whitefly species on a host plant shown to be relatively poor for that whitefly species would have negative consequences for the adults that emerge subsequently, with negative effects on reproductive parameters (even if they are transferred to a relatively good host plant species). We interpret the results in the context of assessing the host plant relationships of each whitefly species in the B. tabaci species complex, which allows a reconsideration of interpretations of competitive exclusion amongst these whiteflies.
Materials and methods
Field surveys
Field surveys were carried out in Australia, between July 2017 and February 2018, in Darwin (Northern Territory), Kununurra (Western Australia), Brisbane and Emerald (Queensland), and Coleambally, Darlington Point and Narrabri (New South Wales). Sampling sites included agricultural fields, research stations, and community gardens. As many species of host plants and weeds as possible were sampled at each locality because B. tabaci sensu lato whiteflies have been recorded on such a wide range of plant species. Multiple adult whiteflies per host plant were collected and multiple host plants of each species were sampled at each site. Sampling was conducted by two individuals for a maximum of two hours per location. Samples were stored in 95% ethanol before sequencing the mtCOI gene.
DNA extraction, mitochondrial DNA sequencing and analysis
DNA was extracted from B. tabaci specimens using a modified Chelex extraction, adapted from White et al. (2009). Single whiteflies were homogenized using zirconium beads in 1.5 ml tubes containing 6 µl of 10 mg/ml Proteinase K and 50 µl of Chelex solution (10% Chelex in 10 mM Tris HCl and 1 mM EDTA pH 8.0), then incubated at 37 °C for 1 h, followed by incubation at 96 °C to inactivate the Proteinase K.
PCR amplification of an 819 bp region of the mtCOI gene was achieved using the primers C1-J-2195 (5′-TTGATTTTTTGGTCATCCAGAAGT-3′) and L2-N-3014 (5′-TCCAATGCACTAATCTGCCATATTA-3′) (Simon et al. 1994). The 3′ COI region has been used in almost all previous Bemisia studies (and not the 5′ end used in The Barcode of Life Data System), so screening this 3′ region meant that these new sequences could be compared with most Bemisia sequences available on GenBank.
Each 30 µl reaction contained 2 µl DNA template, 1U MyTaq Polymerase (Bioline, Australia), 0.2 µM of each PCR primer, and 1 × buffer. PCR reaction conditions consisted of an initial denaturation at 95 °C for 3 min, followed by 10 cycles of 30 s at 95 °C, annealing at 45 °C for 30 s, and 1 min extension at 72 °C, then 30 cycles of 30 s at 95 °C, annealing at 50 °C for 30 s, and 1 min extension at 72 °C, and the final extension was at 72 °C for 10 min. PCR products were verified by agarose gel electrophoresis and cleaned using 1U of Exonuclease I and Antarctic Phosphatase (New England Biolabs, Ipswich, Mass., USA) by incubating at 37 °C for 20 min followed by 10 min enzyme denaturation at 80 °C. The clean products were sequenced using the same forward and reverse primers used for PCR by Macrogen Inc. (Seoul, Republic of Korea). Sequences were aligned with known samples of the B. tabaci species complex mtCOI haplotypes (available from GenBank plus some new sequences (van Brunschot, unpublished)), using MAFFT method alignment, and also checking for internal stop codons (an indicator of pseudogenes). The alignment was trimmed to 654 bp and a neighbour-joining phylogenetic tree was constructed using a bootstrap analysis of 10,000 replications in Geneious version 9.1.8 (http://www.geneious.com) (Kearse et al. 2012). Those whiteflies whose DNA did not amplify with the primers C1-J-2195 and L2-N-3014 were presumed to represent other whitefly species, such as Trialeurodes vaporariorum. For these latter individuals, the LCO1490 and HCO2198 primers (Folmer et al. 1994) were used, and their identity confirmed by searching GenBank.
Tests for host plant-associated differentiation across AUSII and B. argentifolii samples
Microsatellite loci were used to investigate whether any genetic differentiation is associated with host plant species within AUSII and B. argentifolii in Australia. AUSI could not be included because too few samples were collected in surveys. The microsatellite loci used for AUS II were those developed by Wongnikong et al. (2020). Those used for B. argentifolii had been developed for assessing gene flow in B. argentifolii (Wongnikong et al. 2021). The PCR and genotyping protocols are the same as those of Wongnikong et al. (2020).
The peaks were analysed using the microsatellite plugin in Geneious version 9.1.8 (http://www.geneious.com) (Kearse et al. 2012). The basic population genetics statistics, including Hardy–Weinberg probability tests, were calculated in Genepop version 4.6 (Rousset 2008) with 100 batches (10,000 iterations per batch). Null allele frequencies were estimated with the EM algorithm (Dempster et al. 1977) implemented in FreeNA with 5000 replications (Chapuis and Estoup 2007). The locus-specific statistics across samples on each host plant species were calculated using GenAlEx 6.5 (Peakall and Smouse 2006, 2012), and included the number of different alleles (Na), Shannon’s Information Index (I), observed heterozygosity (HO), expected heterozygosity (HE), and fixation index (F). The population assignment of AUSII and B. argentifolii was analysed using Structure version 2.3.4 (Pritchard et al. 2000; Falush et al. 2003, 2007; Hubisz et al. 2009). Structure runs were performed using the admixture model with a burn-in of 50,000 iterations followed by 500,000 iterations. K values were set from one to four with the same parameters as above. Then 10 runs were conducted, and these were permuted and plotted using CLUMPAK server (http://clumpak.tau.ac.il/). To estimate the most likely K value in the data set, Structure Harvester (http://taylor0.biology.ucla.edu/structureHarvester/) (Earl and vonHoldt 2012) and the method of Evanno et al. (2005) was used. A Principal Component Analysis (PCA) was performed using the adegenet package (Jombart 2008; Jombart and Ahmed 2011) in R version 3.6.1 (R Core Team 2019).
Whitefly colonies and experimental insects
Each laboratory colony of whiteflies was established from field collections as follows: (i) AUSI from painted spurge at Bundaberg (coastal Queensland), (ii) AUSII from lilac tasselflower at Kununurra (northern inland region of West Australia), and (iii) B. argentifolii from Hibiscus trionum (Malvaceae, bladder ketmia) at Emerald (Central Highlands Region, Queensland), with all sites being in Australia. Each species was maintained independently on Solanum melongena (Solanaceae, eggplant Black Beauty variety) in separate cages with fine mesh nylon netting (150 × 150/160 µm aperture), to avoid cross contamination. All colonies had been maintained for multiple generations. Environmental conditions were 26 ± 1 °C, 14 h:10 h L:D photoperiod, and 60 ± 4% RH. The purity of each culture was monitored regularly every 8 weeks, and was checked again before conducting each experiment, by taking four female adult whiteflies randomly from each colony and checking their identity by mtCOI sequencing.
Host plant tests
To test the responses of the three whitefly species to different host plants, five plant species were selected. The ‘best’ host plant species was selected for each species of whitefly, based on the survey results (see results).
Golden crownbeard was selected for AUSI, chia for AUSII, and tomato (Money Maker variety) was selected for B. argentifolii. In addition, Manihot esculenta (Euphorbiaceae, cassava) was included because it is a well-defended plant that produces compounds involved in direct defence against herbivory and is a good host for cassava-adapted cryptic species of B. tabaci (Malka et al. 2018). Lastly, cotton was included given it is an economically important crop that is attacked by whiteflies in Australia.
Test plants were grown from seed, except for cassava which was vegetatively propagated from stem cuttings. All host plants were maintained in a glasshouse in cages with fine mesh nylon netting to prevent insect infestation.
Initial attraction of whiteflies and their subsequent settling
This experiment tested which of the five host plant species attracts and retains most whiteflies of each cryptic species. One plant of each of the five host plant species, each with at least four or five true leaves, was placed in the same cage in a randomized position and 5 cm from one another (whitefly-proof screen cages (32.5 × 32.5 × 32.5 cm), MegaView Science, Taichung, Taiwan). Fifty adult whiteflies of a particular cryptic species, with about 1:1 sex ratio (one to two days post-emergence), were released in the centre of the cage above the plant canopy. The pattern of whitefly settling across the different plant species was counted at intervals on each leaf, with the counts being at 0.25 h, 1 h, 6 h, 12 h, 24 h, 48 h, 72 h, and 96 h after the whiteflies had been released. The adult whiteflies on each leaf were counted by flipping the leaf gently under dim light so as not to disturb the whiteflies. Four replicate cages of each whitefly species (treatments) were conducted each of the three times the experiment was run, to control for any time-related variables that might influence the results, until a sample size of 12 replicate cages had been run for each whitefly species. For B. argentifolii only 11 replicate cages were used in the analysis because one replicate contained more than 50 individuals and was removed from the analysis.
Adult lifespan and survival rate
To test the duration for which recently emerged AUSI, AUSII, and B. argentifolii adults survive on each host plant species, 10 adult whiteflies (about 1:1 sex ratio, and one to two days post-emergence) were introduced into a clip cage (Muñiz and Nombela 2001) on each host plant species (12 replicates/plant species/whitefly species). Adult survival was recorded daily after introduction until all whiteflies had died. Replicates of all three species of whiteflies were conducted on each day of the experiment. However, some tomato (n = 7) and golden crownbeard (n = 6) plants died during the experiment, so replicates on these host plants were performed at a different time from the remaining tests.
Oviposition rate across host plant species in no-choice tests
The number of eggs laid by AUSI, AUSII, and B. argentifolii females on each of the five host plant species was quantified as follows. One newly emerged pair of whiteflies (all of which had developed on eggplant) was introduced into a clip cage (12 replicates/plant species/whitefly species). Females were allowed to lay eggs for 96 h, then the leaf was removed, and the number of eggs counted using a stereomicroscope. This exposure period was used to even out variance across days, but without the arena becoming crowded with eggs, and without them hatching before being counted. After counting, the leaf was placed in a Petri dish with a moistened cotton ball to keep the leaf fresh and then, after 10 days, the numbers of eggs that hatched were counted and converted to a hatching rate. Replicates of all three species of whiteflies were conducted on each day of the experiment.
Nymphal viability and development time in no-choice tests
This experiment tested the impact of each host plant species on nymphal viability and the nymphal development time of each of the three whitefly species. Ten adult whiteflies (about 1:1 sex ratio, and 1 to 2 days post-emergence), which had been reared on eggplant, were introduced into a clip cage on a particular host plant of each of the five host plant species, with 12 replicates per host plant species per whitefly species. All adult whiteflies were removed 48 h after introduction to ensure enough eggs had been deposited for this test. The eggs and hatched offspring were monitored daily until all had emerged as adults, and development time from egg to adult was recorded. This was determined as the time when the first individual became an adult in each replicate. The adults were also counted, as were the numbers of third instar nymphs before that, because they are big enough to count accurately in situ using a hand lens (with smaller ones too easily miscounted without damaging the plant). Replicates of all three species of whiteflies were conducted on each day of the experiment.
Effect of developmental host species on subsequent oviposition and hatch rates
This experiment was designed to test whether the host species on which adults develop have a long-term impact on their subsequent oviposition and hatch rates. Individuals of each of the three whitefly species were reared to the adult stage on each of the test plant species used in the experiments above (the ‘Developmental’ host plant). However, cassava was not used in any test because none of the whiteflies reproduced on this host in earlier tests. Likewise, tomato was not used for AUSI because nymphs of this species did not survive on this plant in an earlier test. Thus, the Developmental host plants of AUSI included three species: golden crownbeard, chia, and cotton, whereas the Developmental host plants for AUSII and B. argentifolii included four plant species, those used for AUSI plus tomato. The nymphal production experiment had shown that the host plant on which most nymphs were produced by AUSI was cotton.
Some of the adults produced were then transferred, on eclosion (n = 12 pairs per host plant species), to the same (Developmental) host species and others (n = 12 pairs) were transferred to the host plant that had been determined in the previous experiment to support the highest nymphal production for that whitefly species, and which is referred to as the ‘Best’ host species (see results). For those insects for which the Developmental host species was also the Best host species, 12 pairs were simply placed on the same plant species to assess egg production. That plant species, for each whitefly species, constituted the control plants. These females were allowed to lay eggs for 96 h (n = 12 replicates/treatment/whitefly species), which were then counted. The exposure time was the same as that for the test of oviposition rate across host plant species in single-host tests.
Statistical analysis
Initial attraction of whiteflies and their subsequent settling
The numbers of adult whiteflies that settled on the different host plant species were statistically evaluated using two models. First, differences in whitefly numbers on each host plant species across observation periods (time) were tested using a generalized linear mixed model (GLMM). Next, differences in whitefly numbers within each period were tested using a generalized linear model (GLM) with the baseline for comparison being the numbers of adult whiteflies on each host species with the numbers on cassava (which had the lowest response) at 0.25 h after release of the insects into the cage. Subsequently, the pairwise multiple comparisons were made with Tukey’s Honestly Significant Difference test (Hothorn et al. 2008). All analyses and visualizations (including those in the following sections) were performed in R version 3.6.1 (R Core Team 2019).
The evaluation of whitefly numbers across observation periods included the fixed effects of host species and observation period, as well as the interaction across host species and observation period. The specific cage in which whiteflies were counted across repeated periods was included as a random effect. The analysis was run using the ‘nlme’ package in R version 3.6.1 (Pinheiro et al. 2020).
Adult lifespan and survival rate
The duration (in days) for which AUSI, AUSII, and B. argentifolii adults survived on each host species was evaluated using a GLM. The host species and observation period, as well as the interaction between host species and observation period, were included as explanatory variables. Differences across host species and observation periods were compared statistically by excluding the intercept. When statistical significance was detected, pairwise multiple comparisons were made across host species using Tukey’s Honestly Significant Difference test at P = 0.05.
The adult survival for the three whitefly species on each host plant species was evaluated every 7 days across a 63 day period. The percentage of survival for each whitefly species was evaluated separately in relation to host species using GLMMs followed by post hoc pairwise comparisons using Tukey’s Honestly Significant Difference tests. GLMMs were run using the glmmTMB package in the R statistical software (Brooks et al. 2017). Whitefly species were included as a fixed effect, observation period as a random effect, and a negative binomial error distribution (“nbinom2”) for overdispersion. GLMMs for the host plants chia, tomato, and cotton also included a log link function. Fitting the model for the golden crownbeard host plant evaluation required including a zero-inflation term, and a logit link function. All whiteflies on cassava died within 7 days and were not included in statistical evaluations.
Oviposition rate across host plant species in no-choice tests
The oviposition rate (the number of eggs laid in the 96-h exposure period) across all five host plant species for each of the three whitefly species was statistically analysed using a GLM with a quasipoisson error distributions (to account for overdispersion). Host plant species was included as an explanatory variable. Oviposition rates across host plant species were compared, for each whitefly species, against the plant species that supported the fewest eggs for that particular whitefly species. When statistical significance was detected in comparisons across the plant species with the lowest oviposition rate and those with relatively higher rates, pairwise multiple comparisons across host plant species were conducted using Tukey’s Honestly Significant Difference test at P = 0.05.
Nymphal viability and development time in no-choice tests
The number of nymphs produced in this no-choice test, and their development time (from egg to adult), were evaluated with independent GLMs as described for the oviposition rate analysis (see above). Those host plant species on which no whiteflies developed were excluded (for AUSI, this involved cassava and tomato, and cassava was excluded for AUSII and B. argentifolii).
Effect of developmental host species on subsequent oviposition and hatch rates
The percentage egg hatch (proportional data) was analysed. Each whitefly species was evaluated separately in relation to host species, using a GLM with quasibinomial error distributions with a logit link function. This analysis investigated the interaction between percentage egg hatch and ~ host. Statistical differences were evaluated across all five host plant species. When statistical significance was detected, pairwise multiple comparisons were conducted using Tukey’s Honestly Significant Difference test at P = 0.05. For the experiment on the effect of developmental host species (number of eggs and hatch rate), the statistical significance was determined by comparison with the ‘best’ host. The comparisons across all ‘best’ host plants and across all “developmental” hosts for each whitefly species were evaluated. Analyses and visualizations were performed using the ggplot package (Wickham 2009).
Results
Field survey and whitefly identities in Australia
Only few whitefly nymphs were found during the field surveys, and densities of adults were low. In Darwin most adult whiteflies were found on Abelmoschus esculentus (Malvaceae, okra) and in Kununurra they were relatively numerous on various cultivated hosts in different families. All were identified as AUSII and B. tabaci Asia II (unclassified as to subgroup), with both usually collected from the same host species (Table 1). The latter was, however, always in low numbers. No specimens of AUSI or B. argentifolii were collected in either locality.
In Queensland and New South Wales, the whiteflies were mostly B. argentifolii and were found on various host plants in several families (Table 2). Populations of AUSI, AUSII, and Asia II (unclassified as to subgroup) were also found in Emerald, but in low numbers. Most individuals of the native species AUSI were found on only one host, golden crownbeard. Few hosts harboured mixed populations of whiteflies, but common sowthistle and Ipomoea plebeian (Convolvulaceae, bellvine) did so in Emerald (Table 2). In the other localities, only B. argentifolii was collected (Table 2).
Tests for host plant-associated differentiation across AUSII and B. argentifolii samples
In total, 96 AUSII individuals were genotyped across 11 loci from the four host plant species that had sufficient samples for analysis, including okra, tomato, Cucurbita maxima (Cucurbitaceae, pumpkin, Jap variety), and chia. Only seven loci were used in the analysis (four microsatellite loci were excluded, for high null allele frequencies). In general, the mean number of alleles per locus ranged from 7.1 to 8.3. The observed heterozygosity (HO) ranged from 0.551 to 0.604, whereas the expected heterozygosity (HE) ranged from 0.648 to 0.682 (Supplementary Table 1). For B. argentifolii, 115 individuals from three host plant species (cotton, common sowthistle and Abutilon sp.) were genotyped at 11 loci. The mean number of alleles per locus ranged from 4.1 to 5. The observed heterozygosity (HO) ranged from 0.488 to 0.554, whereas the expected heterozygosity (HE) ranged from 0.512 to 0.548 (Supplementary Table 2).
The PCA across seven microsatellite loci for AUSII (96 individuals) collected at Darwin and Kununurra indicated no genetic variation across host plant species, with all samples grouped in one genetic cluster (Fig. 1). Similarly, B. argentifolii collected in Queensland and New South Wales showed no evidence of population structuring associated with host plant species in the PCA or the structure analysis (Fig. 2). These results suggest there is high gene flow across populations of each of these species, even across distances as great as 400 kms (the direct distance between Darwin and Kununurra) for the former species, and no genetic differentiation was associated with host plant species.
Microsatellite testing of patterns of genetic variation across host plant-associated populations of Bemisia tabaci AUSII (96 individuals) collected in Darwin (Northern Territory), and Kununurra (Western Australia) in 2017 across four host plant species, namely Abelmoschus esculentus (okra), Solanum lycopersicum (tomato), Cucurbita maxima (Jap pumpkin), and Salvia hispanica (chia). (Top) Bayesian clustering analysis performed in Structure, based on data from seven microsatellite loci. The results are shown for K = 2. Each vertical line represents a single individual. The results suggest no pattern of genetic differentiation across host plant species and showed high gene flow across the two sampling areas: Darwin and Kununurra. (Bottom) A Principal Coordinates Analysis of data from seven microsatellite loci from 96 individuals. The first and second axes accounted for 5 and 4.12% of the variance, respectively, indicating that AUSII collected across four host plants all belong to the same genetic grouping
Microsatellite testing of patterns of genetic variation amongst host plant-associated populations of Bemisia argentifolii (155 individuals) collected in 2018 across three host plant species, namely Gossypium sp. (cotton) (collected in Emerald, Queensland), Sonchus oleraceus (common sowthistle) (collected in Emerald and New South Wales (Coleambally, Darlington Point and Narrabri), and Abutilon sp. (Indian mallow) (collected in Brisbane, Queensland)). (Top) Bayesian clustering analysis performed in Structure, based on data from 11 microsatellite loci. The results are shown for K = 2. Each vertical line represents a single individual. The results indicate no pattern of genetic differentiation across host plant species and showed high gene flow across Queensland and New South Wales samples. (Bottom) A Principal Coordinates Analysis of data from 11 microsatellite loci from 155 individuals. The first and second axes accounted for 5.66 and 5.27% of the variance, respectively, indicating that B. argentifolii collected across the three host plant species all belong to the same genetic grouping
Host plant tests
Initial attraction of whiteflies and their subsequent settling
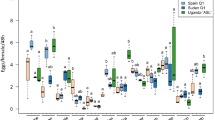
The settling patterns of AUSI, AUSII, and B. argentifolii, after their simultaneous exposure to the same five host plant species, were different from one another in their initial settling pattern, and also subsequent to that across the time-related counts (Fig. 3). Significant differences in numbers, of each whitefly species and at each time interval, with respect to their distribution across the different host plant species, were first detected at 6 h, and then again at each remaining time interval. Nevertheless, all three whitefly species showed the strongest association with tomato, and very few of any of the species settled on cassava. These associations were generally statistically significant, and the species-by-species statistical comparisons across the host plants, for each whitefly species and each time interval, are given in Supplementary Tables 3–10.
The numbers (\(\overline{x}\)±1SE) of adult whiteflies of three species in the Bemisia tabaci complex that settled on five host plant species presented simultaneously (namely, Manihot esculenta (cassava), Salvia hispanica (chia), Gossypium hirsutum (cotton), Verbesina encelioides (golden crownbeard), and Solanum lycopersicum (tomato)), at intervals after their release into the cage: a AUSI, b AUSII, and c Bemisia argentifolii. Adult whitefly numbers were statistically evaluated (within species and within time intervals) with a generalized linear mixed model (GLMM) (AUSI and AUSII, n = 12, and B. argentifolii, n = 11 (50 whitefly individuals per replicate)). Whitefly numbers on each species were compared against those on cassava because this species hosted the fewest insects of each whitefly species at 0.25 h after their introduction into cages (the earliest period in which insects were counted). The asterisks associated with each set of bars indicate statistical significance within that time interval and for that particular species of whitefly (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001)
AUSI associated with all host plants throughout the test, but mostly (and significantly so) settled on tomato. AUSII was most strongly, and statistically so, associated with tomato and golden crownbeard, with relatively few on chia. Bemisia argentifolii was very strongly associated with tomato, more so than the other two whitefly species, with relatively few on other host plant species, and this was consistent (and mostly statistically significant) through time.
Adult lifespan and survival rate
Host plant species had an effect on the adult life span of all three whitefly species (Table 3). As expected, the life span of adult AUSI, AUSII, and B. argentifolii whiteflies on cassava was extremely short relative to that on the other host plants, and mostly significantly so. That of AUSI on tomato was also short, at about three days, and statistically was no different from that on cassava. All three whitefly species lived significantly longer on cotton than on any other host plant species, at 12.3, 17.3, and 30.8 days for AUSI, AUSII, and B. argentifolii, respectively, with the latter being significantly different from the other two (Table 3). Also, B. argentifolii and AUSII lived significantly longer on tomato than did AUSI (Table 3).
For all three whitefly species, there were statistically significant differences in whitefly survival rates across host plant species (Fig. 4, Supplementary Table 11). During the 63-day experiment, the survival rate on cotton was significantly higher than on the other host plant species for all three whitefly species through time. The relative survival across whitefly species differed for each host species, except for AUSI and AUSII on cotton. For example, AUSI survival on chia was higher than that of AUSII individuals, but on cotton AUSI had lower survival. All whiteflies on cassava died within seven days, and were not included in the statistical analyses.
Adult survival (\(\overline{x}\)±1SE) through time of three species in the Bemisia tabaci complex: a AUSI, b AUSII, and c Bemisia argentifolii on each of five host plant species (Manihot esculenta (cassava), Salvia hispanica (chia), Gossypium hirsutum (cotton), Verbesina encelioides (golden crownbeard), and Solanum lycopersicum (tomato)). See text for statistical comparisons
Oviposition rate across host plant species in no-choice tests
All three whitefly species laid eggs on each of the five host plant species. The oviposition and hatch rates of AUSI were not statistically significant across host species (Table 4). Nevertheless, the mean hatch rate was low on cotton (36.9%) relative to that on the other species (59.1–76.0%) (Table 4).
The oviposition rate of AUSII was not significantly different across host species (Table 4). The highest rate was on chia at 16.3 eggs/female, followed by tomato at 15.1 eggs/female. A significantly greater hatch rate was observed on tomato at 84.5%, followed by cassava (66.3%), than on golden crownbeard (23.8%) (Table 4).
For B. argentifolii, significant differences were detected across host species in both oviposition and hatch rate. Significantly more eggs were laid on tomato at 25.6 eggs/female and cotton at 19.6 eggs/female than on cassava (at 5.3 eggs/female) (Table 4). A significantly greater hatch rate was recorded on tomato at 84.3% than on golden crownbeard (43.7%) and cassava (45.9%) (Table 4).
The main differences across the whitefly species were as follows. With respect to tomato AUSI laid significantly fewer eggs than the other two species. On chia plants, the number was significantly highest for AUSII, and on cotton plants B. argentifolii laid more eggs than AUSI, and had a significantly higher hatch rate than the other two species.
Nymphal viability and development time in no-choice tests
For all three whitefly species, no nymphs developed on cassava and no AUSI nymphs developed on tomato. Significantly more AUSI nymphs were produced on cotton (10.7 individuals) and chia (9.1 individuals) than on golden crownbeard at 2.7 individuals (Table 5). AUSI took significantly more time to develop from egg to adult on cotton, at 33.2 days, than on golden crownbeard (24.4 days) or chia (27 days), with these last two each being significantly different from cotton in this respect (Table 5).
AUSII produced more nymphs on chia (14.9 individuals), followed by golden crownbeard (12.3 individuals), then tomato (5.2 individuals), but the differences were not statistically significant (Table 5). The development time of AUSII was similar across the host species (except for cassava), but development time was longest on cotton at 32.1 days and shortest on golden crownbeard (25.7 days) (Table 5).
The highest number of B. argentifolii nymphs, on average, was found on tomato (36.6 individuals) and cotton (31.5 individuals) and these were not significantly different from one another, but chia (17.1 individuals) and golden crownbeard (5.6 individuals) were so (Table 5). Bemisia argentifolii took significantly longer to develop on golden crownbeard (30.8 days) than on the other host plant species, although the others were not much lower.
Effect of developmental host species on subsequent oviposition and hatch rates
The females of all three whitefly species generally (with a few exceptions) laid more eggs and had a better hatch rate on the Best host plant for that species, regardless of which host plant species on which they had developed (Table 6). AUSI females laid more eggs on golden crownbeard (7.3 eggs/female) than chia (1.4 eggs/female), but these results were not significantly different from those on the Best host (cotton) (Table 6). However, AUSII did show some inconsistencies with the females laying more eggs and having a better hatch rate on the Developmental host plant (tomato) than on what had been deemed to be the Best host (Table 6). Bemisia argentifolii laid more eggs and had a higher hatch rate on its Best host plant than was achieved by the other whitefly species on their respective Best host plant species (Table 6).
Discussion
We found at least four species belonging to the B. tabaci species complex in Australia, namely AUSI, AUSII, B. argentifolii, and B. tabaci Asia II (unclassified as to subgroup) (Tables 1 and 2). The genetic divergence of AUSII and B. argentifolii is such that the microsatellite markers did not cross-amplify well across these two species, and we had to use a different suite of microsatellite markers for each species. The microsatellite data revealed no evidence of genetic structuring associated with host plant species in either B. argentifolii or AUSII (Figs. 1 and 2). High levels of gene flow evidently occurs across populations within each of these two species and no genetic differentiation associated with their host plants is evident in either.
Host plant relationships across whitefly species
The results demonstrate comprehensively that the three cryptic species of whiteflies (AUSI, AUSII, and B. argentifolii) differ significantly from one another in their host plant relationships. Also, each of the three whitefly species interacted in its own way with each of the five host plant species in the laboratory, and no single measure of their interaction really gives complete insight into their host relationships. Nevertheless, several general conclusions can be drawn (before we consider each species independently).
The extent to which the different host plant species support reproduction of each whitefly species in the field cannot yet be fully assessed, as our knowledge is not yet sufficient. The primary host plant species of each therefore cannot be specified with any confidence. But it is clear that the two species believed to be indigenous to Australia (AUSI and AUSII) are limited in the range of host species they use, relative to B. argentifolii, and possibly also in their geographic distribution within Australia, so structured field sampling needs to be designed for further quantification of these aspects (Rafter & Walter 2020).
Most published host records do not specifically mention the presence (or otherwise) of nymphs, and our field sampling returned few nymphs. The host lists that are available may thus be inflated by the inclusion of non-reproductive or incidental host plant species, with the latter being plants on which the insects are found only sporadically and in relatively low numbers. Nevertheless, incidental hosts may well play a significant role in the survival of adults when their primary host species are not available (Rafter & Walter 2020).
We recommend that all future field host records be associated with mtCOI sequence data of the insects recorded, so it is possible to relate material (including nymphs) to species in the B. tabaci complex and thus determine the relative significance of different plant species to the ecology of each whitefly species (e.g. Rafter et al. 2013; Silva et al. 2018). Molecular techniques also allow analysis of the gut contents of existing collections in ethanol (Hereward and Walter 2012). Combined with mtCOI barcode information, this technique should help provide a more complete picture of the host plant relationships of each species.
None of the three whitefly species we tested could use cassava as a reproductive host (Table 5), and adult life span on this plant was extremely short relative to that on the other host plants (Table 3). Indeed, few individuals of any of the three whitefly species even settled on cassava when other hosts were available (Fig. 3). However, in single-host assays, oviposition on cassava was not significantly different from that on the other host plants (except for B. argentifolii), and hatch rate on this plant was not the lowest (Table 4). Perhaps the nymphs cannot deal with the secondary metabolites of cassava, which include cyanogenic glucosides and flavonoids (Alves 2002; Douglas 2003; Prawat et al. 1995). By contrast, the cassava-adapted cryptic species of B. tabaci (e.g. SSA1-SG3) have a broad reproductive host range (Sseruwagi et al. 2006), and their settling and reproductive rates are higher on cassava than on other host species (Omondi et al., 2005; Malka et al. 2018). This contrast clearly reveals, even further, the extent of the differential adaptations to host plants by members of the B. tabaci species complex.
Many reports mention that B. argentifolii can use a wide range of host plant species, but quantified data are few. Of particular relevance from our results is that B. argentifolii settled predominantly on tomato (Fig. 3), which is consistent with the results of Jiao et al. (2012). Watanabe et al. (2019) showed, also, that this species does well on tomato, so it may have a strong association with this crop species. If so, we predict this will hold across its broad geographical distribution. Also, it performs better on cabbage (var. Jingfeng1) than on poinsettia and cotton (Jiao et al. 2013). A full understanding of the host plant relationships of this species clearly demands a lot more work. Nevertheless, our results and other reports of broad host plant use by B. argentifolii (Oliveira et al. 2001; Simmons et al. 2008; Abd-Rabou and Simmons 2010) help explain why this species has been able to establish widely across different continents, and thus become an invasive pest on many commercial crops, including tomato, cotton, and cabbage (Oliveira et al. 2001; Watanabe et al. 2019).
For AUSI, no host species really stood out, but it is noticeable that these insects settled readily on tomato (on which they cannot develop). Further, AUSI was found only on golden crownbeard and only in Emerald in our surveys (Table 2), despite it not reproducing well on this species in the laboratory (Table 5), and despite past records from cotton, sunflower, Euphorbia heterophylla (Euphorbiaceae, wild poinsettia), soybean, and common sowthistle (van Brunschot, unpublished data). Even though AUSI performed well on cotton as a reproductive host (Table 5) we did not find it on cotton in the field survey (Table 2). Indeed, this species has never been considered a pest of cotton in Australia (unlike B. argentifolii) (De Barro et al. 2000). Our results therefore suggest there must be one or more native host plant species to which AUSI is adapted, and which we did not encounter.
AUSII settled most frequently on tomato and golden crownbeard (Fig. 3) and seems able to use multiple host species, although these insects did not do as well as B. argentifolii across the five host species tested, and especially not on the economic crops tomato and cotton (Tables 4 and 5). These results do, however, support field-based observations in the early 1990s that indigenous B. tabaci populations in northern Australia (likely AUSII) used tomato primarily as a feeding host, and did not reproduce on it (Stonor et al. 2003).
Further field sampling and laboratory tests are clearly needed to pinpoint the primary host species of these whiteflies.
Competitive exclusion by invading whiteflies
The invasive B. argentifolii is said to displace native populations in the B. tabaci complex. For example, Liu et al. (2007) reported that B. argentifolii was widespread and displaced native species in Zhejiang (China; B. tabaci Asia II 3) and Queensland (Australia; AUSI). Mating interactions between invasive and native species were suggested as the means by which the change in species composition occurred (Liu et al. 2007).
The results of the field surveys presented in our study suggest that these conclusions need to be tested more rigorously. Our data indicate that the focus on cotton (in China) and common sowthistle (in Australia) is unlikely to reveal the real cause(s) of the temporal pattern documented on one plant species in each of these countries. It is clear that other variables could have been influential in the changing patterns recorded, mainly the host plant species that were sampled for whiteflies and the number of whiteflies sequenced for their mtCOI identity. Moreover, B. argentifolii is known to have greater insecticide resistance than indigenous species (Costa et al. 1993; Horowitz et al. 2005, 2020; Wang et al. 2010), so pesticide applications could have eliminated whiteflies other than B. argentifolii. Therefore, an interpretation based on sampling focussed on an agricultural crop (and its associated weeds), and which does not consider insecticide resistance, could lead to misinterpretation of changing patterns of distribution across host species.
In Emerald, we found the indigenous AUSI and the invasive B. argentifolii alongside one another, but on different host plant species (Table 2), and it may well be common for them to have different host plant relationships even when found at the same location. The results are similar to those of Delatte et al. (2006), who found that B. argentifolii (an invasive species) and B. tabaci Indian Ocean (a native population referred to as the Ms biotype) occurred together in the same localities on the island of La Réunion, but had different patterns of host use. Bemisia argentifolii was found on crops such as eggplant and cabbage, whereas B. tabaci Indian Ocean was predominant on weeds, including painted spurge and Lantana camara (Verbenaceae, lantana) (Delatte et al. 2006). Recently, B. tabaci Indian Ocean was found on La Réunion only in low numbers, mainly on weeds in non-cultivated areas and was found to be sensitive to insecticides (acetamiprid and pymetrozine) (Taquet et al., 2020). This is a pattern that may well be common across the various B. tabaci sensu lato populations globally, and this implies that competitive interactions are unlikely to be influencing the spatio-temporal dynamics of these cryptic species significantly.
Conclusion
Surprisingly little is known about the host plant relationships of species within the B. tabaci species complex. This should improve now we have reliable behavioural (Wongnikong et al. 2020) and population genetics (see above) methods to resolve species accurately, and to ascertain their host associations more realistically. It is clear, though, that different species within the complex have different host associations from one another, and evolutionary shifts in diet breadth have taken place within this complex of species. This is likely to be a common phenomenon in cryptic species complexes of herbivorous insects and highlights the importance of determining species limits accurately and investigating the host relationships of each of the species involved (Rafter & Walter 2020). Only then will we be able to understand the spatio-temporal dynamics of each species and interpret how they diversified. This demands structured field sampling associated with hypothesis testing in the laboratory on each of them (Rafter & Walter 2020). In summary, each species in such a complex presents us with its own ecological problems.
References
Abd-Rabou S, Simmons AM (2010) Survey of reproductive host plants of Bemisia tabaci (Hemiptera: Aleyrodidae) in Egypt, including new host records. Entomol News 121:456–465
Ahmed MZ, Naveed M, Noor Ul Ane M, Ren SX, De Barro P, Qiu BL (2014) Host suitability comparison between the MEAM1 and AsiaII 1 cryptic species of Bemisia tabaci in cotton-growing zones of Pakistan. Pest Manag Sci 70:1531–1537
Alves AA (2002) Cassava: biology, production and utilization. In: Hilocks RJ, Tresh JM, Bellotti A (eds) Cassava: biology, Production and Utilization. CABI, Wallingford, pp 67–89
Bellows TS, Perring TM, Gill RJ, Headrick DH (1994) Description of a species of Bemisia (Homoptera: Aleyrodidae). Ann Entomol Soc Am 87:195–206
Bird J (1957) A whitefly-transmitted mosaic of Jatropha gossypifolia. Agric Exp Station Univ Puerto Rico 22:1–35
Brooks ME, Kristensen K, van Benthem KJ, Magnusson A, Berg CW, Nielsen A, Skaug HJ, Maechler M, Bolker BM (2017) glmmTMB balances speed and flexibility among packages for zero-inflated Generalized Linear Mixed Modeling. The R Journal 9:378–400
Chapuis M-P, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24:621–631
Costa HS, Brown JK, Sivasupramaniam S, Bird J (1993) Regional distribution, insecticide resistance, and reciprocal crosses between the A and B biotypes of Bemisia tabaci. Int J Trop Insect Sci 14:255–266
De Barro PJ, Hart PJ (2000) Mating interactions between two biotypes of the whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae) in Australia. Bull Entomol Res 90:103–112
De Barro PJ, Driver F, Naumann ID, Schmidt S, Clarke GM, Curran J (2000) Descriptions of three species of Eretmocerus Haldeman (Hymenoptera: Aphelinidae) parasitising Bemisia tabaci(Gennadius) (Hemiptera: Aleyrodidae) and Trialeurodes vaporariorum (Westwood) (Hemiptera: Aleyrodidae) in Australia based on morphological and molecular data. Aust J Entomol 39:259–269
Delatte H, David P, Granier M, Lett JM, Goldbach R, Peterschmitt M, Reynaud B (2006) Microsatellites reveal extensive geographical, ecological and genetic contacts between invasive and indigenous whitefly biotypes in an insular environment. Genet Res 87:109–124
Dempster AP, Laird NM, Rubin DB (1977) Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc Series B Stat Methodol 39:1–38
Douglas A (2003) The nutritional physiology of aphids. Adv in Insect Phys 31:73–140
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Elbaz M, Lahav N, Morin S (2010) Evidence for pre-zygotic reproductive barrier between the B and Q biotypes of Bemisia tabaci (Hemiptera: Aleyrodidae). Bull Entomol Res 100:581–590
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299
Han E-J, Choi B-R, Lee J-H (2013) Temperature-dependent development models of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) Q biotype on three host plants. J Asia Pac Entomol 16:5–10
Hereward JP, Walter GH (2012) Molecular interrogation of the feeding behaviour of field captured individual insects for interpretation of multiple host plant use. PLoS One 7:e44435
Hopkinson J, Pumpa S, van Brunschot S, Fang C, Frese M, Tay WT, Walsh T (2020) Insecticide resistance status of Bemisia tabaci MEAM1 (Hemiptera: Aleyrodidae) in Australian cotton production valleys. Austral Entomol 59:202–214
Horowitz AR, Kontsedalov S, Khasdan V, Ishaaya I (2005) Biotypes B and Q of Bemisia tabaci and their relevance to neonicotinoid and pyriproxyfen resistance. Arch Insect Biochem Physiol 58:216–225
Horowitz AR, Ghanim M, Roditakis E, Nauen R, Ishaaya I (2020) Insecticide resistance and its management in Bemisia tabaci species. J Pest Sci 93:893–910
Hothorn T, Bretz F, Westfall P (2008) Simultaneous inference in general parametric models. Biom J 50:346–363
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332
Iida H, Kitamura T, Honda K (2009) Comparison of egg-hatching rate, survival rate and development time of the immature stage between B-and Q-biotypes of Bemisia tabaci (Gennadius) (Homoptera: Aleyrodidae) on various agricultural crops. Appl Entomol Zool 44:267–273
Jiao X, Xie W, Wang S, Wu Q, Zhou L, Pan H, Liu B, Zhang Y (2012) Host preference and nymph performance of B and Q putative species of Bemisia tabaci on three host plants. J Pest Sci 85:423–430
Jiao X, Xie W, Wang S, Wu Q, Pan H, Liu B, Zhang Y (2013) Differences in host selection and performance between B and Q putative species of Bemisia tabaci on three host plants. Entomol Exp Appl 147:1–8
Jiao X, Xie W, Guo L, Liu B, Wang S, Wu Q, Zhang Y (2014) Differing effects of cabbage and pepper on B and Q putative species of Bemisia tabaci. J Pest Sci 87:629–637
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jombart T, Ahmed I (2011) adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27:3070–3071
Kakimoto K, Inoue H, Yamaguchi T, Ueda S, Honda K-i, Yano E (2007) Host plant effect on development and reproduction of Bemisia argentifolii Bellows et Perring (B. tabaci [Gennadius] B-biotype) (Homoptera: Aleyrodidae). Appl Entomol Zool 42:63–70
Kanakala S, Ghanim M (2019) Global genetic diversity and geographical distribution of Bemisia tabaci and its bacterial endosymbionts. PLoS ONE 14:e0213946
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Liu S-S, De Barro PJ, Xu J, Luan J-B, Zang L-S, Ruan Y-M, Wan F-H (2007) Asymmetric mating interactions drive widespread invasion and displacement in a whitefly. Science 318:1769–1772
Malka O, Santos-Garcia D, Feldmesser E, Sharon E, Krause-Sakate R, Delatte H, van Brunschot S, Patel M, Visendi P, Mugerwa H, Seal S, Colvin J, Morin S (2018) Species-complex diversification and host-plant associations in Bemisia tabaci: A plant-defence, detoxification perspective revealed by RNA-Seq analyses. Mol Ecol 27:4241–4256
McKenzie CL, Bethke JA, Byrne FJ, Chamberlin JR, Dennehy TJ, Dickey AM, Gilrein D, Hall PM, Ludwig S, Oetting RD, Osborne LS (2012) Distribution of Bemisia tabaci (Hemiptera: Aleyrodidae) Biotypes in North America After the Q Invasion. J Econ Entomol 105:753–766
Muñiz M (2000) Host suitability of two biotypes of Bemisia tabaci on some common weeds. Entomol Exp Appl 95:63–70
Muñiz M, Nombela G (2001) Bemisia tabaci: A new clip-cage for biological studies. European Whitefly Studies Network (EWSN), Norwich
Nava-Camberos U, Riley DG, Harris MK (2001) Temperature and host plant effects on development, survival, and fecundity of Bemisia argentifolii (Homoptera: Aleyrodidae). Environ Entomol 30:55–63
Oliveira MRV, Henneberry TJ, Anderson P (2001) History, current status, and collaborative research projects for Bemisia tabaci. Crop Prot 20:709–723
Omondi AB, Obeng-Ofori D, Kyerematen RA, Danquah EY (2005) Host preference and suitability of some selected crops for two biotypes of Bemisia tabaci in Ghana. Entomol Exp Appl 115:393–400
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Perring TM, Cooper AD, Rodriquez RJ, Farrar CA, Bellows TS (1993) Identification of a whitefly species by genomic and behavioral studies. Science 259:74–77
Pinheiro J, Bates D, DebRoy S, Sarkar D, R Core Team (2020) nlme: Linear and Nonlinear Mixed Effects Models. R package version 3.1–150, https://CRAN.R-project.org/package=nlme
Prawat H, Mahidol C, Ruchirawat S, Prawat U, Tuntiwachwuttikul P, Tooptakong U, Taylor WC, Pakawatchai C, Skelton BW, White AH (1995) Cyanogenic and non-cyanogenic glycosides from Manihot esculenta. Phytochemistry 40:1167–1173
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/
Rafter MA, Walter GH (2020) Generalising about generalists? A perspective on the role of pattern and process in investigating herbivorous insects that use multiple host species. Arthropod Plant Interact 14:1–20
Rafter MA, Hereward JP, Walter GH (2013) Species limits, quarantine risk and the intrigue of a polyphagous invasive pest with highly restricted host relationships in its area of invasion. Evol Appl 6:1195–1207
Rousset F (2008) GENEPOP’007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8:103–106
Sequeira RV, Reid DJ (2019) Numerical host plant relationships of Bemisia tabaci MEAM1 (Hemiptera: Aleyrodidae) within and among major Australian field crops. Austral Entomol 58:370–381
Silva R, Hereward JP, Walter GH, Wilson LJ, Furlong MJ (2018) Seasonal abundance of cotton thrips (Thysanoptera: Thripidae) across crop and non-crop vegetation in an Australian cotton producing region. Agric Ecosyst Environ 256:226–238
Simmons AM, Harrison HF, Ling K-S (2008) Forty-nine new host plant species for Bemisia tabaci (Hemiptera: Aleyrodidae). Entomol Sci 11:385–390
Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P (1994) Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am 87:651–701
Simón B, Cenis JL, De La Rúa P (2007) Distribution patterns of the Q and B biotypes of Bemisia tabaci in the Mediterranean Basin based on microsatellite variation. Entomol Exp Appl 124:327–336
Sseruwagi P, Maruthi MN, Colvin J, Rey MEC, Brown JK, Legg JP (2006) Colonization of non-cassava plant species by cassava whiteflies (Bemisia tabaci) in Uganda. Entomol Exp Appl 119:145–153
Stonor J, Hart P, Gunther M, DeBarro P, Rezaian M (2003) Tomato leaf curl geminivirus in Australia: occurrence, detection, sequence diversity and host range. Plant Pathol 52:379–388
Sun DB, Xu J, Luan JB, Liu SS (2011) Reproductive incompatibility between the B and Q biotypes of the whitefly Bemisia tabaci in China: genetic and behavioural evidence. Bull Entomol Res 101:211–220
Tahiri A, Halkett F, Granier M, Gueguen G, Peterschmitt M (2013) Evidence of gene flow between sympatric populations of the Middle East-Asia Minor 1 and Mediterranean putative species of Bemisia tabaci. Ecol Evol 3:2619–2633
Taquet A, Delatte H, Barrès B, Simiand C, Grondin M, Jourdan-Pineau H (2020) Insecticide resistance and fitness cost in Bemisia tabaci (Hemiptera: Aleyrodidae) invasive and resident species in La Réunion Island. Pest Manag Sci 76:1235–1244
Tsueda H, Tsuchida K (2011) Reproductive differences between Q and B whiteflies, Bemisia tabaci, on three host plants and negative interactions in mixed cohorts. Entomol Exp Appl 141:197–207
Vyskočilová S, Seal S, Colvin J (2019) Relative polyphagy of “Mediterranean” cryptic Bemisia tabaci whitefly species and global pest status implications. J Pest Sci 92:1071–1088
Wang Z, Yan H, Yang Y, Wu Y (2010) Biotype and insecticide resistance status of the whitefly Bemisia tabaci from China. Pest Manag Sci 66:1360–1366
Watanabe LFM, Bello VH, De Marchi BR, da Silva FB, Fusco LM, Sartori MM, Pavan MA, Krause-Sakate R (2019) Performance and competitive displacement of Bemisia tabaci MEAM1 and MED cryptic species on different host plants. Crop Prot 124:104860
White JA, Kelly SE, Perlman SJ, Hunter MS (2009) Cytoplasmic incompatibility in the parasitic wasp Encarsia inaron: disentangling the roles of Cardinium and Wolbachia symbionts. Heredity 102:483–489
Wickham H (2009) ggplot2: Elegant graphics for data analysis. Springer, New York
Wongnikong W, van Brunschot SL, Hereward JP, De Barro PJ, Walter GH (2020) Testing mate recognition through reciprocal crosses of two native populations of the whitefly Bemisia tabaci (Gennadius) in Australia. Bull Entomol Res 110:328–339
Wongnikong W, Hereward JP, van Brunschot SL, Walter GH (2021) Multiple invasions of Bemisia argentifolii into Australia and its current genetic connectivity across space. J Pest Sci 94:1331–1343
Xu J, Lin KK, Liu SS (2011) Performance on different host plants of an alien and an indigenous Bemisia tabaci from China. J Appl Entomol 135:771–779
Acknowledgements
This research was funded by the Cotton Research and Development Corporation, Australia. We are grateful to Mary Finlay-Doney, Richard Sequeira, David Parlato, the community garden staff and growers in Kununurra, Darwin, and Emerald for their time and their permission to collect whiteflies on their properties. We would like to thank Komal Gurdasani, Leela Rizal, Ana Barcos, and Hannah Tibbetts for their help on the host plant test experiments and whitefly culture maintenance. We appreciate the time and effort put into the manuscript by the anonymous reviewers.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Kerry Mauck.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wongnikong, W., Hereward, J.P., van Brunschot, S.L. et al. Assessment of relative host plant quality for three cryptic species of the Bemisia tabaci species complex in Australia. Arthropod-Plant Interactions 15, 845–859 (2021). https://doi.org/10.1007/s11829-021-09863-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11829-021-09863-z