Abstract
Sweetpotato [Ipomoea batatas (L.) Lam] is one of the most important crops for food, feed, industrial material, and bio-energy resource. However, since it originated in tropical regions, sweetpotato exhibits limited cold tolerance, hindering its cultivation in low-temperature areas. To comprehend the sweetpotato response to chilling stress, de novo transcriptome assembly was conducted on leaves from two sweetpotato cultivars: the cold-tolerant Liaohanshu 21 and the cold-sensitive Shenshu 28, exposed to low-temperature stress for 3 h and 24 h. 787.24 million raw reads and 763.44 million clean reads were obtained, 60.60–66.06% of clean reads successfully mapped to the reference database across all 18 libraries. 2524 unigenes were identified as transcript factors and classified into 55 families in Plant Transcription Factor Database (PlantTFDB) and 4671 unigenes were matched to 24 groups in Plant Resistance Gene Database (PRGdb). In Liaohanshu 21, 3618 genes and 8391 genes exhibited significant differential expression 3 h and 24 h of low-temperature exposure, respectively. In Shenshu 28, 13,394 genes and 11,627 genes displayed substantial differential expression after 3 h and 24 h of low-temperature exposure, respectively. The differentially expressed genes (DEGs) were further analyzed using Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genome (KEGG) database. DEGs related to “active oxygen scavenging systems”, “lipid metabolism”, and “plant hormone signal transduction” were potentially associated with sweetpotato’s cold resistance. This research offers insights into the mechanisms and genes involved in cold stress in sweetpotato’s response to cold stress and will contribute valuable information for cold-tolerant breeding of sweetpotato.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Low temperature stress, categorized into chilling (0–15 °C) and freezing (< 0 °C), constitutes a major abiotic challenge affecting physiological activity, biochemical metabolism, growth, development, crop yield, and quality, particularly for tropical and subtropical plants (Sanghera et al. 2011; Rinalducci et al. 2011). Numerous plants enhance their cold tolerance through exposure to chilling temperatures, known as cold acclimation (Thomashow et al. 1999). However, many tropical and subtropical crops like tomato, maize, and rice lack cold acclimation capacity. When faced with cold stress, plants undergo a series of signal transduction events, triggering or suppressing the expression of genes related to cold stress. The C-repeat-binding factor/dehydration responsive element-binding factor (CBF/DREB) signaling pathway has been extensively studied in plants, with CBFs primarily functioning in the early stages of cold stress to enable rapid plant responses (Kurepin et al. 2013). The ICE1 (Inducer of CBF expression 1) transcription factor directly binds to the CBF promoter region, activating CBF transformation (Zuo et al. 2019; Liu et al. 2009; Miura et al. 2007; Chinnusamy et al. 2003). Subsequently, CBF binds directly to the promoters of downstream target genes, such as COR genes, and contributes to regulating plant cold tolerance through target gene transcription modulation (Liu et al. 2018). When exposed to stress, reactive oxygen species was rapidly accumulated leading to metabolic disorders in plants and efficient defense mechanisms including the expression and activation of genes related to intracellular and extracellular ROS scavengers have developed to cope with oxidative stress (Kim et al. 2013). Biological membranes participate in various physiological activities such as signal transduction, energy conversion, metabolic regulation, and transport, which are thought to be the primary sites for the development of chilling injury (Rui et al. 2010). The membranes of chilled tissues undergo a phase transition from a flexible liquid-crystalline to a solid-gel structure, which significantly increases the risk of loss of cell membrane semi-permeability (Xie et al. 2019). Meanwhile, cold transiently activates calcium-permeable channels in Arabidopsis mesophyll cells (Carpaneto et al. 2007). Phytohormones work as chemical messengers to regulate a variety of cellular processes in plants in very low concentrations and play key roles in abiotic stress (Peleg and Blumwald 2011). ABA, SA, and JA trigger phosphoprotein cascade pathways, leading to expression of genes associated with cold stress tolerance (Kolaksazov et al. 2013).
Sweetpotato is a vital crop for food, feed, and energy production, offering high yields and nutritional value. However, as a typical tropical plant, it is highly sensitive to low temperatures (Ji et al. 2017, 2019; Xie et al. 2019). Prolonged exposure to temperatures below 10℃ can lead to the death of stems and leaves, and storage roots can decay if kept below 9℃ for more than ten days, increasing susceptibility to fungal infections (Xie et al. 2019). Cold stress is a significant challenge for sweetpotato cultivation in China, particularly in the northeastern region. A deeper understanding of the molecular mechanisms underlying sweetpotato's response to low temperatures could provide valuable genetic information for enhancing cold tolerance through genetic improvement and molecular breeding. Sweetpotato's autohexaploid nature (2n = 6 × = 90) and high heterozygosity across six chromosome groups result in limited genomic data sources (Zhang et al. 2017). Consequently, transcriptome sequencing (RNA-seq) has been employed to investigate specific biological processes in sweetpotato. RNA-seq studies have focused on various molecular regulatory mechanisms in sweetpotato under low temperatures, with two studies examining tuberous roots and one analyzing leaves (Ji et al. 2017, 2019; Xie et al. 2019). Transcriptome sequencing comparing tuberous roots stored at optimal (13 ℃) and low temperature (4 ℃) for six weeks revealed that up-regulated differentially expressed genes (DEGs) are involved in the phenylalanine metabolism, pathogen defense, and biosynthesis of unsaturated fatty acids, while downregulated DEGs are associated with glycerophospholipid, carbohydrate, and energy metabolism (Ji et al. 2017). Another transcriptome study on Xushu 18 cultivar tuberous roots indicated that numerous DEGs are related to hormone metabolism, carbohydrate metabolism, antioxidant enzymes, and cell membrane systems under low temperatures (Xie et al. 2019). An investigation into leaf transcriptomes during exposure to low temperature and subsequent recovery revealed an exploration into genes associated with antioxidant enzymatic processes. These genes play a significant role in managing reactive oxygen species (ROS) reactions, particularly under the influence of cold stress conditions (Ji et al. 2019).
To comprehend the cold response in sweetpotato leaves, de novo transcriptome assembly was conducted under cold stress, comparing the distinct low-temperature responses of two sweetpotato cultivars: cold-tolerant variety Liaohanshu 21 and cold-sensitive variety Shenshu 28. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genome (KEGG) analyses suggested that DEGs related to antioxidant enzymes, lipid metabolism, and hormone signal transduction might play crucial roles in combating low temperatures in sweetpotato. This study offers insights into the molecular mechanisms of low-temperature response in sweetpotato leaves, contributing to the development of enhanced cold tolerance in sweetpotato cultivation.
Materials and methods
Plant materials and stress treatment.
For the RNA-seq analysis, two sweetpotato varieties were chosen: Liaohanshu 21, which exhibits cold tolerance, and Shenshu 28, a cold-sensitive variant, both developed by the Liaoning Academy of Agricultural Sciences. These plants, cultivated in pots, were housed in a light incubator for a fortnight at 25 °C, under a light–dark cycle of 16 h light and 8 h dark, with a cool-white fluorescent light intensity of 150 μmol m−2s−1 and 75% humidity. Subsequently, these plants were relocated to a 4 °C environment for periods of 3–24 h, respectively. The third fully expanded leaf from the top was harvested, promptly immersed in liquid nitrogen, and then stored at -80°C for future analysis. Control plants were those unexposed to the low-temperature condition. Each treatment was replicated in three separate biological instances.
Antioxidant enzyme assays
Superoxide dismutase (SOD), peroxidase (POD) and catalase (CAT) were determined as previously described (Wang et al. 2016).
For the SOD activity determination, 1 g sweetpotato leaves were homogenized on ice with a mortar in 5 mL 0.05 M sodium phosphate buffer containing 2% PVP (pH 7.8), centrifuged for 15 min at 12,000 rpm at 4 ℃. The supernatant was collected and stored before use. Then the SOD activity was assayed using the photochemical nitro blue tetrazolium (NBT) method. Sodium phosphate buffer was instead of SOD extract as a control, incubated for 20 min at 25 ℃, and then SOD activities were determined at 560 nm.
For the CAT activity determination, 1 g sweetpotato leaves were homogenized on ice with a mortar in 10 mL 0.1 M sodium phosphate buffer containing 5 mmol/L dithiothreitol and 5% PVP (pH 7.8), centrifuged for 30 min at 12,000 rpm at 4 ℃. The supernatant was collected and stored before use. The enzyme catalysis system was composed of 2.9 mL H2O2 and 100 μL enzyme extract, and ddH2O was as the control. The absorbance at 15 s and 2 min at 240 nm were recorded, respectively.
For the POD activity determination, 1 g sweetpotato leaves were homogenized on ice with a mortar in 20 mL phosphoric acid buffer, centrifuged for 15 min at 4000 rpm at 4 ℃. The supernatant was collected and stored before use. The reaction mixture was consisted of 30% H2O2 solution, 20 mmol/L KH2PO4 solution and 100 mmol/L phosphoric acid buffer. phosphoric acid buffer was instead of POD extract as a control. The absorbance at 1 min and 2 min at 470 nm were recorded, respectively.
SOD activity, CAT activity, and POD activity were all determined by Molecular Devices SpectraMax ABS plus Multiskan Spectrum (Molecular Devices, CA, USA) according to the operating manual.
Biochemical assays
Hydrogen peroxide (H2O2) content, malondialdehyde (MDA) content, and the production rate of superoxide anion (•O2−) (Zhang et al. 2019) were determined as previously described.
For the H2O2 content determination, 1 g sweetpotato leaves were homogenized on ice with a mortar in 5 mL pre-cooled acetone, centrifuged for 10 min at 3000 rpm at 4 ℃. The supernatant was collected and stored before use. The reaction mixture was consisted of 100 μmol/L H2O2-acetone solution, acetone at − 20 ℃, 10% HCl-Titanium tetrachloride solution, and stronger ammonia water. 1 mL H2O2 extract was added into the reaction mixture, centrifuged for 10 min at 12,000 rpm at 4 ℃, The supernatant was abandoned, 3 mL 2 mol/L H2SO4 was added to dissolve the precipitate. Then H2O2 content were determined at 412 nm.
For the MDA content determination, 1 g sweetpotato leaves were homogenized on ice with a mortar in 10 mL 5% trichloroacetic acid, centrifuged for 10 min at 3000 rpm at 4 ℃. The supernatant was collected and stored before use. 2 mL MDA extract was mixed with 2 mL 6% 2-thiobarbituric acid solution, incubated for 15 min at 100 ℃, then absorbance at at 450 nm, 532 nm, and 600 nm were recorded.
For the production rate of •O2− determination, 1 g sweetpotato leaves were homogenized on ice with a mortar in 3 mL 65 mmol/L potassium phosphate buffer (pH 7.8), centrifuged for 20 min at 12,000 rpm at 4 ℃. The supernatant was collected and stored before use. 0.5 mL of •O2− extract was mixed with 0.1 mL of hydroxylamine hydrochloride, incubated for 20 min at 25 ℃. 1 mL sulfanilic acid solution and 1 mL α-naphthylamine were then added, incubated for 20 min at 25 ℃. At last, 3 mL chloroform was added and then centrifuged for 3 min at 10,000 rpm at 4 ℃. The supernatant was collected and stored before use.
H2O2 content, MDA content, and the production rate of •O2− were all detected by Molecular Devices SpectraMax ABS plus Multiskan Spectrum (Molecular Devices, CA, USA) according to the operating manual.
RNA extraction, library construction, and RNA-Seq
Total RNA extraction from the leaves was performed using the RNAprep Pure Plant Kit (Tiangen Biotech, Beijing, China). The extracted RNA's purity was assessed using a NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA), and RNA integrity was verified via an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Poly-A mRNA was subsequently enriched from these total RNA samples using poly-T oligo-attached magnetic beads. A DNA probe was utilized to hybridize with rRNA, after which RNaseH was deployed to selectively digest the DNA/RNA hybridization chain, followed by the use of DNaseI to digest the DNA probe, yielding purified RNA. This RNA was then fragmented using an interrupt buffer, and a random N6 primer was used for reverse transcription. This process was followed by synthesizing the cDNA second strand to form double-stranded DNA. The 5' end of this synthesized double-stranded DNA was phosphorylated, and the 3' end was adenylated, after which a bubble-like adaptor with a 3' thymine overhang was ligated. The ligated products were then amplified using PCR with specific primers. The PCR product underwent thermal denaturation into a single strand, then circularized with a bridge primer to create a single-stranded circular cDNA library. These constructed cDNA libraries were sequenced using a DNBSEQ platform. The raw sequence reads were submitted to the National Genomics Data Center (NGDC) Biological Project Library (BioProject) database.
Preprocessing and de novo transcriptome assembly
The filtration of total raw sequencing data was accomplished using SOAPnuke (v1.4.0), a software developed by Huada (Huada, Beijing, China), using a Phred quality score threshold (Q ≥ 20). The filtered reads were aligned to the assembled unigenes using HISAT (Hierarchical Indexing for Spliced Alignment of Transcripts) software (Kim et al. 2015a, b). Moreover, Bowtie2 (Langmead and Salzberg 2012) was utilized to map the clean reads to the assembled unigenes. Following the quantification of the mapped reads for each unigene, the expression value was calculated using RSEM (Li and Dewey 2011). The determination of DEGs was based on a Qvalue (Adjusted Pvalue) ≤ 0.05 as per the method outlined by (Love Michael et al. 2014), implemented using DEseq2 (Anders and Huber 2010).
Functional annotation
The open reading frame (ORF) of the unigenes was pinpointed by employing getorf (http://emboss.sourceforge.net/apps/cvs/emboss/apps/getorf.html), after which hmmsearch (http://hmmer.org) was utilized for the alignment of the ORFs with the transcription factor protein domain. Subsequently, the unigenes were categorized based on the transcription factor family traits outlined by Plant Transcription Factor Database (PlantTFDB). This allowed for determining the transcription factor families to which the unigenes belonged.
For annotation, the unigenes were aligned with Plant Resistance Gene Database (PRGdb) using DIAMOND software (Buchfink et al. 2015; Sanseverino et al. 2010) (https://github.com/bbuchfink/diamond). The parameters set for this alignment included a query coverage of ≥ 50% and an identity of ≥ 40%. The annotation results were further refined based on the query coverage and identity.
Quantitative real-time PCR (qRT-PCR) analysis
The first-strand cDNA for qRT-PCR was synthesized utilizing Fastking gDNA Dispelling RT SuperMix FastKing (Tiangen Biotech, Beijing, China). The qRT-PCR was conducted on an ABI Step One Plus Real-time PCR system, employing an SYBR green Realtime PCR Master (Toyobo, Beijing, Shanghai). The primer pairs used for qRT-PCR amplification were specifically designed with the Primerblast online tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). The design parameters included amplification products ranging from 80 to 150 bp, an optimal primer melting temperature of 58–62 ℃, and GC contents between 40 and 60%. The PCR followed a thermal cycling program of 40 cycles of 95 °C for 15 s, 60 °C for 15 s, and 72 °C for 45 s, after an initial denaturation stage for 1 min at 94 °C. The 2−ΔΔCT method was employed for gene expression calculations (Livak and Schmittgen 2001). The β-actin gene of sweetpotato was used as an internal control. The primers, designed according to the sequencing data, are provided in Table S1.
Results
Different physiological responses of sweetpotato leaves between Shenshu 28 and Liaohanshu 21 under cold stress
To identify the effect of low temperature on sweetpotato, cold-tolerant sweetpotato variety Liaohanshu 21 and cold-sensitive sweetpotato variety Shenshu 28 were treated at low-temperature (4℃) for 3 h and 24 h, respectively. Shenshu 28 and Liaohanshu 21 grew normally at room temperature as control (Fig. S1). After 3 h of low-temperature treatment, the leaves of Shenshu 28 are slightly wilted, while there is no significant change on Liaohanshu 21. After 24 h, the leaves of Shenshu 28 were all wilted, while the leaves of Liaohanshu 21 were moderately wilted (Fig. 1A).
Phenotypic and biochemical analysis of changes in sweetpotato leaves under low temperature. A Sweetpotato leaves under low temperature. B production rate of •O2− in leaves C H2O2 levels in leaves. D MDA levels in leaves. E CAT activities in leaves. F POD activities in leaves. D SOD activities in leaves. Each experiment included three independent replicates. The error bars indicate ± SD (n = 3). Different lowercase letters indicate a significant difference at P < 0.05 based on Student’s t test
To investigate the physiological responses of leaves under low temperature conditions, the production rate of •O2−, H2O2 content, and MDA content were measured. When exposed to low-temperature, the production rate of •O2−, H2O2 content, and MDA content continued to accumulate in Liaohanshu 21, while in Shenshu 28, the production rate of •O2− continued to accumulate over time, but MDA and H2O2 decreased firstly in 3 h and then increased in 24 h (Fig. 1B–D). The production rate of •O2−, H2O2 content, and MDA content in Liaohanshu 21 were significantly lower than that in Liaohanshu 21 under normal condition (Fig. 1B–D). Under low temperature conditions, the production rate of •O2− in Shenshu 28 was continuous lower than that in Shenshu 28 (Fig. 1B), while the H2O2 content in Liaohanshu 21 was high than that in Shenshu 28 (Fig. 1C). The MDA content in Liaohanshu 21 was lower than that in Shenshu 28 at 3 h but higher in 24 h under low temperature conditions (Fig. 1D).
CAT, POD, and SOD activity were measured to investigate the antioxidant capacity of leaves under low temperature conditions. The activity of CAT and POD continued to increase over time both in Liaohanshu 21 and Shenshu 28 (Fig. 1E–F). SOD in Liaohanshu 21 came to its highest at 3 h under low temperature and then decreased at 24 h, while in Shenshu 28, SOD activity decreased firstly at 3 h and then increased at 24 h (Fig. 1G). CAT and POD activity in Liaohanshu 21 were higher than that in Shenshu 28 both in normal and low temperature conditions (Fig. 1E–F). However, the SOD activity in Liaohanshu 21 was firstly lower than that in Shenshu 28 under normal conditions, but then higher than that in Liaoshu 28 after exposed to low temperature for 3 h, and finally became lower than that in Liaoshu 28 again at 24 h (Fig. 1G).
Transcriptome sequencing and de novo transcriptome assembly
The DNBSEQ platform was deployed to investigate the transcriptome in three biological replicates of various samples. These included samples under normal temperature (ss_ck and lhs_ck), those exposed to a low temperature for durations of 3 h (L_ss_3h and L_lhs_3h), and 24 h (L_ss_24h and L_lhs_24h). On average, each sample generated 6.36 Gb of data. The study gathered a total of 787.24 million raw reads, and after filtering out reads with adaptors, unknown base N content ≥ 5%, and low-quality reads, 763.44 million clean reads were retained (Table 1). From all 18 libraries, 60.60% to 66.06%, with an average of 65%, of the clean reads successfully aligned with the reference database (Table 1). The process identified 82,513 unigenes, comprising 71,157 known genes and 11,356 predicted new genes. Moreover, 38,207 new transcripts were detected, 26,784 of which were new variable splicing subtypes of known protein-coding genes, and 11,423 were transcripts of newly predicted protein-coding genes. All clean reads were deposited in the NGDC and can be accessed under BioProject accession number PRJNA987163.
Annotation and classification of sweetpotato unigenes
To identify the function of the unigenes, functional annotation was carried out using two databases, PlantTFDB and PRGdb. PlantTFDB is a plant transcription factor database that provides a web link to access the transcription factor collection of several plants, such as Arabidopsis, poplar, and rice, providing protein sequences, coding regions, genomic sequences, and expression sequence tags for each transcription factor family. A total of 2524 unigenes were identified as transcript factors and classified into 55 families. The MYB family (323 genes, 12.80%), bHLH family (225 genes, 8.91%), WRKY family (177 genes, 7.01%), and AP2-ERFBP family (170 genes, 6.73%) formed the four largest clusters (Fig. 2A).
PRGdb is a comprehensive and regularly updated database that contains known and predicted disease-resistant genes in many plants, providing information such as nucleic acid and protein sequences, structural domains, Genbank numbers, disease-resistant hosts, pathogens, and disease descriptions of disease-resistant genes. A total of 4671 unigenes were matched to 24 groups. Genes containing RLP (1161 genes, 24.86%), NL (726 genes, 15.54%), and CNL (701 genes, 15.01%) were the three largest groups (Fig. 2B).
Analysis of differentially expressed unigenes
Nine comparison groups obtained from the samples were identified using DEseq2 with Qvalue (Adjusted Pvalue) ≤ 0.05 for the filter parameter (Table 2).
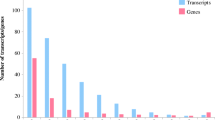
In the Liaohanshu 21 samples, significant expression differences were observed in 3618 genes in the L_lhs_3h samples compared to the lhs_ck samples. Among these, 1883 genes were up-regulated, and 1735 genes were down-regulated. In the L_lhs_24h samples, 8391 genes exhibited notable expression deviations from lhs_ck, with 4035 genes up-regulated and 4356 genes down-regulated. Additionally, 2781 genes in the L-lhs_24h samples demonstrated substantial expression differences from the L-lhs_3h samples, with 1234 genes up-regulated and 1547 genes down-regulated (Fig. 3) (Table 3).
In the Shenshu 28 samples, the L-ss-3h sample set demonstrated significant expression changes in 13,394 genes compared to ss-ck, with 6737 genes up-regulated and 6657 genes down-regulated. In the L-ss-24h samples, 11,627 genes showed notable expression variations from L_ss_3h, with 6032 genes up-regulated and 5595 genes down-regulated. Also, 211 genes in the L-ss_24h samples revealed significant expression differences from the L_ss_3h samples, with 154 genes up-regulated and 57 genes down-regulated (Fig. 3).
When comparing the gene expression of Liaohanshu 21 and Shenshu 28 at the same sampling time, DEGs were analyzed between the two. In the L_lhs_3h vs. L_ss_3h comparison, 1030 and 1145 genes were concurrently up-regulated and down-regulated, respectively; 20 genes were up-regulated in L_lhs_3h but down-regulated in L_ss_3h, and conversely, 7 genes were up-regulated in L_ss_3h but down-regulated in L_lhs_3h. In the L_lhs_24h vs. L_ss_24h comparison, 1805 and 1890 genes were simultaneously up-regulated and down-regulated, respectively; 21 genes were up-regulated in L_lhs_24h but down-regulated in L_ss_24h, while 36 genes were up-regulated in L_ss_24h, but down-regulated in L_lhs_24h.
Relative to lhs_ck samples, 13,547 genes showed significant expression differences in ss_ck samples, with 6460 genes up-regulated and 7087 genes down-regulated. Relative to L-lhs_3h samples, 16,385 genes showed significant expression differences in L-ss_3h samples, with 8188 genes up-regulated and 8267 genes down-regulated. Compared to L-lhs_24h samples, 13,556 genes showed significant expression differences in L-ss_24h samples, with 6826 genes up-regulated and 6728 genes down-regulated (Fig. 3).
GO annotation and KEGG classification of DEGs
The GO analyses were conducted on the DEGs in nine datasets to understand their biological functions better (Table S3). The DEGs were categorized into three main groups: biological process (BP), cellular component (CC), and molecular function (MF). Within the BP category, the two largest subclasses across all nine datasets were "cellular process" and "metabolic process." In the CC category, only "cellular anatomical entity" and "protein-containing complex" were identified across all groups. Within the MF category, "catalytic activity" was the largest subclass in groups 1, 2, 3, 5, and 9, followed by "binding." However, in groups 4, 6, 7, and 8, "binding" was the largest subclass, with "catalytic activity" as the second largest.
To further understand the significant biological pathways involved in the sweetpotato's response to cold stress, the DEGs from the nine datasets were annotated to the reference pathways in the KEGG (Table S4). Across all nine datasets, the two most abundantly represented pathways were "global and overview maps" and "carbohydrate metabolism." The third most represented pathway varied: it was "environmental adaptation" in groups 1, 2, 3, and 8; "translation" in groups 4, 7, and 9; and "signal transduction" in groups 5 and 6. These findings may indicate the diverse functional and metabolic adaptations of sweetpotato under cold stress conditions.
Validation of RNA-seq results
To validate the RNA-seq results, 4 genes from the sequencing data (encoding SwPN1, ICE1-like, ABC transporter F family member 4-like, and ERF109-like) were selected for qRT-PCR analysis. All the 4 genes showed the same expression profiles as those in the transcriptome profiling, although the fold changes were inconsistent with those in sequencing data (Fig. 4).
Expression patterns of selected mRNAs obtained by qRT-PCR and sequencing. mRNA expression levels during qRT-PCR analysis were normalized to the level of β-actin. All qRT-PCRs were repeated three times for each sample. The error bars indicate ± SD (n = 3). Different lowercase letters indicate a significant difference at P < 0.05 based on Student’s t test
Discussion
Different responses to cold between Shenshu 28 and Liaohanshu 21
Our transcriptomic investigation revealed divergent patterns in the number of DEGs between the sweetpotato varieties Liaohanshu 21 and Shenshu 28 as the duration of low-temperature treatment extended. Shenshu 28 exhibited a considerably higher tally of DEGs across all phases of the cold stress, with 13,394 DEGs emerging after a mere three hours of low-temperature exposure, contrasted by only 3,618 DEGs in Liaohanshu 21. These figures underscore Shenshu 28's rapid responsiveness to cold stress, while Liaohanshu 21 demonstrated a more gradual adaptation. Further analysis revealed a minor count of 211 DEGs in L_lhs_24h compared to L_lhs_3h, as opposed to the substantial 2,781 DEGs in L_ss_24h versus L_ss_3h, hinting at Shenshu 28's stability in gene expression during the latter phase of cold stress. This underpins the notion that the cold-sensitive variety, Shenshu 28, experiences pronounced early-stage reactions that persist, whereas the cold-tolerant Liaohanshu 21 undergoes less severe changes. Examining the gene expression of both varieties at identical sample times revealed significant disparities between Liaohanshu 21 and Shenshu 28 under normal conditions. The quantity of DEGs escalated at the 3-h mark and subsequently declined at 24 h, possibly reflecting their distinct cold response mechanisms.
Tables 3 and 4 elucidate the principal 20 KEGG pathways enriched among the DEGs in L_lhs_24h versus lhs_ck and L_ss_24h versus ss_ck. Shared pathways across both varieties, encompassing "Metabolic pathway", "Biosynthesis of secondary metabolites", "Plant-pathogen interaction", "Plant hormone signal transduction", "Starch and sucrose metabolism", "Carbon metabolism", "Biosynthesis of cofactors", "Amino sugar and nucleotide sugar metabolism", "Pentose and glucuronate interconversions", "Glycolysis/Gluconeogenesis", "Galactose metabolism", and "Biosynthesis of nucleotide sugars", suggest a common cold stress response. Notably, 19 out of 20 pathways in Liaohanshu 21 were implicated in "Metabolism". In contrast, Shenshu 28's DEGs were not only implicated in "Metabolism" but also in "Organic Systems", "Mental Information Processing" and "Genetic Information Processing" pathways, indicate a more substantial response from Shenshu 28 to the cold stress.
CBF cold response pathway
The CBF transcriptional cascade plays a crucial role in responses to low temperature in diverse plant species. CBF can directly bind to the promoters of downstream target genes (such as COR) and regulate plant cold tolerance by regulating the transcription of target genes. At the same time, the transcriptional activity of CBF genes is regulated by upstream regulatory factors (Liu et al. 2018). Under cold stress, ICE1 can bind to the MYC binding site of the CBF1/2/3 promoter, competing with MYB15 (negatively regulated CBF transcription) for this site, inhibiting the negative regulatory effect of MYB15, thereby promoting the expression of CBF and downstream target genes (Kim et al. 2015a, b). Cold tolerance could be enhanced by overexpressing ICE1 to regulate CBF genes transcriptional activity in crops such as rice, maize, and tomato (Deng et al. 2017; Lu et al. 2017; Nosenko et al. 2016). ICE1 genes (G11642_TU19184 in Liaohanshu 21, G11642_TU19184 and G12016_TU19749 in Shenshu 28) were up-regulated in plants exposed to low-temperature stress. However, no other transcription factors positively regulate CBF genes, such as CAMTAs and CESTAs (Doherty et al. 2009), were identified in our analysis.
Our transcriptomic data revealed numerous factors associated with negative regulation. Plant Intracellular Factor (PIF) proteins, known for their specific binding to the G-box in gene promoters, regulate target gene expression (Leivar and Monte 2014). PIF4 and PIF7 have been identified as inhibitors of the CBF signaling pathway under warm temperatures, thereby conserving energy and nutrients to expedite plant growth and development. However, at lower temperatures, the repression of CBF transcription by PIF4 and PIF7 diminishes, enabling plants to adapt more effectively to the forthcoming cold conditions (Lee and Thmashow 2012). In Liaohanshu 21, four PIF4-encoding genes (G23568_TU38558, G24019_TU39314, G24019_TU39315, and G24019_TU39317) were found up-regulated under low temperature, while only two such genes (G23568_TU38558 and G24019_TU39314) were up-regulated in Shenshu 28. Three PIF3-encoding genes (G14363_TU23442, G15876_TU25939, and G46233_TU74501) showed increased expression in Liaohanshu 21, whereas no differential expression was observed in Shenshu 28. The role of bZIP transcription factors in plant development, environmental signaling, and stress response has been well-established (Wolfgang et al. 2018). Specifically, bZIP68 inhibits cold tolerance in maize by binding to the A-box element of the DREB1s (CBFs) gene promoter (Li et al. 2022). An up-regulation of a bZIP68-encoding gene (G12592_TU20673) was observed in Shenshu 28 under low-temperature stress, whereas no change was detected in Liaohanshu 21. Besides, PIF16 (G1516_TU2558), PIF17 (G3557_TU5832) and PIF60 (G13483_TU22051) exhibited the same expression pattern as bZIP68. EIN3 (ethylene insensitive 3) can negatively regulate the expression of CBF by binding to the promoters of CBFs. The cold tolerance was reduced when EIN3 overexpressed in Arabidopsis, while in ein3-1 mutants, CBF gene expression was significantly up-regulated (Shi et al. 2012). Three genes (G26606_TU43719, G27348_TU4485, and G27348_TU44856) in Liaohanshu 21 and 2 genes in Shenshu 28 (G27348_TU44853 and G24019_TU39314) encoding EIN3 were up-regulated under low-temperature stress.
CBF genes, known for their rapid induction in response to low temperatures, bind to the CCGAC sequence found in the promoters of COR genes. This interaction results in forming a CBF-COR regulatory network, which boosts plant resilience to low temperatures (Park et al. 2015). In Arabidopsis, COR gene expression has been higher in cold-tolerant leaves than in cold-sensitive pollen (Lee and Lee 2003). Upon exposure to low-temperature stress, six COR-encoding genes (G14164_TU23132, G14640_TU23898, G17721_TU28981, G17751_TU29024, G26475_TU43482, and G8901_TU146600) in Liaohanshu 21 and another six genes (G14640_TU23898, G17721_TU28981, G17751_TU29024, G19198_TU31379, G26475_TU43482, and G8901_TU14660) in Shenshu 28 displayed down-regulation. Notably, our DEG analysis did not identify any putative CBF genes. This suggests that the CBF cold response pathway may have a limited function in sweetpotato under low-temperature stress. Therefore, enhancing cold tolerance in sweetpotato might be achievable by activating the plant's CBF pathway.
Antioxidant genes
During exposure to environmental stressors, plants rapidly accumulate ROS such as •O2− and H2O2, disrupting membrane structures and nucleic acid integrity (Waszczak et al. 2018). To counteract this, plants have evolved systems to scavenge active oxygen species, including enzymes like SOD, CAT, POD, ascorbate peroxidase (APX), and glutathione peroxidase (GPX) (Gill and Tuteja 2010). Following 24 h of low-temperature exposure, the expression of 20 and 28 antioxidant enzyme-encoding genes differed in Shenshu 28 and Liaohanshu 21, respectively.
SOD, the primary enzymatic antioxidant system in plants combatting ROS, catalyzes the conversion of •O2− into H2O2 and O2 (Finkel 2011). Given that more •O2− was generated in Shenshu 28 than in Liaohanshu 21 during cold stress (Fig. 1B), the greater SOD activity in Shenshu 28 (Fig. 1G) is justifiable. However, our findings deviate from previous studies (Ji et al. 2019), wherein FeSOD (G46877_TU75463) and CuZnSOD (G33204_TU54407) were found to be down-regulated in both Shenshu 28 and Liaohanshu 21, and in Shenshu 28, respectively, after 24 h of cold stress. CAT can keep H2O2 at a low level by catalyzing the decomposition of H2O2 into H2O and O2. Previous studies showed that catalase-related genes were downregulated during cold stress in sweetpotato (Ji et al. 2017), while in our study, expression of CAT1 (G27995_TU45897) was up-regulated concurrently in Shenshu 28 and Liaohanshu 21. Another CAT1 coding unigene (G27995_TU45897) was also up-regulated in Shenshu 28, but CAT3 (G29753_TU48752, G29753_TU48753) and putative CAT (G33052_TU54145, G33052_TU54148) were down-regulated in Liaohanshu 21. The relationship between SOD and CAT expression may vary with varieties by the type, cellular location, and mechanisms of action of SODs and CATs. In terms of POD genes, 21 and 14 showed different expression patterns in Liaohanshu 21 and Shenshu 28, respectively. Notably, cationic peroxidase 2-like (G30887_TU50643) and POD21 (G8404_TU13835) demonstrated an increased expression in both Shenshu 28 and Liaohanshu 21, whereas the expression of POD47-like (G6059_TU10000, G6057_TU9998), POD4-like (G37128_TU60871, G37121_TU60862), and POD3-like (G23304_TU38123) decreased in both plants. A particularly interesting finding was that SwPN1 (G31194_TU51163 and G31292_TU51304 in Shenshu 28, G31292_TU51304 in Liaohanshu 21) was less expressed in Shenshu 28 but more expressed in Liaohanshu 21, suggesting a potential role in cold stress defense.
APX serves as a key enzyme in converting H2O2 into H2O. Past studies found that overexpression of OsAPXa in rice increased the plant's cold tolerance at the booting stage (Sato et al. 2011), and overexpression of LeAPX in tomato resulted in enhanced chilling resistance (Duan et al. 2012). GPX, the main enzyme responsible for cellular lipid peroxidation repair (Kühn and Borchert 2002), showed that overexpression led to increased cold tolerance in Arabidopsis thaliana and tomato (Gaber et al. 2004, 2010). The present study found that APX (G46795_TU75338) and GPX (G9028_TU14900) were more expressed in Shenshu 28, while only one APX (G46795_TU75338) exhibited increased expression in Liaohanshu 21.
Upon comparing the gene expression levels of antioxidant enzymes in Shenshu 28 and Liaohanshu 21 post-24-h exposure to low temperature, no variation in the expression of SOD genes was observed. While the expression of CAT3 (G29753_TU48752) and CAT (G21007_TU34349) was elevated in Shenshu 28, higher expression levels of CAT1 (G27995_TU45897 2) and CAT3 (G29753_TU48753) were noted in Liaohanshu 21. Out of 12 differentially expressed POD orthologs in Shenshu 28 and Liaohanshu 21, POD15 (G30329_TU49715) and POD15-like (G30329_TU49714, G30329_TU49716) showed enhanced expression in Shenshu 28, while the expression of POD P7-like (G25807_TU42358, G25835_TU42401) was reduced in the same. Interestingly, SwPN1, being more expressed in Liaohanshu 21 and less expressed in Shenshu 28, exhibited higher expression levels in Liaohanshu 21 than in Shenshu 28 (G31198_TU51167). This finding suggests a potential role of SwPN1 in cold stress defense. Furthermore, APX (G3714_TU6097) demonstrated higher expression in Liaohanshu 21, while GPX (G9025_TU14894, G9028_TU14900) showed enhanced expression in Shenshu 28.
These gene expressions were consistent with the changes of H2O2 content (Fig. 2C), CAT enzyme activity (Fig. 2E), and POD enzyme activity (Fig. 2F) in Liaohanshu 21 and Shenshu 28.
Lipid metabolism
Biological membranes, which play vital roles in transportation, signal transduction, and metabolic regulation, are regarded as primary sites susceptible to chilling injury (Rui et al. 2010). During exposure to low temperatures, plants generally increase the content of unsaturated fatty acids to enhance their cold stress resistance (Hazel 1995; Wongsheree et al. 2009). In Liaohanshu 21, differential expression was observed in 199 genes associated with glyceride metabolism and 170 genes linked with glycerophosphate metabolism.
Lysophospholipid acyltransferase (LPEAT) is critical for acyl modification processes such as fatty acid desaturation and contributes to membrane biosynthesis (Shindou et al. 2013). Phosphatidic phosphatase (PAH) catalyzes the conversion of phospholipid into diacylglycerol (DAG), serving as a key enzyme in controlling the levels of PA and DAG in organisms. This enzymatic activity can regulate lipid metabolism and signal transduction, significantly influencing plant stress responses (Eastmond et al. 2010; Csaki et al. 2013). Phospholipase D (PLD) hydrolyzes phospholipids into inositol triphosphate (IP3), DAG, phosphatidic acid (PA), and acetylcholine. DAG, IP3, PA, and acetylcholine molecules function as secondary messengers in intracellular signal transduction and alter cell structure and physiological function. Unigenes encoding LPEAT2 (G38146_TU62498 and G38146_TU62500), PAH4-like (G8614_TU14174), and PLD-like (G19676_TU32200) were found to be overexpressed in Liaohanshu 21, but no differential expression was observed in Shenshu 28. These genes may be crucial in maintaining membrane integrity and protecting Liaohanshu 21 from the deleterious effects of low temperatures.
Plant hormone signal transduction
In the plant varieties, Shenshu 28 and Liaohanshu 21, 342 and 258 DEGs were found to participate in the plant hormone signal transmission pathway. Plant hormones, acting as chemical messengers, enable the efficient adaptation of plants to abiotic stress (Peleg and Blumwald 2011; Shabir et al. 2016).
Auxin, a crucial plant hormone, regulates the growth and development of many plants (Shivani et al. 2013). Among the DEGs identified in the two varieties, a significant number of auxin-responsive genes were found, primarily consisting of auxin transporters and auxin-responsive proteins. Most of these genes were down-regulated, although a few were up-regulated. OsSAUR45, which influences auxin synthesis and transport, has been found to be dependent on PIF4 under heat-stress conditions in rice (Xu et al. 2017). However, in the present study, the unigenes encoding PIF4-like proteins (G23568_TU38558, G24019_TU39314, G24019_TU39315, and G24019_TU39317 in Liaohanshu 21; G23568_TU38558 and G24019_TU39314 in Shenshu 28) were up-regulated when exposed to low temperature. This suggests that the interaction between PIF4 and auxin-related genes might be an adaptive mechanism for plants to cope with low-temperature stress.
Abscisic acid (ABA) is an important plant hormone whose endogenous level can be increased by salinity, drought, and low temperature (Roychoudhury et al. 2013). As ABA receptors, oyrabactin resistance-like (PYL) proteins were the upstream regulators of the ABA signal pathway and then activated SnRK2 by inhibiting PP2C phosphatase activity in activating the ABA signal pathway (Hao et al. 2011). Overexpressing of a rice PYL3 enhanced cold and drought tolerance in Arabidopsis thaliana (Lenka et al. 2018). In our study, unigenes encoding PYL3-like (G18318_TU29932 and G18361_TU29998 in Shenshu 28 and Liaohanshu 21, G18385_TU30035 in Liaohanshu 21) and PYL4-like (G3894_TU6426 in Liaohanshu 21) were up-regulated when exposed to low temperature.
As a pivotal regulator of plant–microbe interactions under abiotic stress conditions, ethylene plays a crucial role (Shekhawat et al. 2022). Ethylene response factors containing AP2 domains are transcription factors that orchestrate many processes. These include the regulation of gene expression related to stress responses, developmental pathways, growth processes, and mechanisms associated with hormone interactions (Cheng et al. 2013). Unigenes encoding EIN3-LIKE (G26606_TU43719, G27348_TU44853 and G27348_TU44856 in Liaohanshu 21, G35436_TU58103,G27348_TU44853 and G27348_TU44856 in Shenshu 28) were up-regulated; Unigenes encoding ethylene response factor ERF109 (G14664_TU23931, G16250_TU26531, G16257_TU26539, G16257_TU26541, G24812_TU40712 in Shenshu 28, G14664_TU23931, G16257_TU26539, G16257_TU26541, G16250_TU26531 in Liaohanshu 21) were up-regulated.
Conclusion
The molecular mechanisms governing sweetpotato's response to low temperatures were investigated by acquiring transcriptomic data from cold-tolerant and cold-sensitive sweetpotato leaf samples, resulting in the identification of 82,513 unigenes. The DEGs observed when exposed to low temperatures for 3 and 24 h were subjected to GO and KEGG analysis. The cold-tolerant variant, Liaohanshu 21, exhibited fewer DEGs than the cold-sensitive variant, suggesting its greater resilience against cold stress. Upon exposure to low temperatures, an ICE1-encoding gene was found to be up-regulated, while three COR-encoding genes were downregulated in both variants. The absence of DEGs encoding CBF indicated a potentially limited role of the CBF cold response pathway in sweetpotato under cold stress. Greater importance was attributed to genes associated with antioxidant enzymes, lipid metabolism, and plant hormone signal transduction in enhancing sweetpotato's resistance to low temperatures. This study shed light on the molecular dynamics of sweetpotato under cold stress, offering valuable insights for the breeding of cold-tolerant sweetpotato varieties.
References
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 2010:11
Buchfink B, Xie C, Huson DH (2015) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60
Carpaneto A, Ivashikina N, Levchenko V, Krol E, Jeworutzki E, Zhu JK, Hedrich R (2007) Cold transiently activates calcium-permeable channels in Arabidopsis mesophyll cells. Plant Physio 143:487–494
Cheng MC, Liao PM, Kuo WW, Lin TP (2013) The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol 162:1566–1582
Chinnusamy V, Ohta M, Kanrar S, Kanrar S, Lee BH (2003) Hong XH (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. G & D 17:1043–1054
Csaki SL, Dwyer RJ, Fong GL, Tontonoz Z, Young SG, Reue K (2013) Lipins, lipinopathies, and the modulation of cellular lipid storage and signaling. Prog Lipid Res, 52:305–316
Deng CY, Ye HY, Fan M, Pu TL, Yan GB (2017) The rice transcription factors OsICE confer enhanced cold tolerance in transgenic Arabidopsis. Plant Signal Behav 2017:12
Doherty CJ, Van BHA, Myers SJ, Thomashow MF (2009) Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21:972–984
Duan M, Feng HL, Wang LY, Li D, Meng QW (2012) Overexpression of thylakoidal ascorbate peroxidase shows enhanced resistance to chilling stress in tomato. J Plant Physiol 169:867–877
Eastmond PJ, Quettier AL, Kroon JTM, Craddock C, Adams N, Slabas AR (2010) Phosphatidic acid phosphohydrolase 1 and 2 regulate phospholipid synthesis at the endoplasmic reticulum in Arabidopsis. Plant Cell 22:2796–2811
Finkel T (2011) Signal transduction by reactive oxygen species. J C Biol 194:7–15
Gaber A, Yoshimura K, Tamoi M, Takeda T, Nakano Y, Shigeoka S (2004) Induction and functional analysis of two reduced nicotinamide adenine dinucleotide phosphate-dependent glutathione peroxidase-like proteins in Synechocystis PCC 6803 during the progression of oxidative stress. Plant Physio 136:2855–2861
Gaber A, Yoshimura K, Yamamoto T, Yabata Y, Takeda T, Miyasaka H, Nakano Y, Shigeoka S (2010) Glutathione peroxidase-like protein of Synechocystis PCC 6803 confers tolerance to oxidative and environmental stresses in transgenic Arabidopsis. Physiol Plantarum 128:251–262
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Bioch 48:909–930
Hao Q, Yin P, Li W, Wang L, Yan CY, Lin ZH, Wu ZJ, Wang JW, Yan SF, Yan N (2011) The molecular basis of ABA-independent inhibition of PP2Cs by a subclass of PYL proteins. Mol Cell 42:662–672
Hazel RJ (1995) Thermal adaptation in biological membranes: is homeoviscous adaptation the explanation? Annu Rev Physiol 57:19–42
Ji YC, Chung WH, Kim SH, Jung WY, Kang L, Jeong JC, Kwak SS (2017) Transcriptome profiling of sweetpotato tuberous roots during low temperature storage. Plant Physiol Bioch 112:97–108
Ji YC, Bian X, Lee C, Kim HS, Kim SE, Park SC, Xie YZ, Guo XD, Kwak SS (2019) De novo transcriptome sequencing and gene expression profiling of sweet potato leaves during low temperature stress and recovery. Gene 700:23–30
Kim YH, Jeong JC, Lee HS, Kwak SS (2013) Comparative characterization of sweetpotato antioxidant genes from expressed sequence tags of dehydration-treated fibrous roots under different abiotic stress conditions. Mol Biol Rep 40:2887–2896
Kim D, Langmead B, Salzberg SL (2015a) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360
Kim YE, Lee MY, LEE JH, Lee HJ, Park CM, (2015b) The unified ICE-CBF pathway provides a transcriptional feedback control of freezing tolerance during cold acclimation in Arabidopsis. Plant Mol Biol 89:187–201
Kolaksazov M, Laporte F, Ananieva K, Dobrev P, Herzog M, Ananiev ED (2013) Effect of chilling and freezing stresses on jasmonate content in Arabis Alpina. Bulg J Agric Sci 19:15–17
Kühn H, Borchert A (2002) Regulation of enzymatic lipid peroxidation: the interplay of peroxidizing and peroxide reducing enzymes. FRBM 33:154–172
Kurepin L, Dahal K, Savitch L, Singh J, Bode R, Ivanov A, Hurry V, Huner N (2013) Role of CBFs as integrators of chloroplast redox, phytochrome and plant hormone signaling during cold acclimation. IJMS 14:12729–12763
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Lee JY, Lee DH (2003) Use of serial analysis of gene expression technology to reveal changes in gene expression in Arabidopsis pollen undergoing cold stress. Plant Physiol 132:517–529
Lee CM, Thmashow MF (2012) Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. PNAS 109:15054–15059
Leivar P, Monte E (2014) PIFs: systems integrators in plant development. Plant Cell 26:56–78
Lenka SK, Muthusamy SK, Chinnusamy V, BANSAL KC, (2018) Ectopic expression of rice PYL3 enhances cold and drought tolerance in Arabidopsis thaliana. Mol Biotechnol 60:350–361
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323
Li Z, Fu D, Wang X et al (2022) The transcription factor bZIP68 negatively regulates cold tolerance in maize. Plant Cell 34:2833–2851
Liu L, Duan L, Zhang J, Zhang Z, Ren H (2009) Cucumber (Cucumis sativus L.) over-expressing cold-induced transcriptome regulator ICE1 exhibits changed morphological characters and enhances chilling tolerance. Sci Hortic-Amsterdam 124:29–33
Liu JY, Shi Y, Yang S (2018) Insights into the regulation of CBF cold signaling in plants. J Integr Plant Biol 60:780–795
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using RealTime quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408
Love MI, Huber W (2014) Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550
Lu X, Yang L, Yu MY, Lai J, Wang C, Mcneil D, Zhou M, Yang C (2017) A novel Zea mays ssp. mexicana L. MYC-type ICE-like transcription factor gene ZmmICE1, enhances freezing tolerance in transgenic Arabidopsis thaliana. Plant Physiol Bioch 113:78–88
Miura K, Jin JB, Lee J, Yoo CY, Stim V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM (2007) SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19:1403–1414
Nosenko T, Böndel KB, Kumpfmüller G, Stphan W (2016) Adaptation to low temperatures in the wild tomato species Solanum chilense. Mol Ecol 25:2853–2869
Park S, Lee CM, Doherty CJ, Gimour SJ, Kim YS, Thomashow MF (2015) Regulation of the Arabidopsis CBF regulation by a complex low temperature regulatory network. Plant J 82:193–207
Peleg Z, Blumwald E (2011) Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol 14:290–295
Rinalducci S, Egidi MG, Karimzadeh G, Jazil FR, Zolla L (2011) Proteomic analysis of a spring wheat cultivar in response to prolonged cold stress. Electrophoresis 32:1807–1818
Roychoudhury A, Paul S, Basu S (2013) Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell Rep 32:985–1006
Rui HJ, Cao SF, Shang HT, Jin P, Wang KT, Zheng YH (2010) Effects of heat treatment on internal browning and membrane fatty acid in loquat fruit in response to chilling stress. JSFA 90:1557–1561
Sanghera GS, Wani SH, Hussain W, Singh NB (2011) Engineering cold stress tolerance in crop plants. Curr Genomics 12:30–43
Sanseverino W, Roma G, De SM, Faino L, Melito S, Stupka E, Frusciante L, Ercolano MR (2010) PRGdb: a bioinformatics platform for plant resistance gene analysis. Nucleic Acids Res 38:D814–D821
Sato Y, Masuta Y, Saito K, Murayama S, Ozawa K (2011) Enhanced chilling tolerance at the booting stage in rice by transgenic overexpression of the ascorbate peroxidase gene, OsAPXa. Plant Cell Rep 30:399–406
Shabir HW, Vinay K, Varsha S, Saroj KS (2016) Phytohormones and their metabolic engineering for abiotic stress tolerance in crop plants. Crop J 4:162–176
Shekhawat K, Fröhlich K, GarcíaRamírez GX, Trapp MA, Hirt H (2022) Ethylene: A master regulator of plant-microbe interactions under abiotic stresses. Cells 12:31–31
Shi YT, Tian SW, Hou LY, Huang XZ, Zhang XY, Guo HW, Yang SH (2012) Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-a ARR genes in Arabidopsis. Plant Cell 24:2578–2595
Shindou H, Hishikawa D, Harayama T, Eto M, Shimizu T (2013) Generation of membrane diversity by lysophospholipid acyltransferases. JBC 154:21–28
Shivani S, Isha S, Navdeep K, PATI PK, (2013) Auxin: a master regulator in plant root development. Plant Cell Rep 32:741–757
Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Phys 50:571–599
Wang B, Zhai H, He S, Zhang H, Ren ZT, Zhang DD, Liu QC (2016) A vacuolar Na+/H+ antiporter gene, IbNHX2, enhances salt and drought tolerance in transgenic sweetpotato. Sci Horti-Amsterdam 201:153–166
Waszczak C, Carmody M, Kangasj J (2018) Reactive oxygen species in plant signaling. Annu Rev Plant Biol 69:209–236
Wolfgang DL, Snoek BL, Berend S, Christoph W (2018) The Arabidopsis bZIP transcription factor family: an update. Curr Opin Plant Biol 45:36–49
Wongsheree T, Ketsa S, Doorn VGW (2009) The relationship between chilling injury and membrane damage in lemon basil (Ocimum×citriodourum) leaves. Postharvest Biol and Tec 51:91–96
Xie ZY, Zhou ZL, Li HM, Yu JJ, Jiang JJ, Tang ZH, Ma DF, Zhang BH, Han YH, Li ZY (2019) High throughput sequencing identifies chilling responsive genes in sweetpotato (Ipomoea batatas Lam.) during storage. Genomics 111:1006–1017
Xu YX, Xiao MZ, Liu Y, Fu JL, He Y, Jiang DA (2017) The small auxin-up RNA OsSAUR45 affects auxin synthesis and transport in rice. Plant Mol Biol 94:97–107
Zhang H, Zhang Q, Zhai H, Li Y, Wang XF, Liu QC, He SZ (2017) Transcript profile analysis reveals important roles of jasmonic acid signalling pathway in the response of sweet potato to salt stress. Sci. Rep 7. https://doi.org/10.1038/srep40819
Zhang H, Gao XR, Zhi YH, Li X, Zhang Q, Niu JB, Wang J, Zhai H, Zhao N, Li JG, Liu QC, He SZ (2019) A non-tandem CCCH-type zinc-finger protein, IbC3H18, functions as a nuclear transcriptional activator and enhances abiotic stress tolerance in sweet potato. New Phytol 223:1918–1936
Zuo ZF, Kang HG, Park MY, Jeong H, Sun HY, Yang DH, Lee YE, Song PS, Lee HY (2019) Overexpression of ICE1, a regulator of cold-induced transcriptome, confers cold tolerance to transgenic Zoysia japonica. J Plant Biol 62:137–146
Acknowledgments
This work was supported by the Research and application on creation of cold tolerant germplasm resources and molecular genetic breeding of sweet potato (22-318-2-18), Shenyang Expert Workstation (20220105), President's Fund Project of Liaoning Academy of Agricultural Sciences (2023QN2423), National Natural Science Foundation Project (32101787) and Jiangsu Province Seed Industry Revitalization Announcement and Leading Project (JBGS[2021]010-2).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pan, J., Zhang, Q., Liu, S. et al. De novo transcriptome sequencing and gene expression profiling of sweetpotato leaves during low temperature stress. Plant Biotechnol Rep 17, 875–888 (2023). https://doi.org/10.1007/s11816-023-00851-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-023-00851-8