Abstract
Soybean [Glycine max (L.) Merr.] is a major economic crop and is used as food, animal feed, and biofuel because of its high contents of oil and proteins. Improvements in soybean quality and yield have so far been achieved using traditional and molecular breeding, including hybridization, mutagenesis, and the insertion of transgenes. However, the breeding of new soybean varieties was unexpectedly prolonged by genetic limitations, such as genomic duplication and redundancy, and complicated social issues caused by the insertion of transgenes. The use of genome-editing technologies for the genetic manipulation of soybean has revolutionized the study of genetic variations, as well as soybean improvement, over the past few years. Here, we summarize the applications of genome editing technologies including zinc finger nuclease (ZFN), transcription activator-like effector nuclease (TALEN), and clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein nuclease 9 (Cas9), in soybean. We discuss targeted gene mutagenesis using current genome-editing technologies, with a focus on the implementation and potential availability of new technical developments in soybean. The accurate, user-friendly approach afforded by CRISPR/Cas9 technology will not only facilitate functional studies of soybean genes but also accelerate soybean breeding for future soybean improvement.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soybean [Glycine max (L.) Merr.] is an important crop that can be used to prevent and treat human health problems because soybean seeds contain abundant nutrients, such as proteins, oils, sugars, and minerals (Bellaloui et al. 2015). Soybeans contain approximately 40% protein and 20% oil (Clemente and Cahoon 2009). Soybean protein is used mostly in human and animal food. Soybean oil is utilized in food, feed, and some industrial applications, such as biodiesel (Clemente and Cahoon 2009). However, to further expand the utilization of soybean, some problems need to be addressed, including the identification of the appropriate ratio of fatty acids, pest resistance, and allergy issues.
Soybean oil consists of the following five fatty acids: palmitic acid (16:0), stearic acid (18:0), oleic acid (18:1), linoleic acid (18:2), and linolenic acid (18:3). In the past, soybean breeding has focused on the improvement of the overall yield of soybean, including increases in total protein and oil (Mahmoud et al. 2006). The variability of soybean germplasm has contributed to critical progress in the improvement of the quality of the oil for food, feed, and industrial applications, using molecular biology and biotechnology tools (Damude and Kinney 2008). Soybean genetics and breeding programs have been developed to improve both oil content and quality, because of increasing demand for vegetable oils and an explosion of interest in the health issues around dietary fats (Clemente and Cahoon 2009). These technologies have led to improved soybean oil quality with high oxidative stability, to enhance the amount of omega-3 fatty acids and avoid the generation of trans fats (Damude and Kinney 2008; Clemente and Cahoon 2009). Some issues related to improvement in the yield and quality of soybeans still need to be addressed. Loss of productivity of soybean crops can be produced by the delayed root growth and consumption of plant nutrients by the soybean cyst nematode (SCN) (Wrather and Koenning 2006). Current SCN-resistant soybean cultivars were mainly developed using germplasms including PI88788, Peking (PI548402), and PI437654, in the United States (Mitchum 2016). However, it is necessary to identify novel genetic resources involved in SCN resistance, because of reduced genetic variability. Soybean is a major food causing allergic reactions and has the high immunological cross-reactivity with almost all other legumes, peanuts, birch pollens, and cow milk proteins (Watanabe et al. 2017). At least 16 potential proteins causing allergies have been identified in soybeans. The immunodominant protein Gly m Bd 30 K (P34) has been identified and characterized, and introgression of low-P34 soybean germplasm has been attempted in order to develop hypoallergenic soybeans for use in food (Herman et al. 2003; Sewekow et al. 2008). Two natural P34-null soybean germplasms with a low expression of P34 protein were isolated via screening of approximately 16,266 accessions by the US Department of Agriculture (Joseph et al. 2006). The breeding of hypoallergenic soybean germplasm provides a basis for the development of a valuable raw material for soybean food production (Herman et al. 2003; Joseph et al. 2006). The optimum yield of soybean is influenced by agricultural decisions, including planting dates, row spacing, seeding rates, and crop rotation (Iqbal et al. 2019). The yield and seed quality of soybean are significantly affected by various environmental conditions, such as biotic and abiotic stresses (Li et al. 2020). Even though traditional breeding and gene insertion approaches have contributed to the development of soybean cultivars with high yield potential and strong stress tolerance, we still need efficient and effective tools for soybean improvement (Hyten et al. 2006; Li et al. 2020). Recently developed genome editing technology is a promising tool for accelerating the genetic engineering of agronomically important traits in soybean.
Genome editing is a useful technology not only for the identification of gene function but also for improving crop traits. Genome editing has advanced by major three technologies: the use of zinc finger nuclease (ZFN), transcription activator-like effector nuclease (TALEN), and the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein nuclease (Cas) system (Wada et al. 2020). These technologies generate double-stranded breaks (DSBs) in the DNA sequence of the target genome, using a nuclease, which are then corrected using homology-directed repair or nonhomologous end joining (NHEJ) (Gaj et al. 2013). Both ZFN and TALEN edit the genome using Fok I nuclease, which has high specificity but different specific sequence DNA binding modules. ZFNs induce both deletions and insertions, whereas TALENs are more effective in inducing deletions than insertions (Ul Ain et al. 2015). CRISPR/Cas is a genome-editing system that involves the generation of DSBs by interacting with Cas nuclease and a guide RNA (gRNA), based on DNA sequences complementary to the gRNA (Jacobs et al. 2015). CRISPR/Cas technology is the most widely used genome-editing tool because it is simpler, faster, and more efficient than ZFN and TALEN (Sun et al. 2015). The CRISPR/Cas system is classified into three categories—types I, II, and III—according to the Cas nucleases and CRISPR sequences used (Sun et al. 2015). Type I and III systems are composed of multiple Cas nucleases (e.g., Cas3 and Cas6) for editing foreign DNA/RNA, whereas the type II system directly cleaves the targeted sequence of genomic DNA using a single Cas nuclease, such as Cas9 (Makarova et al. 2011). The CRISPR/Cas9 system progresses through the formation of mature dual CRISPR RNA (crRNA) with trans-encoded small crRNA. When Cas9 loads with a short crRNA sequence near a typical sequence (NGG) in a proto-spacer-adjacent motif (PAM), Cas9 directly cleaves the target sequence (Makarova et al. 2011). A synthetic single gRNA (sgRNA) has been used in place of the mature crRNA in the CRISPR/Cas9 system (Sun et al. 2015). Thus, sgRNA and Cas9 are important components of simple and efficient genome editing, and the CRISPR/Cas9 system is widely used in plant genetic studies and crop biotechnology. This system is used in many crop species, including rice, sorghum, wheat, maize, soybean, tomato, sweet orange, apple, and grapevine (Feng et al. 2013; Jiang et al. 2013; Shan et al. 2013; Brooks et al. 2014; Jia and Wang 2014; Xing et al. 2014; Li et al. 2015; Osakabe et al. 2018). However, the CRISPR/Cas9 system in crops can lead to the generation of unintended alleles after editing via the double-strand cleavage of DNA. To address this problem and decrease the effect of off-target changes caused by Cas9, A-to-G base editing (BE) by adenine base editors and C-to-T BE by cytosine base editors were developed using Cas9 nickase (nCas9) (Kang et al. 2018; Yan et al. 2018). Crop genome editing and sequencing technologies are being continually and rapidly developed, as important tools for improving crop traits.
In this review, we classify and summarize the existing applications of genome-editing systems, including ZFN, TALEN, and CRISPR/Cas9, in soybeans. We analyze and compare the genetic variation caused by genome editing with natural variations in soybeans. We also discuss future directions for the optimization and development of new genome-editing tools in soybeans.
Soybean genome editing using ZFN
In soybeans, genetic tools, such as RNAi-based mutagenesis (Flores et al. 2008) and random mutagenesis using radiation mutagens (Men et al. 2002), chemical mutagens such as ethylmethanosulfonate (Cooper et al. 2008), and transposon-based mutagens (Mathieu et al. 2009), have been used for increasing genetic variability. However, in soybeans with highly duplicated genomes, these mutagenesis methods can produce problems such as the silencing of individual gene copies with no phenotype (Curtin et al. 2011). Site-directed mutagenesis using ZFN has been useful for solving these problems. After ZFN activity tests using GFP transgenes, ZFN-induced mutagenesis began in endogenous soybean genes such as Dicer-like (DCL), RNA-dependent RNA polymerase (RDR), and HUA enhancer 1 (HEN1), using hairy root transformation methods (Curtin et al. 2011; Sander et al. 2011). ZFN using NHEJ-mediated repair was used for the targeted mutagenesis of the Fatty acid desaturase 2-1A (FAD2-1A) gene in soybean (Bonawitz et al. 2019). Although mutagenesis using ZFN has a high success rate, it should be improved for additional applications in soybeans, to create allelic replacements such as similar site-directed approaches in paralogous genome loci.
Soybean genome editing using TALEN
TALEN has the advantage of highly efficient genome modification compared with ZFN (Bedell et al. 2012). In TALEN, the TAL effector (TALE), which has a 13–28 tandem repeat domain, recognizes a single DNA base pair and binds to specific DNA sequences by repeat-variable diresidues (RVDs) (Graham et al. 2020). The duplicated genes of Fatty acid desaturase 2–1 (FAD2-1A and FAD2-1B), which are major regulators of the soybean fatty acid pathway, have been edited with high efficiency using TALEN (Haun et al. 2014). Targeted mutagenesis of fad2-1A, fad2-1B, and fad2-1A fad2-1B in soybeans has led to improved oil quality (Haun et al. 2014). Additional targeted mutagenesis of FAD3A, which is the gene with the greatest effect on the linolenic acid pathway in soybeans, in fad2-1A fad2-1B soybean plants using TALEN significantly reduces linolenic acid levels compared to fad2-1A fad2-1B plants (Demorest et al. 2016). Although TALEN is a simple, efficient, and widely used method, the DNA binding ability of TALE is poor, because of the presence of 5-methylated cytosine (5mC) in endogenous targeted genes (Valton et al. 2012). TALEN also has problems with transfection in cells because the molecular size of TALE is larger than that of ZFN (Park et al. 2019). Thus, the use of TALEN for trait improvement and plant mutagenesis has been restricted (Weeks et al. 2016).
Soybean genome editing using CRISPR/Cas9
CRISPR/Cas9 technology is the most recent platform for genome editing and is a novel alternative to the Fok I-mediated ZFN and TALEN technologies (Jacobs et al. 2015; Du et al. 2016; Curtin et al. 2018). The efficiency of targeted mutagenesis in soybeans using the TALEN and CRISPR/Cas9 systems was recently compared (Du et al. 2016). Mutation by TALEN using the Arabidopsis AtU6-26 promoter was more efficient than that by CRISPR/Cas9. However, targeted gene mutation using CRISPR/Cas9 with the soybean GmU6-16 g-1 promoter was more efficient than that using TALEN (Du et al. 2016). Soybean has a highly duplicated genome, in which approximately 75% of the genes exist as multiple copies (Schmutz et al. 2010). Mutation by CRISPR/Cas9 technology is more appropriate for the editing of multiple target genes in soybeans (Du et al. 2016). Gene knockout using CRISPR/Cas9 can create null alleles of multiple genes because of the use of one common gRNA with common target sequences. CRISPR/Cas9 using Streptococcus pyogenes Cas9 (SpCas9) and gRNAs of nine targeted genes were used with the particle bombardment methods in soybean embryogenic cultures and showed editing efficiencies ranging from 8 to 76% in all nine genes (Li et al. 2015). The soybean Acetolactate synthase 1 (ALS1) gene is a major metabolic enzyme in branched chain amino acid biosynthesis and is involved in resistance to chlorsulfuron herbicides (Walter et al. 2014). CRISPR/Cas9 with SpCas9 and gRNA based on the unique PAM sequence of ALS1 was used to edit proline to serine at position 178 of the amino acid by changing the DNA sequence from CCC to AGC. Simultaneously, other mutations of the ALS1 gene by CRISPR/Cas9 occurred the 5-bp deletion near the PAM sequence on the cleavage site of the gRNA (Li et al. 2015).
CRISPR/Cas9 technology has been successfully applied to both the identification of gene functions and the improvement of agriculturally important traits, including flowering time, maturity, plant architecture, seed quality, and tolerance to environmental stresses in soybeans.
Application of CRISPR/Cas9-mediated gene editing to the regulation of soybean growth and development
Soybean yield is often influenced by changing agricultural traits such as flowering time, maturity, and plant height (Kantolic and Slafer 2007). Twelve genes in soybean play important roles as major regulators in flowering and maturity responses, including E1 to E11 and J (Cao et al. 2017; Lin et al. 2021). The soybean E1, E2, and E3 genes delay the flowering time when dominant and accelerate the flowering time when recessive (Miranda et al. 2020). The mutant allele Long juvenile trait (J) gene influences the flowering response with the E1 gene under short-day (SD) condition (Miranda et al. 2020). Soybean height affects the number of nodes and pods as well as yields, due to its influence on stem architecture (Kilgore-Norquest and Sneller 2000). The Dt1 gene in soybean is a major regulator of stem architecture, and dt1 recessive alleles reduce both the height and the number of nodes by promoting the termination of apical stem growth (Liu et al. 2010; Tian et al. 2010). Targeted mutagenesis of the soybean E1 gene using CRISPR/Cas9 resulted in two types of deletion mutants 11- and 40-bp in the coding region and produced an early flowering phenotype under long-day (LD) conditions (Han et al. 2019).
Targeted mutagenesis of four Late elongated hypocotyl (LHY) genes, a key component of the central oscillator regulating circadian clock in Arabidopsis (Genoud et al. 1998), by CRISPR/Cas9 produced a reduction in soybean height and elongation of internodes (Cheng et al. 2019). The Squamosa promoter binding protein-like 9 (SPL9) transcription factors are involved in the determination of plant architecture and yields in soybean (Bao et al. 2019). CRISPR/Cas9-mediated mutagenesis of four GmSPL9 genes resulted in a 1-bp deletion in the target site of GmSPL9a and GmSPL9b, and spl9a spl9b homozygous double mutants had the shorter plastochrons. Soybean plants with various combinations of GmSPL9s mutations had increased numbers of nodes (Bao et al. 2019). The quadruple mutagenesis of four Apetala 1 (AP1) genes, which function in the determination of floral meristem and floral organs in Arabidopsis (Mandel et al. 1992), using CRISPR/Cas9, changed the development of soybeans, producing effects including delayed flowering, changes in flower morphology, and increased node numbers and internode lengths (Chen et al. 2020b). Site-directed mutagenesis of the soybean floral activators, Flowering locus T 2a (GmFT2a) and Flowering locus T 5a (GmFT5a) genes, using CRISPR/Cas9 resulted in a late flowering phenotype compared with the flowering time of wild-type plants (Cai et al. 2018, 2020b). Null mutant alleles of the GmFT2b gene produced using CRISPR/Cas9 showed the late flowering phenotype under floral repressing LD conditions (Chen et al. 2020). BE mutagenesis of the GmFT2a and GmFT4 genes using CRISPR/Cas9 changed nucleotides, such as C to T or C to G mutations, at their target sites (Cai et al. 2020a). Two independent T1‐ft2a‐BE alleles showed the late flowering phenotype under both SD and LD conditions (Cai et al. 2020a). The soybean Pseudo-response regulator 37 (GmPRR37) gene was mutated using CRISPR/Cas9 in the late flowering soybean cultivar Zigongdongdou (ZGDD) (Wang L. et al., 2020). CRISPR/Cas9-mediated mutagenesis of GmPRR37 in ZGDD produced an early flowering phenotype and higher expression of GmFT2a and GmFT5a, under natural LD conditions (Wang L. et al. 2020).
The Kinase-inducible domain interacting 8–1 (GmKIX8-1) gene in soybean acts as a negative regulator of cell proliferation by controlling the transcriptional expression of D3-type cyclins (CYCLIN D3) (Baekelandt et al. 2018; Nguyen et al. 2021). CRISPR9/Cas9-induced GmKIX8-1 deletion mutants had larger seeds and leaves than those of wild-type plants, produced by increased cell proliferation (Nguyen et al. 2021).
Application of CRISPR/Cas9-mediated gene editing to the improvement of soybean seed quality
CRISPR/Cas9 technology has been also used to improve soybean seed quality by reducing the content of undesirable factors such as allergenic proteins and indigestible sugars. Mutation of three (Glyma.20g148400, Glyma.03g163500, and Glyma.19g164900) of nine major genes for allergenic conglycinin and glycinin proteins using CRISPR/Cas9 was performed using hairy root transformation methods in the soybean cultivar Harosoy 63 (Li et al. 2019). Conglycinin and glycinin proteins constitute approximately 70% of the total seed proteins in soybeans and are important biochemical components influencing the quantity and quality of soybean food products (Li et al. 2019). Sugars in soybean seeds are important for seed longevity (ElSayed et al. 2014). Raffinose family oligosaccharides (RFOs; raffinose, stachyose, and verbascose) are ubiquitous storage products and desiccation-resistance components in soybean seeds (Wang et al. 2009; ElSayed et al. 2014; de Souza et al. 2016). Although raffinose and stachyose RFOs have positive influences on metabolizable energy, these components are regarded as anti-nutritional components in soybean food because they are indigestible by animals (Valentine et al 2017). Soybean galactinol synthase is a major enzyme in the biosynthesis of RFO pathway (de Souza et al. 2016). CRISPR/Cas9-mediated double mutagenesis of two soybean Galactinol synthase 1 genes, GmGOLS1A and GmGOLS1B, produced a reduction in the total amount of RFOs in soybean seeds, resulting in improved nutritional quality (Le et al. 2020). The Fatty acid desaturase 2 (GmFAD2) changes monounsaturated oleic acid to polyunsaturated linoleic acid, controlling the amount of monounsaturated fats in soybean seeds (Do et al. 2019). Mutant alleles of the GmFAD2-1A and GmFAD2-1B genes produced using CRISPR/Cas9 produced increased amounts of oleic acid (over 80%) and decreased amounts of linoleic acid (approximately 1.3%–1.7%) in soybean seeds (Do et al. 2019). The mutation of FAD2-2 microsomal omega-6 desaturase (GmFAD2-2) using CRISPR/Cas9 increased the content of oleic acid (approximately 45.08%–65.9%) and reduced the levels of linoleic acid (approximately 16.08%–31.95%) in soybean seeds (Al Amin et al. 2019). Simultaneous mutagenesis of the GmFAD2-1A and GmFAD2-2A genes using CRISPR/Cas9 produced an increase in the oleic acid content of soybean seeds (Wu et al. 2020). Three lipoxygenases—LOX1, LOX2, and LOX3—contribute to the preposterous taste of soybeans (Liavonchanka and Feussner 2006). CRISPR/Cas9-mediated triple gmlox1/gmlox2/gmlox3 and double gmlox1/gmlox2 mutagenesis resulted in the reduction of this taste because the activity of lipoxygenases was reduced (Wang J. et al., 2020).
Application of CRISPR/Cas9-mediated gene editing to the verification of gene function in response to environmental stress
CRISPR/Cas9 technology has been successfully applied to the characterization of the genes involved in soybean responses to biotic and abiotic stresses such as pathogens, salt, heat, and drought. The Arabidopsis Constitutive expression of PR genes 5 (CPR5) gene is involved in disease resistance signaling pathways, and cpr5 mutants belong to the “disease lesion mimics” class because of their reaction tendency to as if continuous pathogen attack (Bowling et al. 1997). Targeted mutagenesis of the soybean GmCPR5 gene using CRISPR/Cas9 produced shorter trichomes in soybean leaves (Campbell et al. 2019). Thus, CRISPR/Cas9-mediated gmcpr5 mutants will be suitable for use in the study of the function of trichomes in plant–pathogen interactions in soybean. The Heat shock protein 90 s (Hsp90s), which acts as a molecular chaperone, plays an important role in plant protective responses to various environmental stresses (Xu et al. 2013). CRISPR/Cas9-mediated mutagenesis of the soybean Hsp90A2 (GmHsp90A2) gene decreased plant thermotolerance to heat stress and increased wilting in soybeans (Huang et al. 2019). Plant Sodium/hydrogen exchangers (NHXs) act as ion exchangers in the membrane by catalyzing the movement of Na+ or K+ ions (Barragán et al. 2012). A soybean gmnhk5 knockout mutant produced using CRISPR/Cas9 decreased the salt tolerance of hairy roots, whereas GmNHK5 overexpressing plants showed improved salt tolerance (Sun et al. 2021). Plant transcription factors such as AP2/EREBP, WRKY, NAC, and MYB are associated with plant resistance to abiotic stresses through the regulation of ABA signaling (Yu et al. 2021). Targeted mutagenesis of soybean NAC06 (GmNAC06) gene using CRISPR/Cas9 decreased plant tolerance to salt stress caused by the accumulation of reactive oxygen species (Li et al. 2021). Nuclear factor Y transcription factors, including NF-YA, NF-YB, and NF-YC, are involved in ABA-dependent responses to abiotic stresses, flowering time, and plant development processes (Yamamoto et al. 2009; Liang et al. 2012; Yu et al. 2021). CRISPR/Cas9-induced mutagenesis of the soybean Nuclear factor-YC subunit 14 (GmNF-YC14) gene decreased plant tolerance to salt and drought stresses (Yu et al. 2021). These results indicate that CRISPR/Cas9-mediated gene editing has provided useful clues for verifying gene functions related to stress signaling and the development of stress-tolerant soybean plants.
Application of a multiplex mutagenesis population using a CRISPR/Cas9 library in soybeans
Recent research using CRISPR/Cas9 technology has indicated that plants can be generated via large-scale transformation using CRISPR/Cas9 libraries, to improve crop genetic variation (Jacobs et al. 2017; Lu et al. 2017; Meng et al. 2017; Bai et al. 2020). In soybeans, a pooled mutagenesis population generated 407 double or triple mutants using a single multiplex sgRNA or combination of multiple sgRNAs (Bai et al. 2020). Among them, double mutagenesis of the rhizobia-induced Clavata3/embryo surrounding region-related (CLE) genes, GmRIC1 and GmRIC2, produced increased numbers of nodules in soybean roots. Triple mutagenesis of the nitrate-induced CLE genes, GmRDN1-1, GmRDN1-2, and GmRDN1-3, produced decreased nodulation in soybean roots (Bai et al. 2020). GmRICs and GmRDNs, CLE peptide-encoding genes, are associated with the regulation of nodulation in soybean roots (Reid et al. 2011). Specific gRNAs and the CRISPR/Cas9 have been used to develop an easy, rapid, and highly efficient technique to simultaneously edit various homologous genes in soybeans, an approach that can facilitate the molecular manipulation and identification of genes in soybeans.
Development of cas nucleases in crops
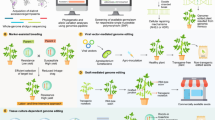
This review summarizes the ways in which the functions of numerous soybean genes have been evaluated using genome-editing technologies (Table 1). In addition, we showed the historical progress of the soybean genome editing to improve growth, seed quality, and stress tolerance (Fig. 1). The ZFN, TALEN, and CRISPR/Cas9 systems have been successfully applied to soybean genome editing, including deletion of DNA fragments, BE, DNA insertion, and gene knockout. Of those technologies, CRISPR/Cas9 is the most efficient. In the CRISPR/Cas9 system, the Cas9 nuclease used is an important component of genome editing (Zhang Y. et al. 2019). For instance, SpCas9, which has a very simple PAM (NGG) has been codon optimized for various species, including human (hCas9; Mali et al. 2013), plant (pcoCas9 and Cas9p; Li et al. 2013; Ma et al. 2015), Arabidopsis (AteCas9; Fauser et al. 2014), maize (zCas9; Lee et al. 2019), and soybean (GmCas9; Michno et al. 2015). Thus, identification of the most efficient type of Cas9 nuclease is important to the development and improvement of this versatile genome-editing tool in soybeans.
Progress and prospects of genome editing-mediated soybean improvement. The target traits and corresponding genes used for soybean genome editing are presented in a chronological order. Prospects for genome editing for further soybean improvement are suggested. The ZFN, TALEN, and CRISPR/Cas9 indicates zinc finger nuclease, transcription activator-like effector nuclease, and clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein nuclease9, respectively
There have been 42 orthologous Cas9 and Cas nucleases identified to date, with various PAMs (Zhang Y. et al. 2019). Recently, the Cpf1 nuclease, known as Cas12a, was identified as a new type of V-A Cas enzyme in class II CRISPR systems (Zetsche et al. 2015). The Cpf1 nuclease targets the T-rich region in the PAM and uses a short guide crRNA, which allows easy synthesis and engineering of crRNA (Zetsche et al. 2015). The Cpf1 nucleases, known as Cas12a, with the PAM TTTV from Lachnospiraceae bacterium (LpCpf1) and Acidaminococcus spp. (AsCpf1) have been used for targeted mutagenesis of the FAD2-1A and FAD2-1B genes in soybean protoplast cells (Kim et al. 2017). The editing efficiency using LpCpf1 and soybean Ubiquitin (GmU) promoter was increased to 91.7%, and eight targeted genes in soybean were simultaneously mutated (Duan et al. 2021). These studies demonstrated that Cpf1 nuclease is as effective and efficient in soybean genome editing as other Cas9 nucleases. Orthologous Cas nucleases from different bacterial species have been identified, including Cpf1, C2c1 (known as Cas12b), Cms1, CasX (known as Cas12e), CasY (known as Cas12d), and Cas13 (Zetsche et al. 2015; Xu et al. 2020). Genome editing using these Cas nucleases has been successful in major crops and model plant species, except soybean (Zetsche et al. 2015; Xu et al. 2020). The applications of various Cas nucleases, and the identification of the most effective Cas nucleases for soybean gene editing, would be valuable for efficient gene editing in the highly duplicated soybean genome.
Prospects for genome editing for soybean improvement
The use of genome editing to produce sophisticated modifications of specific target genes, without altering other traits, can accelerate crop breeding. In soybean, which has duplicated and multiplex genes, genome–editing technology can avert the complicated and time-consuming processes of multiple crossings required by traditional breeding to generate multi-trait soybean cultivars. Thus, it would be valuable to develop new and efficient CRISPR/Cas systems, suitable for simultaneous editing of multiplex gene family members. A multiplex CRISPR/Cas system could be widely applied to the genome editing of various crops with polyploid genomes. In soybeans, gene editing has been mainly applied to the investigation of developmental processes and the improvement of the quality of soybean seeds. In contrast, gene editing for enhancing stress tolerance has not yet been seriously attempted in soybean. There are an increasing number of health-conscious consumers, including vegetarians and vegans, for whom soybean can be used as a plant-based meat substitute. Because of the emergence of the plant-based food industry, the demand for soybeans is expected to grow very rapidly. For the wider utilization of soybeans in the new food industry, the amounts of undesirable factors such as allergenic proteins, phytic acid, and nondigestible saccharides such as melibiose, raffinose, and stachyose should be reduced or removed in soybean seeds. Thus, the elimination of various anti-nutritional factors and the increase of functional materials in soybean seeds using gene editing will lead to advances in the soybean-based food industry and feed manufacturing.
References
Al Amin N, Ahmad N, Wu N, Pu X, Ma T, Du Y, Bo X, Wang N, Sharif R, Wang P (2019) CRISPR-Cas9 mediated targeted disruption of FAD2–2 microsomal omega-6 desaturase in soybean (Glycine max. L). BMC Biotechnol 19(1):9
Baekelandt A, Pauwels L, Wang Z, Li N, De Milde L, Natran A, Vermeersch M, Li Y, Goossens A, Inzé D, Gonzalez N (2018) Arabidopsis leaf flatness is regulated by PPD2 and NINJA through repression of CYCLIN D3 genes. Plant Physiol 178(1):217–232
Bai M, Yuan J, Kuang H, Gong P, Li S, Zhang Z, Liu B, Sun J, Yang M, Yang L, Wang D, Song S, Guan Y (2020) Generation of a multiplex mutagenesis population via pooled CRISPR-Cas9 in soya bean. Plant Biotechnol J 18(3):721–731
Bao A, Chen H, Chen L, Chen S, Hao Q, Guo W, Qiu D, Shan Z, Yang Z, Yuan S, Zhang C, Zhang X, Liu B, Kong F, Li X, Zhou X, Tran LP, Cao D (2019) CRISPR/Cas9-mediated targeted mutagenesis of GmSPL9 genes alters plant architecture in soybean. BMC Plant Biol 19(1):131
Barragán V, Leidi EO, Andrés Z, Rubio L, De Luca A, Fernández JA, Cubero B, Pardo JM (2012) Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24(3):1127–1142
Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG 2nd, Tan W, Penheiter SG, Ma AC, Leung AY, Fahrenkrug SC, Carlson DF, Voytas DF, Clark KJ, Essner JJ, Ekker SC (2012) In vivo genome editing using a high-efficiency TALEN system. Nature 491(7422):114–118
Bellaloui N, Bruns HA, Abbas HK, Mengistu A, Fisher DK, Reddy KN (2015) Agricultural practices altered soybean seed protein, oil, fatty acids, sugars, and minerals in the Midsouth USA. Front Plant Sci 6:31
Bonawitz ND, Ainley WM, Itaya A, Chennareddy SR, Cicak T, Effinger K, Jiang K, Mall TK, Marri PR, Samuel JP, Sardesai N, Simpson M, Folkerts O, Sarria R, Webb SR, Gonzalez DO, Simmonds DH, Pareddy DR (2019) Zinc finger nuclease-mediated targeting of multiple transgenes to an endogenous soybean genomic locus via non-homologous end joining. Plant Biotechnol J 17(4):750–761
Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9(9):1573–1584
Brooks C, Nekrasov V, Lippman ZB, Van Eck J (2014) Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol 166(3):1292–1297
Cai Y, Chen L, Liu X, Guo C, Sun S, Wu C, Jiang B, Han T, Hou W (2018) CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol J 16(1):176–185
Cai Y, Chen L, Zhang Y, Yuan S, Su Q, Sun S, Wu C, Yao W, Han T, Hou W (2020a) Target base editing in soybean using a modified CRISPR/Cas9 system. Plant Biotechnol J 18(10):1996–1998
Cai Y, Wang L, Chen L, Wu T, Liu L, Sun S, Wu C, Yao W, Jiang B, Yuan S, Han T, Hou W (2020b) Mutagenesis of GmFT2a and GmFT5a mediated by CRISPR/Cas9 contributes for expanding the regional adaptability of soybean. Plant Biotechnol J 18(1):298–309
Campbell BW, Hoyle JW, Bucciarelli B, Stec AO, Samac DA, Parrott WA, Stupar RM (2019) Functional analysis and development of a CRISPR/Cas9 allelic series for a CPR5 ortholog necessary for proper growth of soybean trichomes. Sci Rep 9(1):14757
Cao D, Takeshima R, Zhao C, Liu B, Jun A, Kong F (2017) Molecular mechanisms of flowering under long days and stem growth habit in soybean. J Exp Bot 68(8):1873–1884
Chen L, Cai Y, Qu M, Wang L, Sun H, Jiang B, Wu T, Liu L, Sun S, Wu C, Yao W, Yuan S, Han T, Hou W (2020a) Soybean adaption to high-latitude regions is associated with natural variations of GmFT2b, an ortholog of FLOWERING LOCUS T. Plant Cell Environ 43(4):934–944
Chen L, Nan H, Kong L, Yue L, Yang H, Zhao Q, Fang C, Li H, Cheng Q, Lu S, Kong F, Liu B, Dong L (2020b) Soybean AP1 homologs control flowering time and plant height. J Integr Plant Biol 62(12):1868–1879
Cheng Q, Dong L, Su T, Li T, Gan Z, Nan H, Lu S, Fang C, Kong L, Li H, Hou Z, Kou K, Tang Y, Lin X, Zhao X, Chen L, Liu B, Kong F (2019) CRISPR/Cas9-mediated targeted mutagenesis of GmLHY genes alters plant height and internode length in soybean. BMC Plant Biol 19(1):562
Clemente TE, Cahoon EB (2009) Soybean oil: genetic approaches for modification of functionality and total content. Plant Physiol 151(3):1030–1040
Cooper JL, Till BJ, Laport RG, Darlow MC, Kleffner JM, Jamai A, El-Mellouki T, Liu S, Ritchie R, Nielsen N, Bilyeu KD, Meksem K, Comai L, Henikoff S (2008) TILLING to detect induced mutations in soybean. BMC Plant Biol 8:9
Curtin SJ, Zhang F, Sander JD, Haun WJ, Starker C, Baltes NJ, Reyon D, Dahlborg EJ, Goodwin MJ, Coffman AP, Dobbs D, Joung JK, Voytas DF, Stupar RM (2011) Targeted mutagenesis of duplicated genes in soybean with zinc-finger nucleases. Plant Physiol 156(2):466–473
Curtin SJ, Michno JM, Campbell BW, Gil-Humanes J, Mathioni SM, Hammond R, Gutierrez-Gonzalez JJ, Donohue RC, Kantar MB, Eamens AL, Meyers BC, Voytas DF, Stupar RM (2015) MicroRNA maturation and microRNA target gene expression regulation are severely disrupted in soybean dicer-like1 double mutants. G3 (bethesda) 6(2):423–433
Curtin SJ, Xiong Y, Michno JM, Campbell BW, Stec AO, Čermák T, Starker C, Voytas DF, Eamens AL, Stupar RM (2018) CRISPR/Cas9 and TALENs generate heritable mutations for genes involved in small RNA processing of Glycine max and Medicago truncatula. Plant Biotechnol J 16(6):1125–1137
Damude HG, Kinney AJ (2008) Engineering oilseeds to produce nutritional fatty acids. Physiol Plant 132(1):1–10
de Souza VD, Willems L, van Arkel J, Dekkers BJW, Hilhorst HWM, Bentsink L (2016) Galactinol as marker for seed longevity. Plant Sci 246:112–118
Demorest ZL, Coffman A, Baltes NJ, Stoddard TJ, Clasen BM, Luo S, Retterath A, Yabandith A, Gamo ME, Bissen J, Mathis L, Voytas DF, Zhang F (2016) Direct stacking of sequence-specific nuclease-induced mutations to produce high oleic and low linolenic soybean oil. BMC Plant Biol 16(1):225
Do PT, Nguyen CX, Bui HT, Tran LTN, Stacey G, Gillman JD, Zhang ZJ, Stacey MG (2019) Demonstration of highly efficient dual gRNA CRISPR/Cas9 editing of the homeologous GmFAD2-1A and GmFAD2-1B genes to yield a high oleic, low linoleic and alpha-linolenic acid phenotype in soybean. BMC Plant Biol 19(1):311
Du H, Zeng X, Zhao M, Cui X, Wang Q, Yang H, Cheng H, Yu D (2016) Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J Biotechnol 217:90–97
Duan K, Cheng Y, Ji J, Wang C, Wei Y, Wang Y (2021) Large chromosomal segment deletions by CRISPR/LbCpf1-mediated multiplex gene editing in soybean. J Integr Plant Biol 63(9):1620–1631
ElSayed AI, Rafudeen MS, Golldack D (2014) Physiological aspects of raffinose family oligosaccharides in plants: protection against abiotic stress. Plant Biol (stuttg) 16(1):1–8
Fauser F, Schiml S, Puchta H (2014) Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J 79(2):348–359
Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P, Cao F, Zhu S, Zhang F, Mao Y, Zhu JK (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23(10):1229–1232
Flores T, Karpova O, Su X, Zeng P, Bilyeu K, Sleper DA, Nguyen HT, Zhang ZJ (2008) Silencing of GmFAD3 gene by siRNA leads to low alpha-linolenic acids (18:3) of fad3-mutant phenotype in soybean [Glycine max (Merr)]. Transgenic Res 17(5):839–850
Gaj T, Gersbach CA, Barbas CF 3rd (2013) ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31(7):397–405
Genoud T, Millar AJ, Nishizawa N, Kay SA, Schäfer E, Nagatani A, Chua NH (1998) An Arabidopsis mutant hypersensitive to red and far-red light signals. Plant Cell 10(6):889–904
Graham N, Patil GB, Bubeck DM, Dobert RC, Glenn KC, Gutsche AT, Kumar S, Lindbo JA, Maas L, May GD, Vega-Sanchez ME, Stupar RM, Morrell PL (2020) Plant genome editing and the relevance of off-target changes. Plant Physiol 183(4):1453–1471
Han J, Guo B, Guo Y, Zhang B, Wang X, Qiu LJ (2019) Creation of early flowering germplasm of soybean by CRISPR/Cas9 technology. Front Plant Sci 10:1446
Haun W, Coffman A, Clasen BM, Demorest ZL, Lowy A, Ray E, Retterath A, Stoddard T, Juillerat A, Cedrone F, Mathis L, Voytas DF, Zhang F (2014) Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol J 12(7):934–940
Herman EM, Helm RM, Jung R, Kinney AJ (2003) Genetic modification removes an immunodominant allergen from soybean. Plant Physiol 132(1):36–43
Huang Y, Xuan H, Yang C, Guo N, Wang H, Zhao J, Xing H (2019) GmHsp90A2 is involved in soybean heat stress as a positive regulator. Plant Sci 285:26–33
Hyten DL, Song Q, Zhu Y, Choi IY, Nelson RL, Costa JM, Specht JE, Shoemaker RC, Cregan PB (2006) Impacts of genetic bottlenecks on soybean genome diversity. Proc Natl Acad Sci U S A 103(45):16666–16671
Iqbal N, Hussain S, Ahmed Z, Yang F, Wang X, Liu W, Yong T, Du J, Shu K, Yang W, Liu J (2019) Comparative analysis of maize–soybean strip intercropping systems: a review. Plant Prod Sci 22(2):131–142
Jacobs TB, LaFayette PR, Schmitz RJ, Parrott WA (2015) Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol 15:16
Jacobs TB, Zhang N, Patel D, Martin GB (2017) Generation of a collection of mutant tomato lines using pooled CRISPR libraries. Plant Physiol 174(4):2023–2037
Jia H, Wang N (2014) Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS ONE 9(4):e93806
Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 41(20):e188
Joseph LM, Hymowitz T, Schmidt MA, Herman EM (2006) Evaluation of Glycine germplasm for nulls of the immunodominant allergen P34/Gly m Bd 30k. Crop Sci 46(4):1755–1763
Kang BC, Yun JY, Kim ST, Shin Y, Ryu J, Choi M, Woo JW, Kim JS (2018) Precision genome engineering through adenine base editing in plants. Nat Plants 4(7):427–431
Kantolic AG, Slafer GA (2007) Development and seed number in indeterminate soybean as affected by timing and duration of exposure to long photoperiods after flowering. Ann Bot 99(5):925–933
Kilgore-Norquest L, Sneller CH (2000) Effect of stem termination on soybean traits in southern U S production systems. Crop Sci 40(1):83–90
Kim H, Kim ST, Ryu J, Kang BC, Kim JS, Kim SG (2017) CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat Commun 8:14406
Le H, Nguyen NH, Ta DT, Le TNT, Bui TP, Le NT, Nguyen CX, Rolletschek H, Stacey G, Stacey MG, Pham NB, Do PT, Chu HH (2020) CRISPR/Cas9-mediated knockout of galactinol synthase-encoding genes reduces raffinose family oligosaccharide levels in soybean seeds. Front Plant Sci 11:612942
Lee K, Zhang Y, Kleinstiver BP, Guo JA, Aryee MJ, Miller J, Malzahn A, Zarecor S, Lawrence-Dill CJ, Joung JK, Qi Y, Wang K (2019) Activities and specificities of CRISPR/Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnol J 17(2):362–372
Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J (2013) Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol 31(8):688–691
Li Z, Liu ZB, Xing A, Moon BP, Koellhoffer JP, Huang L, Ward RT, Clifton E, Falco SC, Cigan AM (2015) Cas9-Guide RNA directed genome editing in soybean. Plant Physiol 169(2):960–970
Li C, Nguyen V, Liu J, Fu W, Chen C, Yu K, Cui Y (2019) Mutagenesis of seed storage protein genes in soybean using CRISPR/Cas9. BMC Res Notes 12(1):176
Li MW, Wang Z, Jiang B, Kaga A, Wong FL, Zhang G, Han T, Chung G, Nguyen H, Lam HM (2020) Impacts of genomic research on soybean improvement in East Asia. Theor Appl Genet 133(5):1655–1678
Li M, Chen R, Jiang Q, Sun X, Zhang H, Hu Z (2021) GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol Biol 105(3):333–345
Liang M, Hole D, Wu J, Blake T, Wu Y (2012) Expression and functional analysis of NUCLEAR FACTOR-Y, subunit B genes in barley. Planta 235(4):779–791
Liavonchanka A, Feussner I (2006) Lipoxygenases: occurrence, functions and catalysis. J Plant Physiol 163(3):348–357
Lin X, Liu B, Weller JL, Abe J, Kong F (2021) Molecular mechanisms for the photoperiodic regulation of flowering in soybean. J Integr Plant Biol 63(6):981–994
Liu B, Watanabe S, Uchiyama T, Kong F, Kanazawa A, Xia Z, Nagamatsu A, Arai M, Yamada T, Kitamura K, Masuta C, Harada K, Abe J (2010) The soybean stem growth habit gene Dt1 is an ortholog of Arabidopsis terminal flower1. Plant Physiol 153(1):198–210
Lu Y, Ye X, Guo R, Huang J, Wang W, Tang J, Tan L, Zhu JK, Chu C, Qian Y (2017) Genome-wide targeted mutagenesis in rice using the CRISPR/Cas9 system. Mol Plant 10(9):1242–1245
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu YG (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8(8):1274–1284
Mahmoud AA, Natarajan SS, Bennett JO, Mawhinney TP, Wiebold WJ, Krishnan HB (2006) Effect of six decades of selective breeding on soybean protein composition and quality: a biochemical and molecular analysis. J Agric Food Chem 54(11):3916–3922
Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV (2011) Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9(6):467–477
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013) RNA-guided human genome engineering via Cas9. Science 339(6121):823–826
Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360(6401):273–277
Mathieu M, Winters EK, Kong F, Wan J, Wang S, Eckert H, Luth D, Paz M, Donovan C, Zhang Z, Somers D, Wang K, Nguyen H, Shoemaker RC, Stacey G, Clemente T (2009) Establishment of a soybean (Glycine max L Merr) transposon-based mutagenesis repository. Planta 229(2):279–289
Men AE, Laniya TS, Searle IR, Iturbe-Ormaetxe I, Gresshoff I, Jiang Q, Carroll BJ, Gresshoff PM (2002) Fast neutron mutagenesis of soybean (Glycine soja L) produces a supernodulating mutant containing a large deletion in linkage group H. Genome Lett 1(3):147–215
Meng X, Yu H, Zhang Y, Zhuang F, Song X, Gao S, Gao C, Li J (2017) Construction of a genome-wide mutant library in rice using CRISPR/Cas9. Mol Plant 10(9):1238–1241
Michno JM, Wang X, Liu J, Curtin SJ, Kono TJ, Stupar RM (2015) CRISPR/Cas mutagenesis of soybean and Medicago truncatula using a new web-tool and a modified Cas9 enzyme. GM Crops Food 6(4):243–252
Miranda C, Scaboo A, Cober E, Denwar N, Bilyeu K (2020) The effects and interaction of soybean maturity gene alleles controlling flowering time, maturity, and adaptation in tropical environments. BMC Plant Biol 20(1):65
Mitchum MG (2016) Soybean resistance to the soybean cyst nematode Heterodera glycines: An update. Phytopathology 106(12):1444–1450
Nguyen CX, Paddock KJ, Zhang Z, Stacey MG (2021) GmKIX8-1 regulates organ size in soybean and is the causative gene for the major seed weight QTL qSw17-1. New Phytol 229(2):920–934
Osakabe Y, Liang Z, Ren C, Nishitani C, Osakabe K, Wada M, Komori S, Malnoy M, Velasco R, Poli M, Jung MH, Koo OJ, Viola R, Nagamangala KC (2018) CRISPR-Cas9-mediated genome editing in apple and grapevine. Nat Protoc 13(12):2844–2863
Park SR, Park J, Lim SH, Lee JY, Kim BG (2019) Current status of new plant breeding technologies and crop development. Korean J Breed Sci 51(3):161–174
Reid DE, Ferguson BJ, Gresshoff PM (2011) Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Mol Plant Microbe Interact 24(5):606–618
Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, Pierick CJ, Hoffman E, Maeder ML, Khayter C, Reyon D, Dobbs D, Langenau DM, Stupar RM, Giraldez AJ, Voytas DF, Peterson RT, Yeh JR, Joung JK (2011) Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat Methods 8(1):67–69
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu S, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du J, Tian Z, Zhu L, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA (2010) Genome sequence of the palaeopolyploid soybean. Nature 463(7278):178–183
Sewekow E, Kessler LC, Seidel-Morgenstern A, Rothkötter HJ (2008) Isolation of soybean protein P34 from oil bodies using hydrophobic interaction chromatography. BMC Biotechnol 8:27
Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL, Gao C (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol 31(8):686–688
Sun X, Hu Z, Chen R, Jiang Q, Song G, Zhang H, Xi Y (2015) Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci Rep 5:10342
Sun T, Ma N, Wang C, Fan H, Wang M, Zhang J, Cao J, Wang D (2021) A Golgi-localized sodium/hydrogen exchanger positively regulates salt tolerance by maintaining higher K(+)/Na(+) ratio in soybean. Front Plant Sci 12:638340
Tian Z, Wang X, Lee R, Li Y, Specht JE, Nelson RL, McClean PE, Qiu L, Ma J (2010) Artificial selection for determinate growth habit in soybean. Proc Natl Acad Sci U S A 107(19):8563–8568
Ul Ain Q, Chung JY, Kim YH (2015) Current and future delivery systems for engineered nucleases: ZFN TALEN RGEN. J Control Release 205:120–127
Valentine MF, De Tar JR, Mookkan M, Firman JD, Zhang ZJ (2017) Silencing of Soybean Raffinose Synthase Gene Reduced Raffinose Family Oligosaccharides and Increased True Metabolizable Energy of Poultry Feed. Front Plant Sci 8:692
Valton J, Dupuy A, Daboussi F, Thomas S, Maréchal A, Macmaster R, Melliand K, Juillerat A, Duchateau P (2012) Overcoming transcription activator-like effector (TALE) DNA binding domain sensitivity to cytosine methylation. J Biol Chem 287(46):38427–38432
Wada N, Ueta R, Osakabe Y, Osakabe K (2020) Precision genome editing in plants: state-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biol 20(1):234
Walter KL, Strachan SD, Ferry NM, Albert HH, Castle LA, Sebastian SA (2014) Molecular and phenotypic characterization of Als1 and Als2 mutations conferring tolerance to acetolactate synthase herbicides in soybean. Pest Manag Sci 70(12):1831–1839
Wang Z, Zhu Y, Wang L, Liu X, Liu Y, Phillips J, Deng X (2009) A WRKY transcription factor participates in dehydration tolerance in Boea hygrometrica by binding to the W-box elements of the galactinol synthase (BhGolS1) promoter. Planta 230(6):1155–1166
Wang J, Kuang H, Zhang Z, Yang Y, Yan L, Zhang M, Song S, Guan Y (2020) Generation of seed lipoxygenase-free soybean using CRISPR-Cas9. Crop J 8:432–439
Wang L, Sun S, Wu T, Liu L, Sun X, Cai Y, Li J, Jia H, Yuan S, Chen L, Jiang B, Wu C, Hou W, Han T (2020) Natural variation and CRISPR/Cas9-mediated mutation in GmPRR37 affect photoperiodic flowering and contribute to regional adaptation of soybean. Plant Biotechnol J 18(9):1869–1881
Watanabe D, Adányi N, Takács K, Maczó A, Nagy A, Gelencsér É, Pachner M, Lauter K, Baumgartner S, Vollmann J (2017) Development of soybeans with low P34 allergen protein concentration for reduced allergenicity of soy foods. J Sci Food Agric 97(3):1010–1017
Weeks DP, Spalding MH, Yang B (2016) Use of designer nucleases for targeted gene and genome editing in plants. Plant Biotechnol J 14(2):483–495
Wrather JA, Koenning SR (2006) Estimates of disease effects on soybean yields in the United States 2003 to 2005. J Nematol 38(2):173–180
Wu N, Lu Q, Wang P, Zhang Q, Zhang J, Qu J, Wang N (2020) Construction and analysis of GmFAD2-1A and GmFAD2-2A soybean fatty acid desaturase mutants based on CRISPR/Cas9 technology. Int J Mol Sci 21(3):1104
Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14:327
Xu J, Xue C, Xue D, Zhao J, Gai J, Guo N, Xing H (2013) Overexpression of GmHsp90s, a heat shock protein 90 (Hsp90) gene family cloning from soybean, decrease damage of abiotic stresses in Arabidopsis thaliana. PLoS ONE 8(7):e69810
Xu H, Zhang L, Zhang K, Ran Y (2020) Progresses, challenges, and prospects of genome editing in soybean (Glycine max). Front Plant Sci 11:571138
Yamamoto A, Kagaya Y, Toyoshima R, Kagaya M, Takeda S, Hattori T (2009) Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J 58(5):843–856
Yan F, Kuang Y, Ren B, Wang J, Zhang D, Lin H, Yang B, Zhou X, Zhou H (2018) Highly efficient AT to GC base editing by Cas9n-guided tRNA adenosine deaminase in rice. Mol Plant 11(4):631–634
Yu TF, Liu Y, Fu JD, Ma J, Fang ZW, Chen J, Zheng L, Lu ZW, Zhou YB, Chen M, Xu ZS, Ma YZ (2021) The NF-Y-PYR module integrates the abscisic acid signal pathway to regulate plant stress tolerance. Plant Biotechnol J. https://doi.org/10.1111/pbi.13684
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163(3):759–771
Zhang Y, Malzahn AA, Sretenovic S, Qi Y (2019) The emerging and uncultivated potential of CRISPR technology in plant science. Nat Plants 5(8):778–794
Funding
This work was supported by grants from the New Breeding Technologies Development Program (Project No. PJ01476901 and PJ01653302), Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Contributions
D.B. and H.J.C. organized the data and wrote the manuscript. M.C.K. wrote and edited the manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Baek, D., Chun, H.J. & Kim, M.C. Genome editing provides a valuable biological toolkit for soybean improvement. Plant Biotechnol Rep 16, 357–368 (2022). https://doi.org/10.1007/s11816-022-00778-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-022-00778-6