Abstract
The necrotrophic pathogen Valsa mali (Vm) resulting Valsa canker of apple is considered one of the most destructing fungal diseases. To study the resistance mechanism, we investigated gene expression profiles of suspension cells from resistant varieties ‘Dongbeishanjingzi’ (DS, Malus baccata) and susceptible varieties ‘Gala’ (GL, Malus × domestica) in response to Vm metabolism (VmM) using RNA sequencing (RNA-seq). Functional enrichment showed that differentially expressed genes (DEGs) were widely involved in multiple metabolisms or signals, such as “Lipid metabolic process”, “plant hormone signal transduction” and “plant-pathogen interaction”. Further expressional patterns exhibited that induction of genes related to ‘‘xyloglucan biosynthetic process’’ and ‘‘cell wall biogenesis’’ was beneficial for cell wall integrity and tolerance of DS cells. In brassinosteroid signaling, we identified that TCH4 gene MbTCH4-1 positively regulated Vm resistance of ‘Fuji’ fruit, but BSK gene MdBSK1 negatively regulated the resistance. In contrast, cell death associated with hypersensitive response caused by up-regulation of CNGCs and CDPK genes is an important cause of weakened tolerance in GL cells. Our results provide a new insight direction for the molecular mechanism of apple against Valsa canker.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
China, with more than 40% of planting area and output of the whole world in 2020, is the largest apple producer and consumer in the world and plays an important role in the world apple industry. Valsa canker of apple, caused by necrotrophic pathogen Valsa mali (Vm), was considered one of the most destructive fungal diseases. The pathogen mainly damages bark or endangers the branches of the young and old trees in different varieties (Yin et al. 2015). For this disease, wound sites resulted from pruning, frostbite and sunburn are major channels of pathogen invading (Kepley and Jacobi 2000). The pathogen usually results rot, wrinkling, softening and shriveling of tissues, and finally the death of the whole plant (Zhai et al. 2014). The hyphae of Vm could extend along xylem vessels, tracheids and rays, and when surviving in the xylem for at least 3 to 5 years (Chen et al. 2016). At present, little effective strategy was carried out although various management practices (physical, chemical and biological control methods) have been proposed to limit Vm spreading. Additionally, most countries are working to eliminate the destruction of agricultural ecological environment, human, and animal health caused by the excessive use of chemical drugs. For the long run, understanding the mechanism of host–pathogen interaction and resistance breeding takes critical roles in effective control of this disease.
To facilitate colonization, multiple toxic substances could be secreted by necrotrophic pathogens and further resulted in death of host cells. Indeed, abundant pathogenicity-related proteins, such as cellulose, xylanase and pectinase, are widely distributed in Vm genome (Yin et al. 2015). Conversely, cell wall is the dynamic physical barrier to prevent pathogen invasion, and also takes crucial roles in recognition of pathogen signals and induction of immune responses (Ajengui et al. 2017). Plants have a two-layer immune system to block the infection of microbes. Initially, host cell sense signals of pathogen-associated molecular patterns (PAMPs) via cell surface-located pattern-recognition receptors (PRRs) (Man et al. 2022). The first layer of plant defense at the cellular level is the induction of PAMP triggered immunity (PTI) to prevent further infection. Pathogens inhibit the signal transduction of PTI by secreting virulence effectors, and then promote infection (Ortiz-Morea et al. 2022). In the second layer of immune system, the signals of virulence effectors are recognized directly or indirectly by plant resistance (R) proteins, named as effector-triggered immunity (ETI) (Sánchez-Martín and Keller, 2021). The plasmodial exosome effector Avr4 of the tomato (Solanum lycopersicum) leaf mold pathogen Cladosporium fulvum binds to fungal chitin to prevent cell wall degradation by host chitinases (Joosten et al. 1994; Burg et al. 2006). In addition, phytohormones, including jasmonic acid (JA), brassinosteroid (BR), salicylic acid (SA), ethylene (ET), and abscisic acid (ABA), are essential mediators of plant immunity and play major roles in the regulation of plant defense responses (Thaler et al. 2012; Nakashita et al. 2003). Pathogen-induced cell death is effective for plants against biotrophs, but shows the opposite effect against necrotrophs. To combat necrotrophic infections, JA/ET-dependent responses are beneficial for plant resistance (Mao et al, 2021). These signals lead to plant systemic-acquired resistance (SAR) and inhibit secondary infection of healthy tissues.
To accelerate the progress of resistance breeding, a large number of efforts were focused on molecular breeding and discovery of resistance germplasm and related genes. Transgenic plants have been obtained from several apple cultivars or rootstocks, such as ‘Golden Delicious’, ‘Royal Gala’ and ‘eight edged Begonia’. For instance, antisense suppression of Aldose-6-phosphate Reductase (A6PR) in apple reduced the expression of 56 Nucleotide Binding/Leucine-rich Repeat (NLR) genes and compromised its resistance against Alternaria alternata (Meng et al. 2018). Overexpression of a key UDP-glucose gene MdUGT88F1 increased the level of phloridzin but slightly compromised the valsa canker resistance (Zhou et al. 2019). Besides functional genes, it is confirmed that transcription factors MdWHy and MdWRKY15 take a crucial role in apple tolerance to biotic stresses (Zhang et al. 2018; Zhao et al. 2020). Nevertheless, acknowledgment of resistance mechanism and related genes is largely limited until now.
In this study, we obtained suspension cells of apple germplasm ‘Dongbeishanjingzi’ (DS, Malus baccata, resistant to valsa canker) (Yang et al. 2022) and ‘Gala’ (GL, Malus × domestica, highly susceptible) (Zhao et al. 2016). To explain the global expression network of apple responses to Vm signaling, we investigated the gene expression profiles of suspension cells of DS and GL at 0, 1, 3 and 6 h of VmM exposure using RNA sequencing. Disease-resistance-related genes were screened, and the role of several key genes for valsa canker resistance was verified using expression assay and transient expression assay. This provided valuable information for further study of resistance genes and resistant mechanisms.
Materials and methods
Vm metabolism (VmM) collection
To obtain VmM, Vm isolate Vm-A-003 was cultured in PDA medium at 28 °C for 5 days in the dark. Ten mycelial blocks with 5 mm diameter were transferred to 100 mL soluble starch (SS) liquid medium, and were stationary cultured at 25 °C in the dark. Each liter of SS liquid medium contained 10 g Soluble starch, 2 g L-Asparagine, 1 g KH2PO4, 0.5 g MgSO4-7H2O, 0.88 g ZnSO4-7H2O, 1.5 mg Fe (NO3)3-H2O, 0.44 mg MnSO4-5H2O, 0.5 μg vitamin H and 0.1 mg vitamin B1. After 10 days of culture, the VmM was collected by filtration and centrifuging.
Callus induction and treatment of suspension cells
Functional leaves of resistant varieties ‘Dongbeishanjingzi’ (DS) (Yang et al. 2022) and susceptible varieties ‘Gala’ (GL) (Zhao et al. 2016) tissue culture plantlets were collected and used as experimental materials for callus induction (Both DS and GL are grown in the experimental orchard of Gansu Agricultural University.) Leaves were cut into leaf discs with 2.0 cm length and transferred to the induction culture medium. After 30 days, leaf discs were transferred to the proliferation culture medium and cultured in an incubator (25 ± 2 °C, 14 h light and 10 h dark). The best medium for callus induction was selected with different hormone combinations. After several subcultures, when the calluses induced by DS and ‘GL were yellow and loose, put them into liquid culture medium with the same hormone combinations as the respective induction medium, and cultured in shaking table with a rotating speed of 130 r/min. Before VmM treatment, the suspension cells of DS and GL were filtered by 40-mesh filters and pre-cultured at 25 °C for 3 days in the dark. For treatment, 6 mL VmM was mixed with 500 μL cells (dense volume) and 23.5 mL MS liquid medium and shaking co-cultured at 25 °C, 120 rpm in the dark. After co-cultured 6, 12 and 24 h, the viability of cells was determined using MTT method (Meerloo et al. 2011). For RNA sequencing, the cells were collected after co-cultured 1, 3 and 6 h, and recorded as T1, T2 and T3, respectively. For the control (C), 6 mL VmM was replaced with equal volume of optimal liquid medium. Three independent biological replicates were set up for each sample treatment.
Preparation and sequencing of cDNA library
Total RNAs were isolated using the Plant RNAout kit (160,906–50, Tiandz Inc., Beijing, China). The integrity of each RNA sample was examined with 1.5% denatured agarose gel electrophoresis, an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and a NanoDrop 2000 Spectrophotometer (Thermo Scientific). The first cDNA was synthesized using random hexamer primers, and the second cDNA strand was synthesized by RNase H and DNA polymerase I. Poly (A) mRNA was selected using oligo (dT) magnetic beads. The end of cDNA was modified with T4 DNA polymerase, and 3'-terminal single nucleotide adenine (A) was leveled with Klenow 3' to 5' polymerase. T4 ligase is used for connection of the sequenced linker and the fragment. After enrichment of fragment, cDNA was amplified using PCR ABI StepOnePlus (ABI, CA, USA) and cDNA pool is used for high-throughput sequencing (Tarazona et al. 2012; Ye 2006).
Screening and functional annotation of differential genes
After filtering, the high-quality clean reads of DS and GL were mapped on reference genome Malus baccata genome v1.0 and Malus × domestica genome GDDH13 v1.1, respectively (https://www.rosaceae.org/). Gene expression levels were determined using the software RSEM (Dewey and Li. 2011). Differentially expressed genes (DEGs) were screened using software Noiseq (Tarazona et al. 2012). |log2Fold Change|≥ 2 and FDR ≤ 0.01 were adopted to judge the significance of the gene expression differences (Tarazona et al. 2012). To determine the functional annotation of DEGs, a BLAST alignment was performed by searching Kyoto Encyclopedia of Genes and Genomes (KEGG) and Web Gene Ontology Annotation Plot (WEGO) databases with an E-value ≤ 1e-5 (Fukushima et al. 2016).
Quantitative real-time PCR
Expression pattern of DEGs belonging to enriched GO terms or KEGG pathway was validated using qRT-PCR assay. RNA purification and reverse transcription were performed using the PrimeScript™ RT kit with gDNA Eraser (code RR047A, TAKARA Biotechnology Co., Ltd., Dalian, China). Primers were designed by the online website (http://primer3.ut.ee/), and the sequences of the primers used are listed in Table S1. Total RNA extraction, cDNA synthesis and qRT-PCR amplification were performed as described by Zuo et al. (2018). The relative expression levels were normalized according to the 2−ΔΔCt method (Livak and Schmittgen, 2002). Three biological replicates were carried out for each gene.
Vector construction and transient expression assays
We amplified the CDS of the genes MbTCH4-1 (MABA012284) and MdBSK1 (MD06G1030500) in brassinosteroid using specific primers (Table S2). After sequencing, each gene was cloned into the pFGC5941 vector. Based on transformation, positive colonies of agrobacterium GV3101 carrying pFGC5941-MABA012284, pFGC5941-MD06G1030500 and empty vectors (Control) were shaking cultured to OD600 = 0.8 in 15 mL YEP liquid medium-containing appropriate antibiotics. The agrobacterium was suspended in MES buffer (Mao et al, 2021). The suspension was kept at 4 °C for 4 h and 200 µL of the bacterial solution was infiltrated into ‘Fuji’ fruit (150 days after flowering) by injection with a sterile syringe. Vm discs were incubated on each injection site after 3 days of culture at 25 °C. The disease lesions were measured at 24, 36, 48, 60, 72 and 84 h after incubation, respectively. Histograms were also made to analyze their resistance to the Vm.
Statistical analysis
Microsoft Excel 2010 was used for statistical analysis of basic data. The significance of the differences between means was statistically analyzed using t test (*P < 0.05; **P < 0.01).
Results
Selection of medium for GL and DS suspension cells and resistance to VmM
We selected the hormone combinations for callus proliferation: 0.5 mg/L 2,4-D and 1 mg/L 6-BA were suitable for callus of DS, 0.1 mg/L NAA and 1 mg/L 6-BA were for GL. On the MS medium added with above hormone combinations, we obtained the yellowish, sparsely granular callus with high proliferation rate. The callus of each variety was transformed into liquid medium with above hormone combinations and shaking cultured, and the suspension cells were obtained.
The viability of GL and DS suspension cells were calculated when that were treated with VmM for 6 h, 12 h and 24 h, respectively (Fig. 1). Cell viability decreased with increasing treatment time. At the third time point (24 h), the cell viability of GL decreased to 84.80 ± 1.25%, which was much lower than that of DS (89.33 ± 1.71%). These results indicate that the Vm resistance of GL and DS suspension cells could be distinguished by VmM treatment.
Data quality and differential gene analysis
After quality control and filtering, more than 40 million raw reads and 39 million clear reads were detected in each sample (Table S3). The percentage of mapped genome, Clean Data Q20 and Q30 of all samples ranged from 86.60% to 96.02%, 93.55% to 97.55% and 86.69% to 93.14%, respectively. Above results show the high quality of sequence data, which could be used for subsequent analysis.
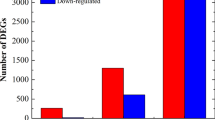
Compared to the control, 1 999, 2 177, and 2 239 DEGs were discovered in DS cells treated with VmM for 1 h (DT1), 3 h (DT2) and 6 h (DT3), respectively. And 1 764 (GT1), 4 947 (GT2) and 3 671 (GT3) DEGs in GL (Fig. 2). More than half of DEGs were up-regulated at the first time point (DT1 and GT1), but down-regulated at the second and third time points (DT2, DT3, GT2 and GT3). No less 60% of DEGs were up-regulated in DT1 and GT1, whereas only 17% and 31% in DT3 and GT3, respectively. These results reveal a distinguish expression pattern of DEGs among different samples.
Enrichment of GO function
The ten most enriched GO terms related to ‘biological process’ were extracted (Fig. 3). Enrichment of GO terms related to cell wall exhibited differences between suspension cells of resistant rootstock DS and susceptible variety GL. For instance, GO terms “lignin catabolic process”, “lipid metabolic process” and “plant type cell wall organization” in GL, while “xyloglucan metabolic process” and ‘cell wall biogenesis’ were enriched only in DT1. Besides that, “Protein dephosphorylation” and “cellular glucan metabolic process” were enriched in GL and DS. The DEGs of GO terms enriched in DT1 were mostly up-regulated, while almost all DEGs were down-regulated in DT2 and DT3. These results displayed the distinct gene expressions between suspension cells of DS and GL in response to VmM signaling.
Enrichment of KEGG pathway
To understand the mechanism of responses to Vm in resistant and susceptible varieties, the ten most enriched KEGG pathways from each sample were carried out (Fig. 4). The results showed that “Plant-pathogen interaction” and “MAPK signaling pathway-plant” were enriched in the early stages of suspension cells from both GL and DS, and most of the DEGs were up-regulated. However, “Plant hormone signal transmission” was enriched in DS cells at all three time points, but not in GL. Additionally, a large number of pathways such as “homologous recombination”, “phosphatidylinositol signal transduction”, “cell cycle” and “DNA replication” were obviously affected by VmM in DS or GL. Overall, obvious differences of enriched KEGG pathways were observed from VmM treated cells between DS and GL.
Expression patterns of “plant-pathogen interaction”-related genes
To explore the biotic resistance related responses of cells to VmM, expressions of DEGs associated with “plant pathogen interaction” were further analyzed (Fig. 5). Compared to DS, more DEGs encoding CNGCs and CDPK proteins, which influence on Ca2+ flux and usually resulting hypersensitive response (HR), were detected in GL. For DEGs encoding FLS2, four members were discovered from DS but only two from GL. On the downstream of FLS2, a few transcription factors (TF) responsible for induction of defense-related genes, such as WRKY22/09, were induced in both DS and GL. In addition, several genes associated with effector-triggered immunity, including Pti5, Pti6, PBS1, and WRKY1/2, were differentially and distinctly expressed in two varieties.
Expression patterns of genes related to “plant hormone signal transduction”
To understanding the roles of hormonal signaling during cells responses to VmM, the expression patterns of DEGs involved in the pathway ‘plant hormone signal transduction’ were further investigated (Fig. 6). In ET and ABA signaling, DEGs exhibited similar expression patterns in DS and GL. In BR signaling pathway, the DEGs were mainly up-regulated in DS, but down-regulated in GL. Furthermore, for disease resistance like genes in SA signaling pathway, three down-regulated TGA genes were discovered in GL but only one in DS. Above all, DEGs related to BR and SA signaling were distinctly expressed in DS and GL.
Two genes in BR signaling regulate the resistance of ‘Fuji’ apple fruits to Vm
In brassinosteroid signaling, we investigated the role of a TCH4 gene (MABA012284, MbTCH4-1) and a BSK gene (MD06G1030500, MdBSK1) on the Vm resistance using agrobacterium infiltration (Fig. 7). Compared to the control (empty vector), overexpression of MdBSK1 decreased the resistance to Vm on ‘Fuji’ fruits. After 84 h of incubation, the diameter of disease lesion was 7.56 ± 0.16 mm in MdBSK1 overexpressed fruits, which was significantly higher than that of control fruit (6.40 ± 0.23 mm). On the contrary, MbTCH4-1 overexpression positively regulated Vm resistance on apple fruit. At the last time point of incubation (84 h), diameter of disease lesion was 5.90 ± 0.29 mm in MbTCH4-1 overexpressed fruits but 6.40 ± 0.23 mm in control fruit. Therefore, both of the above genes were participated in regulation of apple fruit resistance against Vm infection.
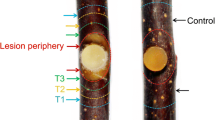
MD06G1030500 and MABA012284 regulate the resistance of ‘Fuji’ apple fruits to Valsa mali (Vm). Control (A, pFGC5941), pFGC5941-MdBSK1 (B), and pFGC5941-MbTCH4-1 (C) were transiently transformed into ‘Fuji’ apples. The lesion diameter (mm) of control and overexpressed fruit were assessed in D (MdBSK1) and E (MbTCH4-1). For each group of bars, asterisk means significantly different at P < 0.05(*) and P < 0.01(**). Vertical bars represent ± SD (standard deviation)
GO terms related to cell wall
To acknowledgement the role of cell wall on cell responses to VmM, the relevant GO terms were listed in Fig. 8. All of the DEGs belonging to “plant-type cell wall organization”, including 4 DEGs in DS and 18 in GL, were down-regulated. Most DEGs associated with “Lignin catabolic process” and “Cell wall modification” were up-regulated in both DS and GL, and more DEGs were found from GL than that from DS. Furthermore, DEGs involved in “xyloglucan biosynthetic process”, “xyloglucan metabolic process” and “cell wall biogenesis” were only observed in DS, and most of these were up-regulated. Above results suggested the important roles of cell wall on cells responses to VmM, and the distinct reactions between DS and GL.
Xylan xylosyltransferases (GUT2, IRX10, UXS6), secondary wall-associated NACs (SND2, ANAC073), LOB domain transcription factor (LBD1), cellulose synthase catalytic subunits (IRX3, CESA7), and fasciclin-like arabinogalactan proteins (FLA12, FLA11) are essential for promoting the formation of cell wall or secondary cell wall (Ajengui et al. 2017). We extracted the homologous genes corresponding to the above proteins in DS and GL. Obviously, most of these genes, such as MABA035166, MABA026316, MABA042192, MABA026005, MABA023682, and MABA043198, were strongly induced in DS, but only weakly induction in GL. These results reveal that induction of these genes is beneficial for cell wall formation and may contribute to resistance of DS (Fig. 9).
qRT-PCR analysis of DEGs
To verify the accuracy of RNA-seq data, expression of 23 DEGs involved in enriched GO terms and/or KEGG pathways was detected using qRT-PCR (Fig. 10 and Fig. 11). These genes included signal transduction, ABA, and ethylene signal pathways. For most genes, the expression patterns from qRT-PCR were similar to those from RNA-seq. These results further confirm the reliability of data from RNA-seq.
Discussion
The gene expression profiles of suspension cells from two germplasm in Malus spp. responses to VmM were investigated using RNA-seq. The results showed that hormone (SA and BR), Ca2+, HR, and cell wall-related signals were participated in the responses of suspension cells to VmM.
In the interactions between plants and pathogens, cell wall plays crucial roles on the resistance against pathogen attack. As a physical barrier consisted of cellulose, hemicellulose, and lignin, cell wall could effectively prevent the transmission of nutrients and control the growth and proliferation of most pathogens (Vorwerk et al. 2004; Chen et al. 2021). Moreover, cell wall is also important for perception of plant to pathogen signals and induction of immune responses. A large number of receptors, such as receptor like kinase and receptor like proteins, located in the cell wall, recognize the signals from pathogen/damage associated molecular patterns (PAMP/DAMP) and take crucial roles in PTI (Fu and Dong, 2013; Stergiopoulos and de Wit 2009). Wall-associated kinases (WAKs) bind to cross-linked pectins as well as pathogen- and damage-induced pectin fragments or oligo-galacturonides (OGs), which further trigger cell expansion and responses to pathogens (Kohorn 2016). Perception of primary and secondary cell walls often results in distinct molecular responses and immune responses in plant cells (Bacete et al. 2017). In Arabidopsis, proteins IRX10, UXS6, IRX3, and CSLC12 are involved in cellulose assembly and its interaction with hemicellulose, whereas protein LBD1 is participated in callus formation (Ajengui et al. 2017). Additionally, FLA11 and FLA12 are involved in maintenance of cell wall integrity, cellulose deposition, and cytoskeleton connection (Ajengui et al. 2017). Indeed, a large number of genes involved in “xyloglucan biosynthetic process” and “cell wall biogenesis” were significantly up-regulated in DS but not in GL. That may be one of the reasons for the strong resistance of DS.
BRs play an important role in stem elongation, pollen tube growth, and root growth, as well as defense responses by activating basic biological systems (Clouse and Sasse 1998). For example, biosynthesis of ET could be activated by BR signaling and contribute to chilling resistance, suggesting the important roles of BRs in the stress response system. Brassinolide, the most biologically active compounds in BR, positively regulate the resistance of tobacco and rice to tobacco mosaic virus (TMV) (Nakashita et al. 2003). Xyloglucan xyloglucosyl transferase TCH4, an important target gene family downstream of BR signaling, was strongly induced by environmental and hormonal stimuli. During cell development, many TCH4 members were expressed in tissues undergoing expansion or cell wall modification (Iliev et al. 2002; Xu et al. 1995). The proteins encoded by TCH4 act as the major components of the plant cell walls and contribute to plant rapid adaption under various environmental conditions (Iliev et al. 2002). In current study, a large number of TCH genes were induced promptly in DS but only one member in GL. Correspondingly, we confirmed that a TCH4 gene positively regulate Vm resistance of ‘Fuji’ fruit. As a key signaling molecule, SA is essential for the establishment of both local and systemic-acquired resistance and plays vital roles in plant responses to a diverse range of pathogens. In the SA signaling pathway, transcription factor TGA regulates plant resistance by manipulating the transcription of downstream target genes, such as PR gene NPR1. SA treatment enhanced the specific interactions between TGA and NPR1, which further improving plant disease resistance (Fan and Dong 2002; Rairdan and Delaney 2002). In addition, exogenous SA is the signaling compound for disease resistance induction of tobacco and the SA-NPR1 signaling pathway in Arabidopsis and rice (Nakashita et al. 2003). After treated with VmM, most differentially expressed TGA genes were down-regulated only in ‘Gala’. Taken together, we suggested that both in SA and BR signaling are participated in cell responses to VmM. Among them, TCH4 in BR and TGA in SA are the key transcription factors to resist VmM infestation.
Cyclic nucleotide-gated channels (CNGCs) are one of important Ca2+-conducting channels involved in plant immunity. The function of plant CNGCs in the autoimmune cascade of early hypersensitivity is manipulated by cyclic nucleotides and Ca2+ carrying cellular signals. Ca2+-dependent protein kinases (CDPKs) are regulated by binding of Ca2+ to EF-hand Ca2+-binding domains and are usually involved in Ca2+ signaling cascades (Boudsocq and Sheen 2013). HvCDPK3 or HvCDPK4 negatively modulates wheat resistance against powdery mildew fungus (Freymark et al. 2007). In addition, Ca2+ flux is considered as a second messenger for induction of cell death and SA, ethylene (ET), and jasmonate (JA) involved defense responses, and contributes to the plant immunity against the pathogen attack. Currently, a large number of CDPK and CNGC genes were up-regulated in GL cells but not DS. Indeed, several CNGC genes activate HR-related cell death and negatively modulate apple tolerance to Vm and Botryosphaeria dothidea (Zhou et al. 2020; Mao et al. 2021). Therefore, we suggested that up-regulation of CDPK and CNGC genes plays important roles in weakening the resistance of ‘Gala’. It is possible that up-regulations of these DEGs also are involved in HR-related cell death.
Many gene families, such as MAPK, WRKY, and FLS2, were also involved in induction of plant immunity. For instance, MAPK–WRKY pathway positively confers resistance to Nicotiana benthamiana against the necrotrophic pathogen Botrytis cinerea (Adachi et al. 2016). FLS2 senses signals produced by pathogens or host damage localized to the extracellular plasma membrane and then enhances plant resistance to most pathogenic bacteria (Yang et al., 2022). In the current study, many genes encoding MAPK, WRKY, and FLS2 were differentially expressed in both DS and GL, thereby suggesting that these genes are participated in cell responses to VmM.
Collection of above results, we suggested that cell wall, BR, SA, and Ca2+-related cell death participated in ‘DS’ and ‘GL’ responses to VmM signaling. Besides as physical barrier, alteration of the cell wall integrity also triggers diverse immune reactions to prevent pathogen infection. SA and Ca2+ were considered as secondary massagers take vital role to active downstream defense responses. In many case, SA induced HR regulating plant resistance against biotrophs positively. Our results exhibited that SA signals contribute to DS resistance to Vm, suggesting that another signal cascades were activated. On the other hand, Ca2+ related HR is lethal factors to compromise the tolerance of GL cells.
Conclusion
In summary, multiple signaling pathways were activated during the treatment of suspension cells with VmM. Among these, induction of a large number of genes associated with “xyloglucan biosynthetic process”, “cell wall biogenesis” and xyloglucan xyloglucosyl transferase TCH4 was beneficial for cell wall integrity and cellulose deposition of DS cells, and contributes to their tolerance to VmM. On the contrary, obvious up-regulation of HR-related genes, such as CNGC and CDPK genes, prompted cell death and compromised the tolerance of GL cells. Our results provided a new insight into molecular mechanisms of resistant and susceptible apple varieties against Vm infection.
References
Adachi H, Ishihama N, Nakano T, Yoshioka M, Yoshioka H (2016) Nicotiana benthamiana mapk-wrky pathway confers resistance to a necrotrophic pathogen botrytis cinerea. Plant Signal Behav 11(6):00–00
Ajengui A, Bertolini E, Ligorio A, Chebil S, Ippolito A, Sanzani SM (2017) Comparative transcriptome analysis of two citrus germplasms with contrasting susceptibility to phytophthora nicotianae provides new insights into tolerance mechanisms. Plant Cell Rep 37(3):483–499
Bacete L, Mélida H, Mie SE, Molina A (2017) Plant cell wall-mediated immunity: cell wall changes trigger disease resistance responses. Plant J 93(4):614–636
Boudsocq M, Sheen J (2013) CDPKs in immune and stress signaling. Trends Plant Sci 18(1):30–40
Burg V, Uvaprd U, Joosten M, Wit D (2006) Cladosporium fulvum avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol Plant Microbe in 19(12):1420–1430
Chen C, Liang W, Li B, Wang C, Dong X (2016) Effects of temperature, humidity, and wound age on valsa mall infection of apple shoot pruning wounds. Plant Dis 100(12):2394–2401
Chen GH, Wang P, Shi L (2021) Review on the dynamic function of cell walls in plant disease resistance. J Inn Mong Agric Univ 42(05):117–120
Clouse SD, Sasse JM (1998) Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Biol 49:427–451
Fan WH, Dong XN (2002) In vivo interaction between npr1 and transcription factor tga2 leads to salicylic acid-mediated gene activation in arabidopsis. Plant Cell 14(6):1377–1389
Freymark G, Diehl T, Miklis M, Romeis T, Panstruga R (2007) Antagonistic control of powdery mildew host cell entry by barley calcium-dependent protein kinases (CDPKs). Mol Plant Microbe In 20(10):1213–1221
Fu ZQ, Dong X (2013) Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol 64(1):839–863
Fukushima A, Nakamura M, Suzuki H, Yamazaki M, Knoch E, Mori T, Saito K (2016) Comparative characterization of the leaf tissue of Physalis alkekengi and Physalis peruviana using RNA-seq and metabolite profiling. Front Plant Sci 7:1883
Iliev EA, Wei X, Polisensky DH, Torisky RS, Braam CJ (2002) Transcriptional and posttranscriptional regulation of arabidopsis tch4 expression by diverse stimuli roles of cis regions and brassinosteroids. Plant Physiol 130(2):770–783
Joosten M, Cozijnsen TJ, Wit P (1994) Host resistance to a fungal tomato pathogen lost by a single base-pair change in an avirulence gene. Nature 367(6461):384–386
Kepley JB, Jacobi WR (2000) Pathogenicity of Cytospora fungi on six hardwood species. J Arboric 26(6):326–333
Kohorn BD (2016) Cell wall-associated kinases and pectin perception. J Exp Bot 2:489–494
Livak KJ, Schmittgen TD (2002) Analysis of relative gene expression data using real-time quantitative pcr. Methods 25(4):402–408
Man N, Ding P, Jones J (2022) Thirty years of resistance: zig-zag through the plant immune system. Plant Cell 5(5):7–17
Mao X, Wang C, Lv Q, Tian YZ, Wang DD, Chen BH, Chu MY, Mao J, Li W, Zuo CW (2021) Cyclic nucleotide-gated channel genes (CNGCs) in rosaceae: genome-wide annotation, evolution and the roles on valsa canker resistance. Plant Cell Rep 40(12):2369–2382
Meerloo JV, Kaspers GJ, Cloos J (2011) Cell sensitivity assays the MTT assay. Humana Press, Totowa, pp 237–245
Meng D, Chunlong L, Park HJ, Jonathan G, Wang J, Dandekar AM, Turgeon BG, Cheng L (2018) Sorbitol modulates resistance to alternaria alternata by regulating the expression of an nlr resistance gene in apple. Plant Cell 30(7):1562–1581
Nakashita H, Yasuda M, Nitta T, Asami T (2003) Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J 33(5):887–898
Ortiz-Morea FA, Liu J, Shan LB, He P (2022) Malectin-like receptor kinases as protector deities in plant immunity. Nat Plants 8(1):27–37
Ping Y, Praz C, Li B, Singla J, Kessel RCB, ScheuermannLüthiOuzunovaErb DLMM (2019) Fungal resistance mediated by maize wall-associated kinase zmwak-rlk1 correlates with reduced benzoxazinoid content. New Phytol 221(2):976–987
Rairdan GJ, Delaney TP (2002) Role of salicylic acid and nim1/npr1 in race-specific resistance in arabidopsis. Genetics 161(2):803–811
Sánchez-Martín J, Keller B (2021) NLR immune receptors and diverse types of non-NLR proteins control race-specific resistance in Triticeae. Curr Opin Plant Biol 62:102053
Stergiopoulos I, de Wit PJ (2009) Fungal effector proteins. Annu Rev Plant Biol 47:233–263
Tarazona S, García F, Ferrer A, Dopazo J, Conesa A (2012) Noiseq: a rna-seq differential expression method robust for sequencing depth biases. Unive Southampt 17(B):18–19
Thaler JS, Humphrey PT, Whiteman NK (2012) Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci 17(5):260–270
Vorwerk S, Somerville S, Somerville C (2004) The role of plant cell wall polysaccharide composition in disease resistance. Trends Plant Sci 9(4):203–209
Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam FJ (1995) Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. Plant Cell 7(10):1555–1567
Yang CM, Li MQ, Chu CL, Li FQ, Shan CL (2022) Preliminary Study on the Grafting of Sorbus aucuparia by Malus baccata. J Jilin For Sci Technol 51(01):1–3
Ye J (2006) Wego: a web tool for plotting go annotations. Nucleic Acids Res 34:W293–W297
Yin Z, Liu H, Li Z, Ke X, Dou D (2015) Genome sequence of valsa canker pathogens uncovers a potential adaptation of colonization of woody bark. New Phytol 208(4):1202–1216
Zhai L, Zhang M, Lv G, Chen X, Jia N (2014) Biological and molecular characterization of four botryosphaeria species isolated from pear plants showing stem wart and stem canker in china. Plant Dis 98(6):716–726
Zhang Q, Ma C, Zhang Yi GuZ, Li W, Duan X, Li H, Wang Y, Wang S, Li T (2018) A single nucleotide polymorphism in the promoter of a hairpin rna contributes to alternaria leaf spot resistance in apple (malus × domestica borkh). Plant Cell 30(8):1924–1942
Zhao DY, Yuan JC, Xu K, Chen CG, Yan S (2016) Tree morphology, accumulation and distribution characteristics of mineral nutrient in root system of Gala apple young tree with different dwarfing interstocks. Acta Agriculturae Boreali-Sinica 31(04):184–191
Zhao X, Qi C, Jiang H, Zhong M, You C, Li Y, Hao Y (2020) Mdwrky15 improves resistance of apple to botryosphaeria dothidea via the salicylic acid-mediated pathway by directly binding the mdics1 promoter. J Integr Plant Biol 62(4):527–543
Zhou K, Hu L, Li Y, Chen X, Ma F (2019) Mdugt88f1-mediated phloridzin biosynthesis regulates apple development and valsa canker resistance. Plant Physiol 180(4):2290–2305
Zhou H, Bai S, Wang N, Sun X, Dong C (2020) Crispr/cas9-mediated mutagenesis of mdcngc2 in apple callus and vigs-mediated silencing of mdcngc2 in fruits improve resistance to botryosphaeria dothidea. Front Plant Sci 11:1640
Zuo C, Mao J, Chen Z, Chu M, Duo H, Chen B (2018) RNA sequencing analysis provides new insights into dynamic molecular responses to Valsa mali pathogenicity in apple ‘Changfu No. 2’. Tree Genet Genomes 14(5):75.1–75.10
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (Grant No. 31860545), and Gansu Science and Technology Program (2021QB-030 and 2019C-11).
Author information
Authors and Affiliations
Contributions
CWZ conceived the idea. CWZ, CW, and XM designed the experiments. CW, XM, and DZ performed experiments. CW, XM, HQY, HD, ES, and YL analyzed the data and wrote the manuscript. CW, CWZ, and DZ revised the manuscript. CWZ supervised the study. All authors read the paper and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
The authors declare that they consent to participate to this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, C., Mao, X., Zhao, D. et al. Transcriptomic analysis reveals that cell wall- and hypersensitive response (HR)-related genes are involved in the responses of apple to Valsa mali. Plant Biotechnol Rep 16, 539–551 (2022). https://doi.org/10.1007/s11816-022-00763-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-022-00763-z