Abstract
OsBAK1 gene belongs to a receptor like kinase gene family in rice and shares a highly conserved gene structure and sequence homology with Arabidopsis thaliana BAK1 gene. To investigate the role of OsBAK1 in rice immunity, the full-length cDNA of OsBAK1 was isolated by RT-PCR and the transgenic rice lines (over expression and RNA-interference lines) were generated using Agrobacterium-mediated transformation. The expression level of OsBAK1 was determined by q-PCR in overexpression and RNAi transgenic rice lines. Based on quantitative polymerase chain reaction (q-PCR) results, two overexpression lines and two RNAi lines were evaluated in bioassays for resistance to Xanthomonas oryzae pv. oryzae PXO99, the causal agent of rice bacterial blight disease. Pathogenicity bioassays showed overexpression OsBAK1 lines exhibited resistance to blight disease whereas OsBAK1 RNAi lines promoted susceptibility. Besides, OsBAK1 can complement the function of AtBAK1 in Arabidopsis bak1 protoplast, activating FRK1 expression, a marker gene in PTI signaling pathway, after treatment by flg22. Furthermore, the transcriptional level of OsBAK1 was induced significantly in rice by defense signaling molecules such as salicylic acid, jasmonic acid, and PXO99 inoculation. Our results illustrated OsBAK1 positively regulates plant defense against rice bacterium pathogen Xanthomonas oryzae pv. oryzae PXO99.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants are exposed to a variety of biotic stresses in natural environment and evolved an elaborate system to defense pathogen attack. The first layer of defense response is the plant innate immune system. Recognition of pathogen- or microbe- associated molecular patterns (PAMPs/MAMPs) is mediated by plant cell surface-localized pattern-recognition receptors (PRRs), typically receptor like kinases or receptor-like proteins with extracellular LRRs (Akira et al. 2006; Chisholm et al. 2006; Zipfel 2008; Boller and Felix 2009; Katagiri and Tsuda 2010). Plants have evolved a large amount of PRRs for recognition of a wide range of PAMPs/MAMPs due to the conserved structure of PAMPs, such as bacterial flagellin, cold shock proteins, elongation factor Tu and fungal chitin, from both pathogenic and non-pathogenic microbes (Sasabe et al. 2000; Peck et al. 2001; Gomez-Gomez et al. 2001; Felix and Boller 2003). More importantly, perception of PAMPs/MAMPs by PRRs activated plant innate immune system, termed PAMP/MAMP-triggered immunity (PTI), which leads to a series of immune response including callose deposition, production of reactive oxygen species (ROS), activation of mitogen-activated protein kinase (MAPK) cascades, induction of defense-related genes, to ward off pathogen attack. Therefore, PRRs have contributed non-host resistance to most pathogens and obtained special attention by crop molecular breeding researchers.
One of the best studied PAMP receptors is the leucine-rich repeat receptor-like kinase (LRR-RLK) FLS2 (Flagellin Sensing 2) (Gomez-Gomez and Boller 2000). FLS2 recognizes bacterial flagellin by direct binding to a conserved 22-amino-acid peptide epitope in the N terminus of flagellin. FLS2 was originally identified in Arabidopsis thaliana and functional orthologs of FLS2 has been found in other plant species such as rice (Oryza sativa) (Takai et al. 2008), tomato (Solanum lycopersicum) (Robatzek et al. 2007), grapevine (Vitis vinifera) (Trda et al. 2014) and tobacco (Nicotiana benthamiana) (Hann and Rathjen 2007), indicating FLS is a convergent component involved in PAMP-triggered immunity among different plant species during evolution.
BAK1, an LRR-RLK, was originally identified as a Brassinosteroid Insensitive-1/BRI1-associated kinase which direct interacts with BRI1 and reciprocal phosphorylation of both proteins, activating brassinosteroid (BR) signaling in a hormone dependent manner (Li et al. 2002; Nam and Li 2002; Wang et al. 2005). Besides participating in brassinosteroid signaling pathway, BAK1 is also involved in multiple PAMP-elicited responses, including flagellin, EF-Tu, bacterial cold-shock protein, and oomycete elicitor INF1 in A. thaliana and Nicotiana benthamiana (Chinchilla et al. 2007; Heese et al. 2007). Upon flg22 perception, FLS2 rapidly recruits BAK1 to form the FLS2-BAK1 complex in a ligand-binding dependent manner and this complex phosphorylates BIK1 and subsequently is phosphorylated by BIK1, initiating innate immunity response (Chinchilla et al. 2007; Lu et al. 2010). It has been reported that Arabidopsis bak1 mutant showed normal flagellin binding but delayed early induction of an oxidative burst and activation of MAPK signaling cascade, indicating BAK1 acts as a positive regulator of PAMP triggered response (Chinchilla et al. 2007). Consistently, the Arabidopsis bak1 mutant displayed enhanced susceptibility to obligate biotrophic oomycete Hyaloperonospora arabidopsidis (Roux et al. 2011) and necrotrophic fungi such as Botrytis cinerea, Alternaria brassicicola (Kemmerling et al. 2007). In addition, plant resistance mediated by BAK1 is a BR-signaling independent manner, bak1 mutant has been shown to have normal binding capacity for BR to BRI1 and flagellin to FLS2, respectively (Kinoshita et al. 2005; Chinchilla et al. 2007). Therefore, except involved in hormone signaling, BAK1 plays an important role in PRR-dependent signaling in plant innate immunity.
Although BAK1-mediated pathogen resistance in A. thaliana has been extensively studied, the function of BAK1 ortholog in Oryza sativa against bacterial blight, caused by Xanthomonas oryzae pv. oryzae (Xoo), remains unclear. Here we investigated whether rice OsBAK1 contributed to PXO99 resistance. We isolated rice OsBAK1 and found OsBAK1 can complement AtBAK1 in bak1 protoplast to activate FRK-LUC expression; OsBAK1 transcript was significantly induced in rice by defense signaling molecules such as salicylic acid, jasmonic acid and PXO99 inoculation. Furthermore, overexpression OsBAK1 in rice exhibited enhanced resistance to PXO99 whereas RNAi-OsBAK1 plants were more susceptible to PXO99. Our results showed OsBAK1 positively regulated rice resistance to PXO99.
Materials and methods
Plant materials, growth conditions and treatments
Oryza sativa L. ssp. japonica cultivar Nipponbare were used for transformation in this study. Arabidopsis thaliana plants used in this study include wild-type Col-0 and bak1-4 mutant (Chinchilla et al. 2007). Transgenic rice plants were grown in square containers with Murashige and Skoog (Duchefa) agar medium in growth chambers set with 16/8 h (L/D) photoperiod at 28/25 °C and 80 % relative humidity with hygromycin selection. Then, 15-day-grown plants were transferred to large containers with rice paddy soil and grown in the same conditions or under sunlight. Chemicals were applied on four-week-old seedlings of Nipponbare by spraying with 0.1 mM salicylic acid (SA), 0.1 mM jasmonic acid (JA).
Gene cloning and plasmid construction
Rice RNA was purified from the frozen tissues by Trizol method and treated with RNase-free RQ1 DNase (Promega). First-strand cDNA synthesis was performed with Superscript III-MLV reverse transcriptase (Invitrogen) using oligo (dT) 15 as a primer. The ten-fold dilution aliquot of first-strand cDNA was then used as a template for the second PCR step. OsBAK1 was amplified by PCR with primer set 5′-ATGGCGGCGCATCGGTGGGC-3′ and 5′-TCACCTCGGCCCTGATAGCTC-3′ using Pyrobest™ DNA polymerase (Takara, Japan) according to manufacturer’s protocol: preheating at 94 °C for 3 min, then 30 cycles of denaturation at 94 °C for 10 s, annealing at 58 °C for 30 s and extension at 72 °C for 2 min, followed by a final extension at 72 °C for 10 min. The PCR product was ligated into pCAMBIA 1300-35S vector to form the OsBAK1 overexpression construct for rice transformation. For complementation of Arabidopsis bak1 mutant in protoplast for FRK1::LUC reporter assay, the OsBAK1 was ligated into pUC19-35S-FLAG-RBS, producing 35Spro:OsBAK1-FLAG overexpression construct for Arabidopsis protoplast transformation. For RNAi, a 438-bp fragment (1-438 bp) of OsBAK1 cDNA was reversely inserted into double-stranded RNA interference vector pTCK303 to generate RNAi plasmid for rice transformation. All plasmids were verified by DNA sequencing.
Rice transformation and expression analysis by q-PCR
Rice transformation was performed with the Agrobacterium-mediated method. The rice Nipponbare was used as recipient to produce more than 30 independent transgenic lines for each construct. Transformants were screened with hygromycin (50 mg/L) in plates and positive transformants were confirmed by genomic DNA polymerase chain reaction using Hpt primer set 5′-AGTCAATGACCGCTGTTATGC-3′ and 5′-CTGATCGAAAAGTTCGACAGC-3′. The expression level of OsBAK1 was determined by q-PCR analysis. The expression level of OsPR1b and OsPOX22.3 was determined by q-PCR analysis as marker genes for SA, JA and PXO99 treatments. Quantitative PCR (q-PCR) was performed using an ABI 7500 Fast Real-Time PCR instrument and SYBR Premix Ex Taq kit (TaKaRa, Otsu, Shiga, Japan). The primer set 5′-CACCCACAGAAAGGTTGCTT-3′ and 5′-CATCATTGGCTAGACGAGCA-3′ was used for OsBAK1 expression analysis. The primer set 5′-GAGGTATCCAAGCTGGCCATT-3′ and 5′-CGTTGTGGAGCCTCACGTAGT-3′ was used for OsPR1b (GenBank accession number: U89895) expression analysis. The primer set 5′-CAGAACTTCAGGGACAGGATCTAC-3′ and 5′-AGGTTGCTGTAGTAGGCGTTGT-3′ was used for OsPOX22.3 (GenBank accession number: AF014467) expression analysis. Gene transcripts were standardized using OsGAPDH (GenBank accession number: GQ848032) with primer set 5′-ACAGGGGAGTTGTGTTTTGC-3′ and 5′-CCCAACCAACCACCATGATA-3′.
Arabidopsis protoplast transfection, FRK1::LUC reporter assay for complementation
The protoplast of Arabidopsis WT (Col-O) and bak1 was transfected with 35Spro:OsBAK1-FLAG together with FRK1pro:LUC (Firefly Luciferase) and 35Spro:RLUC (Renilla Luciferase) for FRK1pro:LUC complementation assay. Protoplast preparation and transfection were performed as described (Asai et al. 2002), with minor modifications. Leaves of 4-week-old plants were sliced into approximately 1-mm strips and were incubated in 15 ml of enzyme solution containing 1 % cellulose R10, 0.2 % macerozyme R10, 0.4 M mannitol, 20 mM KCl, and 20 mM MES, pH 5.7, with gentle shaking (40 rpm) on a rotary shaker for 2 to 3 h. The protoplasts were filtered through a mesh (75 μm) and were centrifuged in a 50 ml tube at 100 × g for 2 min, and the supernatant was carefully removed. The protoplasts were subsequently washed in 10 ml cold W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM morpholine ethane sulfonic acid [MES], pH 5.7), centrifuged at 100×g for 2 min, resuspended in 20 ml W5, and incubated on ice for 30 min. The protoplasts were then collected by centrifugation at 100×g for 2 min and resuspended in a solution containing 0.4 M mannitol, 15 mM MgCl2, and 4 mM MES, pH 5.7, to a final concentration of 2 × 105 cells/ml. For protoplast transfection, 10 μg of plasmid DNA purified by cesium chloride ultracentrifugation was added into 200 μl of protoplasts in a 2 ml microfuge tube. The same amount of 40 % polyethylene glycol (PEG) 4000 solution was then added and thoroughly mixed. The protoplasts were incubated at 23 °C for 8 min, then diluted with 1 ml of W5 solution, centrifuged at 100×g for 2 min to remove PEG. The protoplasts were then resuspended gently in 1 ml of W5, and were incubated overnight under the light. For the FRK1pro:LUC reporter assay, protoplasts were treated with 1 μM flg22 for 3 h before FRK1pro:LUC activity was measured. Proteins was isolated 3 h after flg22 treatment and LUC activity was determined by using the Dual-Luciferase Reporter system (Promega) following the manufacturer’s instructions. The bioluminescence from FRK1pro:LUC expression was recorded by a GLoMax 96 Microplate Luminometer (Promega). Promoter activities are represented by FRK1pro:LUC/35Spro:RLUC activities. All experiments are repeated six times.
Protein exaction and western blotting
Proteins were extracted from Arabidopsis protoplast with a buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.2 % Triton X-100, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 1× protease inhibitor cocktail (Roche, complete, EDTA-free). The lysate was centrifuged at 12,000 rpm at 4 °C for 10 min to remove debris. The proteins were quantified by using Bio-Rad Protein Assay. Protein (10 μg per lane) was loaded for western blot. Monoclonal Anti-Flag M2-peroxidase (HRP) antibody produced in mouse (Sigma) was used in western blot. Horseradish peroxidase was detected by Amersham ECL Prime western blotting detection reagent.
Pathogenicity assessment
The pathogenicity of Xoo strain (PXO99) was evaluated using the leaf clipping method (Kauffman et al. 1973). Xoo strain was grown on PSA plate (10 g/L peptone, 10 g/L sucrose, 1 g/L glutamic acid, 16 g/L bacto-agar, pH 7.0) for 2 days. The bacterial cells of Xoo were suspended in sterile water at an optical density of 0.6 at 600 nm (OD600) and inoculated onto rice leaves of 6-week-old plant. Xoo was inoculated on five fully expanded leaves per plant and 10 individual plants per line. Disease symptoms were scored 2 weeks after inoculation by calculating the lesion length (cm). The results were subjected to statistical analysis using one-way ANOVA. All experiments were repeated three times.
Results
Tissue-specific expression analysis of OsBAK1
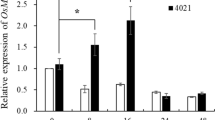
By a blastx search of the whole genome sequence in Rice Annotation Project Database (http://rice.plantbiology.msu.edu/) with the AtBAK1 (At4g33430) amino acids sequence, OsBAK1 (LOC_Os08g07760) was found to be the closest rice homolog of AtBAK1 (72.8 % sequence identity and 78.8 % sequence similarity in their amino acid sequences). The cDNA of OsBAK1 was PCR amplified and sequenced. The expression level of OsBAK1 was then examined in different rice tissues. Q-PCR analysis showed that OsBAK1 gene was expressed in all tested tissues (Fig. 1). The transcript of OsBAK1 was lower in rice stem compared to that of in rice root, leaf, sheath and panicle (Fig. 1). Our expression profiles of OsBAK1 in rice tissues were different as Hu described (Hu et al. 2005), they found OsBAK1 was hardly expressed in root and could not be detected by RNA blot analysis. The transcript level of OsBAK1 in root showed in our experiment was relatively higher (Fig. 1). The difference may be explained by different developmental stage of rice tissue or different rice variety used in OsBAK1 expression analysis. These data indicated that the expression of OsBAK1 may be regulated in rice developmental stage.
Tissue-specific expression analysis of OsBAK1 by q-PCR analysis. Relative expression analysis of OsBAK1 in different rice tissues, including root (R), stem (S), leaves (L), panicle (P) and sheath (H). The rice varieties Nipponbare was used for expression analysis. All experiments were repeated three times using samples from independent treatments. Different letters above the bars indicate values that are significantly different (P < 0.05) from each other as determined by one-way ANOVA (SPSS V20.0) using the Tukey-HSD method
OsBAK1 complements the function of AtBAK1 in FRK1::LUC reporter assay
AtBAK1 was shown to be necessary for flg22 triggered PTI signaling (Chinchilla et al. 2007; Heese et al. 2007). To test whether OsBAK1 can complement the function of AtBAK1 in PTI signaling, the protoplast of Arabidopsis bak1 was co-transformed with 35Spro:OsBAK1-FLAG, FRK1pro:LUC and 35Spro:RLUC plasmids for luciferase reporter assay. FRK1 is a marker gene in PTI signaling and its expression was shown to be upregulated by flg22 (Asai et al. 2002) and Arabidopsis bak1 mutant significant reduced FRK1pro:LUC expression when treated by flg22 (Shan et al. 2008). After induction for 3 h by flg22, the FRK1pro:LUC activities were measured in protoplasts. The FRK1pro:LUC activities of Arabidopsis Col-O, bak1 and bak1/OsBAK1 were 5.5 ± 0.5, 0.8 ± 0.2 and 3.8 ± 0.5, respectively (Fig. 2b). It was shown that over-expression of OsBAK1 in Arabidopsis bak1 protoplast partially rescued the activity of FRK1pro:LUC after flg22 treatment, therefore OsBAK1 can partially complement the function of AtBAK1 during flg22-triggered PTI immunity. This result suggested OsBAK1 may positively regulated flg22-triggered immunity in Arabidopsis. The expression level of 35Spro:OsBAK1-FLAG in Arabidopsis bak1 protoplast was confirmed by western blotting (Fig. 2a).
OsBAK1 complemented Arabidopsis bak1 in FRK1::LUC assay induced by flg22. Western blot showed the expression level of 35Spro:OsBAK1-FLAG in protoplast. Rubisco was used as control. 35Spro:OsBAK1-FLAG expression level was detected by anti-flag antibody. OsBAK1 partially rescued FRK1pro:LUC activity in Arabidopsis bak1 mutant. Protoplasts of Col-O WT and bak1 mutant were co-transfected with/without 35Spro:OsBAK1-FLAG together with FRK1pro:LUC and 35Spro:RLUC plasmids and induced by flg22 treatment for 3 h
Induction of OsBAK1 by defense signaling molecules and PXO99
To assess the effects of defense signaling molecules and PXO99 on the induction of OsBAK1 expression, 4-week-old rice seedlings were treated with salicylic acid (SA), jasmonic acid (JA) and PXO99 infection. Q-PCR analysis showed that OsBAK1 was significantly induced in rice leaves treated with 0.1 mM SA, 0.1 mM JA and PXO99 (Fig. 3). The transcript of OsBAK1 was increased around two folds at 4 h after SA treatment, accumulated to the highest level (around fourfolds) at 12 h and then dropped to two folds (the similar level of 4 h) at 24 h. In the JA treatment, the OsBAK1 transcript was gradually increased during 24 h. The transcript of OsBAK1 was increased by two folds at 12 h and reached the highest level (around fourfolds) at 24 h. The OsBAK1 transcript was also induced by PXO99 at maximum at 4 h (2.5-folds) and maintained the high level of expression until 24 h. As a control, spraying the leaves with water had no effect on induction of OsBAK1 expression. The marker gene, OsPR1b, was shown to be up-regulated by SA and JA treatment and another marker gene, OsPOX22.3, was shown to be up-regulated by PXO99 infection (Chittoor et al. 1997; Agrawal et al. 2000; Mei et al. 2006). Therefore, the inducible expression of OsBAK1 by various defense signaling molecules and PXO99 further suggests that this receptor-like kinase gene, OsBAK1 may participate in host defense responses.
Induction of OsBAK1 by defense signaling molecules and PXO99. Q-PCR analysis of OsBAK1 expression in the four-week-old rice leaves at different time-point after treated with 0.1 mM salicylic acid (SA) (a), 0.1 mM jasmonic acid (JA) (b) and PXO99 (c). Q-PCR analysis of OsPR1b and OsPOX22.3 as maker genes expression after treated with 0.1 mM salicylic acid (SA) (d), 0.1 mM jasmonic acid (JA) (e) and PXO99 (f). Spraying the leaves with water was used as control. All experiments were repeated three times using samples from independent treatments. Different letters above the bars indicate values that are significantly different (P < 0.05) from each other as determined by one-way ANOVA (SPSS V20.0) using the Tukey-HSD method
OsBAK1 confers rice resistance to PXO99
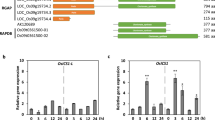
AtBAK1 has been reported to play a positive role in plant immunity (Lin et al. 2014). Arabidopsis bak1 mutant exhibited reduced ability to activated plant defense. In rice, it has also been reported that OsBAK1 positively regulated resistance against rice blast fungus, Magnaporthe grisea (Hu et al. 2005) and Magnaporthe oryzae KJ201 (Park et al. 2011), silencing OsBAK1 by RNA interference resulted in enhanced susceptibility. However, whether OsBAK1 functions in rice immunity against bacterial blight PXO99 remains unclear. We generated OsBAK1-overexpression and RNAi lines and detected its disease resistance to PXO99. Transgenic plants were generated by introducing the overexpression construct (pCAMBIA1300-35S-OsBAK1) and RNAi construct (pTCK303-BAK1-RNAi) into rice cultivar, Nipponbare. The Nipponbare transformed with pCAMBIA1300 empty vector was used as control. A total of 27 and 19 independent transgenic lines with overexpression and RNAi were generated for Nipponbare, respectively. Those positive transgenic lines were confirmed by hygromycin resistance in plates and positive PCR of the Hpt gene. The level of OsBAK1 transcript was then determined by q-PCR analysis. Compared to control plant, the expression level of OsBAK1 in 27 independent overexpression lines were all significantly increased more than two-fold whereas most of OsBAK1 in 19 RNAi independent lines were significantly decreased more than four-fold, respectively, as indicated by the significantly (P < 0.05). Subsequently, two OsBAK1 overexpression T1 lines and two RNAi T1 lines were selected to evaluate PXO99 resistance (Fig. 4a). Ten four-week-old T1 seedlings from each line were subjected to PXO99 infection by the leaf-cutting method at the booting stage. Two overexpression lines named OE6 and OE12 showed significantly enhanced resistance to PXO99 compared to control (Fig. 4b). The average lesion length of two overexpression lines are 3.3 and 3.05 cm, respectively, whereas that of the control is 5.2 cm (Fig. 4c). By contrast, two RNAi lines showed significantly decreased resistance to PXO99 (Fig. 4b). The average lesion length of RNAi-4 and RNAi-14 lines is 9.3 cm and 9.7 cm, respectively (Fig. 4c). Our results indicated that OsBAK1 positively regulated rice resistance to PXO99.
Resistance analysis of over-expression and RNAi lines against PXO99. a Relative expression analysis of OsBAK1 in two overexpression and RNAi rice lines by q-PCR. b Disease symptoms of transgenic rice on 14th day infected by PXO99. c The average lesion length of transgenic rice on 14th day infected by PXO99. WT, wild type; OX6 and OX12, two overexpression OsBAK1 lines, respectively; RNAi-4 and RNAi-14: two RNAi-OsBAK1 lines, respectively. All experiments were repeated three times. Different letters above the bars indicate values that are significantly different (P < 0.05) from each other as determined by one-way ANOVA (SPSS V20.0) using the Tukey-HSD method
Discussion
BAK1 is first identified as a signaling partner of the BR receptor BRI1, and mediates BR-signaling in plant development (Li et al. 2002; Nam and Li 2002; Belkhadir and Chory 2006). Recently, BAK1 has been shown to play dual function in the plant hormone BR signaling and immune response. BAK1 serves as an important player in PAMP signaling where it associates with flagellin receptor FLS2 or other PRRs upon microbial elicitors to initiate plant immune response (Chinchilla et al. 2007; Heese et al. 2007). In A. thaliana and N. benthamiana, BAK1 is required for plant innate immunity to different pathogens (Chinchilla et al. 2007; Heese et al. 2007).
Rice is one of the most economically important crops in the world and also a model species of monocots. In the BR signaling pathway, OsBAK1 was found to play a conserved function with AtBAK1 in rice. OsBAK1 localizes in the plasma membrane and interacts with OsBRI1 in vivo, which is similar to AtBAK1 (Li et al. 2009). Whether rice ortholog of AtBAK1 contributed resistance against rice bacterial blight pathogen, PXO99, and showed conserved function in rice immunity, remains unclear. In our study, we identified a rice receptor like kinase gene, OsBAK1, an ortholog of AtBAK1, by a blast search based on sequence similarity in Rice Annotation Project Database. We found OsBAK1 gene was induced by rice bacterial blight pathogen, PXO99 and defense signaling molecules, assuming OsBAK1 may participate in defense responses in rice. Our further experiments showed overexpression of OsBAK1 in protoplast of Arabidopsis bak1 could rescue the flg22-elicited PAMP immunity, activating FRK1::LUC activity. In addition, overexpression OsBAK1 in transgenic rice showed resistance to PXO99 whereas knock down OsBAK1 in transgenic rice promoted susceptibility. Our results demonstrated OsBAK1 positive regulate rice resistance upon bacterium attack. In line with our results, OsBAK1 also confers resistance to rice blast fungus Magnaporthe oryzae and Magnaporthe grisea, respectively (Hu et al. 2005; Park et al. 2011). These results indicated OsBAK1 might play a conserved function in rice immunity. However, further biochemical evidence such as direct interaction of OsBAK1 with its ligand-binding LRR-RLKs such as OsBIK1 need to be investigated to further understand the mechanism of OsBAK1 mediated resistance in rice.
References
Agrawal GK, Rakwal R, Jwa NS (2000) Rice (Oryza sativa L.) OsPR1b gene is phytohormonally regulated in close interaction with light signals. Biochem Biophys Res Commun 278(2):290–298
Akira S, Uematsu S, Takeuchi O (2006) Pathogen recognition and innate immunity. Cell 124(4):783–801
Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415(6875):977–983
Belkhadir Y, Chory J (2006) Brassinosteroid signaling: a paradigm for steroid hormone signaling from the cell surface. Science 314(5804):1410–1411
Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60:379–406
Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nurnberger T, Jones JD, Felix G, Boller T (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448(7152):497–500
Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124(4):803–814
Chittoor JM, Leach JE, White FF (1997) Differential induction of a peroxidase gene family during infection of rice by Xanthomonas oryzae pv. oryzae. Mol Plant-microbe Interact MPMI 10(7):861–871
Felix G, Boller T (2003) Molecular sensing of bacteria in plants. The highly conserved RNA-binding motif RNP-1 of bacterial cold shock proteins is recognized as an elicitor signal in tobacco. J Biol Chem 278(8):6201–6208
Gomez-Gomez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5(6):1003–1011
Gomez-Gomez L, Bauer Z, Boller T (2001) Both the extracellular leucine-rich repeat domain and the kinase activity of FSL2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 13(5):1155–1163
Hann DR, Rathjen JP (2007) Early events in the pathogenicity of Pseudomonas syringae on Nicotiana benthamiana. Plant J Cell Mol Biol 49(4):607–618
Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104(29):12217–12222
Hu H, Xiong L, Yang Y (2005) Rice SERK1 gene positively regulates somatic embryogenesis of cultured cell and host defense response against fungal infection. Planta 222(1):107–117
Katagiri F, Tsuda K (2010) Understanding the plant immune system. Mol Plant-microb Interact MPMI 23(12):1531–1536
Kauffman HE, Reddy APK, Hsieh SPY, Merca SD (1973) An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis Rep 57:537–541
Kemmerling B, Schwedt A, Rodriguez P, Mazzotta S, Frank M, Qamar SA, Mengiste T, Betsuyaku S, Parker JE, Mussig C, Thomma BP, Albrecht C, de Vries SC, Hirt H, Nurnberger T (2007) The BRI1-associated kinase 1, BAK1, has a brassinolide-independent role in plant cell-death control. Curr Biol CB 17(13):1116–1122
Kinoshita T, Cano-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, Chory J (2005) Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433(7022):167–171
Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110(2):213–222
Li D, Wang L, Wang M, Xu YY, Luo W, Liu YJ, Xu ZH, Li J, Chong K (2009) Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol J 7(8):791–806
Lin W, Li B, Lu D, Chen S, Zhu N, He P, Shan L (2014) Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proc Natl Acad Sci USA 111(9):3632–3637
Lu D, Wu S, Gao X, Zhang Y, Shan L, He P (2010) A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA 107(1):496–501
Mei C, Qi M, Sheng G, Yang Y (2006) Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol Plant-microb Interact MPMI 19(10):1127–1137
Nam KH, Li J (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110(2):203–212
Park HS, Ryu HY, Kim BH, Kim SY, Yoon IS, Nam KH (2011) A subset of OsSERK genes, including OsBAK1, affects normal growth and leaf development of rice. Mol Cells 32(6):561–569
Peck SC, Nuhse TS, Hess D, Iglesias A, Meins F, Boller T (2001) Directed proteomics identifies a plant-specific protein rapidly phosphorylated in response to bacterial and fungal elicitors. Plant Cell 13(6):1467–1475
Robatzek S, Bittel P, Chinchilla D, Kochner P, Felix G, Shiu SH, Boller T (2007) Molecular identification and characterization of the tomato flagellin receptor LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristically different perception specificities. Plant Mol Biol 64(5):539–547
Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tor M, de Vries S, Zipfel C (2011) The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23(6):2440–2455
Sasabe M, Takeuchi K, Kamoun S, Ichinose Y, Govers F, Toyoda K, Shiraishi T, Yamada T (2000) Independent pathways leading to apoptotic cell death, oxidative burst and defense gene expression in response to elicitin in tobacco cell suspension culture. Eur J Biochem/FEBS 267(16):5005–5013
Shan L, He P, Li J, Heese A, Peck SC, Nurnberger T, Martin GB, Sheen J (2008) Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4(1):17–27
Takai R, Isogai A, Takayama S, Che FS (2008) Analysis of flagellin perception mediated by flg22 receptor OsFLS2 in rice. Mol Plant-microb Interact MPMI 21(12):1635–1642
Trda L, Fernandez O, Boutrot F, Heloir MC, Kelloniemi J, Daire X, Adrian M, Clement C, Zipfel C, Dorey S, Poinssot B (2014) The grapevine flagellin receptor VvFLS2 differentially recognizes flagellin-derived epitopes from the endophytic growth-promoting bacterium Burkholderia phytofirmans and plant pathogenic bacteria. New Phytol 201(4):1371–1384
Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J, Asami T, Yoshida S, Huber SC, Clouse SD (2005) Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell 17(6):1685–1703
Zipfel C (2008) Pattern-recognition receptors in plant innate immunity. Curr Opin Immunol 20(1):10–16
Acknowledgments
This work is supported by the Special Fund for Agro-scientific Research in the Public Interest of the People’s Republic of China (Grant No. 201403075), the Natural Science Fundation of Hainan Province (Grant No. 20153071).
Author information
Authors and Affiliations
Corresponding authors
Additional information
H. Liao and X. Xiao contributed equally to this work.
Rights and permissions
About this article
Cite this article
Liao, H., Xiao, X., Li, X. et al. OsBAK1 is involved in rice resistance to Xanthomonas oryzae pv. oryzae PXO99. Plant Biotechnol Rep 10, 75–82 (2016). https://doi.org/10.1007/s11816-016-0387-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-016-0387-6