Abstract
Plant growth and productivity are greatly affected by environmental stresses such as dehydration, high salinity, low temperature and pathogen infection. Plant adaptation to these environmental stresses is controlled by cascades of molecular networks. The dehydration-responsive element-binding (DREB) transcription factors play an important role in the response of plants to environmental stresses by controlling the expression of many stress-related genes. They specifically interact with C-repeat/DRE (A/GCCGAC) sequences present in the promoter regions of target genes. One of the DREB1 cDNA was previously cloned and overexpressed in transgenic potato plants. These transgenic plants displayed an improved tolerance to high salinity and drought stresses. The StDREB1 factor belongs to A-4 group that seem to be involved in biotic stress response. This report investigates the effect of Fusarium solani infection on the StDREB1 transgenic lines. Since a number of pathogenesis-related (PR) proteins are considered as DREB1 target genes, the expression of PR2, PR9 and PR3 genes were tested under biotic stress conditions. The β-1,3-glucanase (PR2) was specifically induced upon infection, whereas the chitinase and the peroxydase were expressed constitutively. The data also show that high levels of DREB1 transcripts accumulated rapidly when wild-type and transgenic plants were infected by F. solani. DREB1 transgenic potato plants accumulated higher levels of pathogenesis-related gene transcripts, such as PR2. These results showed that StDREB1 plays an important role in response to fungal attack in potato.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Environmental stresses, such as drought, high salt and low temperature, as well as pathogen have adverse effects on plant growth and crop yields.
The physiological responses of plants to these stresses arise from changes in the cellular gene expression profiles, via the induction of a number of proteins (Shinozaki and Yamaguchi-Shinozaki 1997). These gene products can be classified into two groups: those that directly protect against environmental stresses and those that regulate gene expression and signal transduction during the stress response (Bray 1997). The first class includes genes encoding proteins that function most likely by protecting cells from dehydration, including enzymes required for the biosynthesis of various osmoprotectants, late embryogenesis–abundant (LEA) proteins, antifreeze proteins, chaperones, and detoxification enzymes (Shinozaki and Yamaguchi-Shinozaki 1997). The second includes transcription factors, protein kinases, and enzymes involved in phosphoinositide metabolism (Shinozaki and Yamaguchi-Shinozaki 1997).
Transcription factors play pivotal functions in signal transduction pathways to activate or suppress defense gene expression. They directly regulate downstream target gene expression by binding to specific elements (cis-elements) in the promoters. Responses and adaptations require differential gene expression that is regulated by specific transcription factors. More than 1500 genes encoding transcription factors have been described in the Arabidopsis genome such as NAC, b-ZIP, Myb/c and AP2/ERF (Xu et al. 2011). Among them, the AP2/ERF are characterized by the presence of AP2/ERF DNA-binding domains of approximately 60 amino acids that directly interact with the GCC-box and/or DRE/CRT (dehydration-responsive element/C-repeat element) cis-acting elements at the promoter sequence of downstream target genes.
The AP2/ERF transcription factors play crucial roles in diverse processes of plant development and stress responses, such as vegetative and reproductive development, cell proliferation, abiotic and biotic stress responses, and plant hormone responses (Nakano et al. 2006).
Sakuma et al. (2002) classified the AP2/ERF transcription factors into four subfamilies: AP2 (APETALA2), RAV (related to ABI3/VP1), DREB (dehydration-responsive element-binding protein) and ERF (ethylene-responsive factor) based on the number and similarities of the DNA-binding domains.
The CBF/DREB and ERF factors are major subfamilies of the AP2/ERF family; these factors contain a single AP2 domain that regulates diverse gene expression.
ERFs display an alanine and aspartic acid residue at position 14 and 19, respectively (Sakuma et al. 2002). However DREBs are characterized by a valine residue at position 14 and glutamic acid at position 19. In addition to these two residues, a conserved alanine at position 37 in the AP2/ERF domain has also been shown to be essential for binding to DRE elements (Liu et al. 2006).
DREB factors have been identified in several plant species including Brassica napus (Gao et al. 2002), barley (Choi et al. 2002), rice (Dubouzet et al. 2003), wheat (Shen et al. 2003) maize (Qin et al. 2004), tomato (Li et al. 2012), potato (Bouaziz et al. 2012, 2013) and soybean (Mizoi et al. 2013; Nasreen et al. 2013).
Transcription factors belonging to the CBF/DREB subfamily are involved in signal transduction pathways involved in plant response to abiotic stress by recognizing the dehydration-responsive or cold-repeat element (DRE/CRT; Zhou et al. 2010). DREB transcription factors also have an upstream regulatory role in mediating signaling pathways for biotic stress responses (Agarwal et al. 2010; Zhou et al. 2010).
Arabidopsis DREB protein DEAR1 (DREB and EAR motif protein 1) seems to play a regulatory role in mediating crosstalk signaling pathways involved in biotic as well as in abiotic stress response (Tsutsui et al. 2009). In this context, Sun et al. (2008) reported that Arabidopsis TINY, a DREB-like factor was capable of binding to both the DRE and the GCC-box with similar affinity and can thus activate the expression of both the DRE and the GCC-box-containing genes. This DREB factor seems to be involved in both abiotic and biotic stress signaling pathways. Moreover several ERF proteins were shown to bind to the DRE/CRT element besides the GCC-box in vitro. Moreover, transgenic tobacco plants overexpressing PgDREB2A (Pennisetum glaucum) showed an upregulation of biotic stress-related genes (Agarwal et al. 2010). Similarly, the overexpression of the OsDREB1B from rice led to enhanced disease resistance against tobacco streak virus (TSV) in transgenic tobacco plants in addition to enhanced tolerance to various abiotic stresses (Gutha and Reddy 2008).
The overexpression of Pti4 in tomato improved resistance to Erysiphe orontii and increased tolerance to Pseudomonas syringae pv. (Wu et al. 2002).
Pathogenesis-related (PR) proteins are considered as ERF/AP2 factors target genes. Indeed, these gene promoters harbor a GCC-box (ERE). The accumulation of these PR proteins can be induced by salicylic acid, jasmonic acid and ethylene treatments. Most PR proteins possess antimicrobial activities through hydrolytic activities on pathogen cell walls, contact toxicity or involvement in plant defense signaling (Van Loon et al. 2006).
PR proteins were first described in tobacco plants infected with TMV (Verwoerd et al. 1989), they were then found in a variety of infected plants including potato and grouped into eleven classes based on homologies in primary structure, enzymatic and biological activities (Van Loon et al. 1994). PR proteins of groups 2 and 3 display a β-1,3-glucanase and chitinase activity, respectively (Sabater-Jara et al. 2010), and their involvement in plant resistance against pathogens has been clearly demonstrated. PR-2 can either directly impair the growth of a fungus by hydrolyzing the β-1,3/1,6-glucans within fungal cell walls (Mauch et al. 1988) or by releasing short glucan fragments from pathogen cell walls, that can also be recognized by plants and further induce plant defense responses (Ebel and Cosio 1994).
PR3 chitinases are capable of degrading chitin within fungal cell walls (Van Loon and Van Strien 1999).
The PR9 class is associated to peroxidase activity (Van Loon et al. 1994), that are believed to be involved in several plant defense responses including cell wall metabolism, wound healing, auxin catabolism (Welinder 1992) and in an antimicrobial response.
The involvement of PR2, PR3 and PR9 in the interaction between potato and Phytophthora. infestans infection has been previously described (Collinge and Boller 2001).
Potato (Solanum tuberosum L.) is one of the most important crops in the world that is ranked fourth behind wheat, rice, and maize. Potato is not only an important food source but it is also a source of animal feed. This vital raw material in the starch-processing industry may be a potentially important resource in medicine owing to the compounds found in its seeds (Ortiz and Watanabe 2004). Potato is relatively vulnerable to abiotic stress but it is also affected by many pathogens leading to substantial economic losses worldwide such as bacterial and fungal pathogens. Phytophthora infestans is the causal agent of potato late blight (LB), one of the most devastating diseases of potatoes (Fry and Goodwin 1997).
Fusarium spp. are ubiquitous fungal pathogens in a wide variety of crops. F. solani causes dry rot on potato tubers. It induces tuber and stolon diseases leading to sprout death, poor plant development, and severe reduction of tuber production (Li et al. 2009).
Dry rot is an important post-harvest disease that affects potato tubers during storage and after planting. F. solani f. sp. eumartii infects tubers at wounded sites causing lesions on the surface and extends deeply into the tuber tissue producing a visible dry rot (D’Ippólito et al. 2010).
In this report, we investigate the effect of F. solani infection on StDREB1 (Solanum tuberosum Dehydration-Responsive Element-Binding 1; Bouaziz et al. 2013) transgenic potato lines. The expression of three StDREB1 putative target PR genes was tested in plants inoculated with F. solani.
Materials and methods
Plant material
Transgenic potato plants overexpressing the StDREB1 gene (Bouaziz et al. 2013) were used in this study. They are designated BF1, BF2, BF3, BF4 and BF5. Two independent commercial potato genotypes are also used i.e., Nicola (Ni) and belle de Fontenay (BF). The latter genotype was used for gene transfer and called (NT).
Potato plants (S. tuberosum L.) were propagated in vitro on an MS basal medium (Murachige and Skoog 1962) supplemented with vitamins (Morel and Wetmore 1951) and 3 % (w/v) sucrose. Culture conditions were 22 °C, 16 h/day illumination at 250 µE m−2/s light intensity.
Fungus strain
Fusarium solani strain was isolated from decaying wood of forests in Northeast Tunisia and kindly provided from Pr. T. Mechichi (University of Sfax). The fungus isolate was cultured on potato dextrose agar (PDA) medium at 25 °C.
Plant inoculation
Spore suspensions were collected from 10–14 day-old F. solani cultures by first washing culture plates with 10 ml sterilized distilled water and the suspensions were adjusted with malassez counting chamber to 5 × 10−1 spores per ml corresponding to 1.38 cfu/ml.
Five plantlets from transgenic and control potato plants of 5-cm height were cultivated on MS liquid medium supplemented with 1 ml of 5 × 10−1 spores per ml F. solani for 20 days. Changes in leaf morphology, plant elongation and leaf alteration index were followed during the treatment period. The drying symptom index on leaves was estimated by score of 0–3 with categories (Carlos, 1987): (0) no symptoms on leaves, (0.5) symptoms on few leaves, (1) symptoms on the 1/3 of leaves, (2) symptoms on the 2/3 of leaves, (3) symptoms on all leaves.
Preparation of total RNA
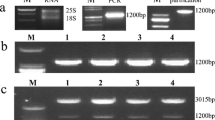
Total RNA was extracted from leaves as described by Verwoerd et al. (1989). The RNA solution was treated by DNase I for 30 min at 37 °C as described by Bouaziz et al. (2012). UV absorbance at 260 nm was used to determine the concentration of RNA samples. The integrity of total RNA was further confirmed by formaldehyde agarose electrophoresis (Sambrook et al. 1989).
Semi-quantitative RT-PCR analyses
Semi-quantitative RT-PCR analyses were carried out using M-MuLV reverse transcriptase (200 U/µl; BIO BASIC INC). Single stranded-cDNAs were synthesized from 2 µg of total RNA of healthy and inoculated leaves. Each cDNA sample was diluted tenfold, and 1 µl of the diluted cDNA was used for PCR amplification (94 °C for 1 min, 60 °C for 1 min, 72 °C for 1 min; in a final volume of 25 µl).
The ef1α (elongation factor) gene (GenBank ID: AB061263) was used as constitutive gene marker. Its specific primers were designed as ef1αF (5′-ATTGGAAACGGATATGCTCCA-3′) and ef1αR (5′-TCCTTACCTGAACGCCTGTCA-3′). Twenty five cycles were performed to amplify the cDNA fragments.
The expression pattern of StDREB1 (GenBank accession no JN125862) and of the PR2, PR3 and PR9 putative target genes (SGN-U01901; SGN-AF024537; SGN-AJ401150) corresponding to PGSC0003DMG400024781, PGSC0003DMG400001528 and PGSC0003DMG400014867 in phytozome ver 9.1, respectively, was carried out using specific primers (Table 1). All the primers used were characterized by a Tm of 60 °C and 35 cycles were performed to amplify cDNA fragments. During the first step of RT-PCR amplification, 25 cycles were performed (supplementary Fig S2).
The RT-PCR amplified products were visualized on ethidium bromide-stained 1.5 % agarose gels and quantified using the Gel DocXR Gel Documentation System (BioRad). This software was used to calculate the average band density. Band density was calculated and graphed using Microsoft Excel.
Analyses of StDREB1 promoter
Cis-element analysis of the promoter sequence of StDREB1 was performed by searching using the PLACE database, and the TSS of the promoter sequence was identified by the FGENESH analysis of www.softberry.com.
Statistical analysis
Data were analyzed by one-way analysis of variance (ANOVA) using graphpad prism v 5. Significant differences among means were determined at P < 0.05.
Results
F. solani inoculation upregulates the expression of StDREB1
Sequence analysis of the StDREB1 gene showed that transcription start site (TSS) is probably located 41 bp upstream of the translation start codon. By searching the StDREB1 promoter in the plant promoter PLACE database, a number of potential regulatory motifs corresponding to cis-acting elements related to tissue specific gene expression, abiotic and biotic stress responses were predicted.
In addition, several consensus cis-acting elements like DRE, MYB, WBOX, and WRKY were also found (Table 2). The GCC core (GCCGCC) cis-element was found in the nucleotide sequence of the promoter. The fact that many cis-elements related to various stresses are present in the promoter region of StDREB1 gene, suggests that this transcription factor is controlled by different mechanisms involved in response to several stresses.
To gain more insight on the stress-induced expression pattern of the StDREB1 potato gene (cv Nicola) were inoculated with F. solani spore suspension and cultured for 15 days.
Semi-quantitative RT-PCR analysis showed that the transcription of the StDREB1 gene increased progressively from 4 days to reach a maximum level on day 7 (Fig. 1a).
To further investigate the putative role of StDREB1 expression in response of potato plants to biotic stress, transgenic plants overexpressing StDREB1 were inoculated by F. solani and StDREB1 transcript accumulation was examined in leaves after 7 dpi. The data showed that StDREB1 mRNA was expressed in leaves of transgenic plants under control conditions while this gene seemed to be silent in untransformed BF control plants genes (Fig. 1b). Moreover, a significant increase of StDREB1 expression was noticed in all plants inoculated with F. solani. This remained much higher in transgenic lines in comparison to BF controls. These data suggest that StDREB1 expression kinetic differs from BF and Ni cultivars. They also showed that StDREB1 factor seems to be involved in the response of potato plants to biotic stress.
Moreover a correlation between StDREB expression and resistance strength was observed for BF2 and BF3 that showed lower growth inhibition in inoculated plants when compared to healthy ones (supplementary Fig S2). These lines also showed significantly higher StDREB1 expression than control WT plant when inoculated with the fungi (Fig. 1b).
Disease resistance analysis of transgenic StDREB1 plants
To confirm the implication of StDREB1 expression in biotic stress response, transgenic plants overexpressing StDREB1 (Bouaziz et al. 2013) were inoculated with F. solani. Potato plants from Ni and BF untransformed were treated similarly and used as controls. Disease symptoms and plant growth were followed for 15 days post-infection (dpi).
Morphological differences were observed between transgenic and control plants inoculated with the fungus (Fig. 2a). Indeed, the leaves of the transgenic lines increased in size and remained green, while the edges of the leaves in the control plants became necrotic after 7 days of treatment. Later, these latter plants displayed visible necrotic lesions in their leaves and the entire plant finally died after 15 days (Fig. 2a).
To better evaluate the response of these transgenic plants to F. solani infection, their growth was examined by measuring the aboveground length after 15 days of fungus treatment. The WT plants were treated under the same conditions and used as controls.
The BF untransformed plants and those belonging to Ni cultivar grew slowly when inoculated with F. solani, while a significantly better elongation of the stems was noticed in BF2, BF4 and BF5 transgenic plants overexpressing StDERB1 (Fig. 2b).
The Log2 (elongation of transgenic lines/elongation of untransformed BF; Supplementary Fig. S2) clearly showed the improved growth of F. solani-inoculated transgenic plants in comparison to untransformed ones. Similarly, the comparison of StDREB1 expression in transgenic and control plants clearly showed a strong correlation between the strength of expression of StDREB1 in transgenic plants and their resistance capacity to F. solani (Supplementary Fig S1). The constitutive expression of StDREB1 in transgenic plants may be responsible for the F. solani resistance observed in these lines.
Estimation of drying symptoms index (Table 3), confirmed the improved resistance of all transgenic lines when compared to control plants. The BF1 line showed the lowest infection symptoms. BF4 and BF5 lines also exhibited very low infection symptoms.
These results suggest that StDREB1 expression in potato can efficiently improve plant resistance to fungi attack. These data corroborated the low incidence of fungus infection on StDREB1 transgenic potato plant elongation.
Expression of target genes in potato F. solani inoculated plants
The PR proteins encoding genes are among AP2/ERF transcription factors target genes. The PR9 Peroxidase group exhibits a protective effect against pathogenic attack.
Indeed the DREB family members play central roles in the regulation of the expression of stress-responsive genes by binding to the CRT/DRE or GCC-box. Cis-regulative elements such as the GCC-box were reported to be present in the PR gene promoters (Ohme-Takagi and Shinshi 1995). We have selected three PR proteins encoding genes (PR2, PR3 and PR9) after confirming by in silico analysis that they harbor a GCC-box in their promoter sequence. To determine the involvement of these genes in response to pathogen attack, potato plants (cv Nicola) were inoculated with 10−1 spores/ml F. solani. Semi-quantitative RT-PCR analysis showed that PR2 gene expression increased to twice the level of control in F. solani-inoculated Nicola potato plants (Fig. 3), while the expression level of the PR3 gene remained constant.
In this Nicola cultivar, the increase in PR2 mRNA seems to reach a maximum level 10 days post inoculation while, for PR9 this maximum level was reached after 24 h of inoculation.
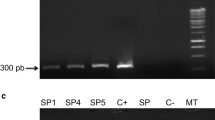
The putative relationship between StDREB1 expression and PR2 gene expression was investigated in transgenic plants overexpressing StDREB1, as well as in wild-type commercial BF and Ni potato plants (Fig. 4). The data showed that under standard conditions, the accumulation of PR2 mRNA as well as that of StDREB1 mRNA was detected in three transgenic lines (BF2, BF3 and BF4). However, low transcription levels of these genes were observed in the other transgenic lines (BF1 and BF5) as well as in the WT BF cultivar. The highest increase in the PR2 transcript was observed in BF2 transgenic line inoculated with F. solani. In contrast, the PR2 gene transcription did not increase significantly in the other transgenic potato plants.
These data suggest that PR2 may be involved in resistance against F. solani in the Nicola potato cultivar. The PR2 gene promoter possesses a GCC-box that can be a putative target for the StDREB1 transcription factor (Bouaziz et al. 2012). However, this increase in PR2 expression may not be sufficient to induce complete resistance against this pathogen.
Discussion
This study aimed to evaluate the response of potato plants overexpressing the StDREB1 factor after inoculation with F. solani. The StDREB1 gene was previously isolated from the Nicola cultivar of potato (Bouaziz et al. 2012). Its amino acid sequence analysis showed that this protein harbors valine in the 14th position and glutamine in the 19th position. Sequence analysis also revealed the presence of serine in the 15th position which was reported to be a crucial amino acid for specific binding to the ERE site in the Arabidopsis TINY factor (Sun et al. 2008). Previous data showed that StDREB1 belongs to the DREB A-4 protein family (Bouaziz et al. 2013). Moreover, this transcription factor seems to be upregulated by ABA, Jasmonic acid and Ethylen treatment (Bouaziz et al. 2013). Sequence analysis suggested also that this transcription factor is involved in biotic stress response.
In this study, potato plants overexpressing StDREB1 and WT plants were inoculated with F. solani. A clear difference between transgenic plants and controls was observed. Indeed, transgenic plants grew normally and their leaves remained green while the growth of WT plants stopped. Similarly Reddy Gutha and Reddy (2008) showed that constitutive expression of OsDREB1B in tobacco plants did not result in any growth retardation or visible phenotypic alterations in transgenic plants. However, transgenic rice plants constitutively expressing OsDREB1 have shown growth retardation (Ito et al. 2006). Moreover, several other transgenic plants constitutively expressing DREB genes have shown growth retardation (Dubouzet et al. 2003; Cong et al. 2008). These data suggest that the growth abnormalities observed in many DREB transgenic plants may be due to the source of the gene, the promoter, the host plant, the growth stage of the transgenic plant, and of the set of target genes.
StDREB1 transcript accumulation was first examined in Nicola potato cultivar by semi-quantitative RT-PCR analysis after 1, 2, 4, 7, 10 and 15 dpi with F. solani and compared to control plants. The StDREB1 expression started to be detectable 4 dpi in Nicola cultivar, it seems to be upregulated by fungal infection. The overexpression of the StDREB1 factor was also observed in transgenic potato plants accompanied by a significant improvement of F. solani tolerance in vitro. These data suggest that StDREB1 may be involved in biotic stress response. Similarly, many reports showed that ERF family members can regulate the expression of a number of stress-related functional genes (Zhou et al. 2010). Among these target genes, the induction of several PR proteins in response to pathogen attack has been extensively demonstrated (Sabater-Jara et al. 2010). Indeed, β-1,3-glucanase (PR-2 group) hydrolyzes glucans, chitinase (PR-3 group), that are major components of fungal cell walls. They may act synergistically as part of the defense strategy activated by plants to fight against the invading pathogen. Such induction may limit colonization of the plant cells by inhibiting fungal growth (Zhou et al. 2010).
Interestingly, OsDREB1B induced the GCC-box cis-elements reported in PR1b, PR2, PR3 and PR5 genes that were shown to be target genes of various ERF transcription factors (Zhou et al. 2010). Moreover, it has been well demonstrated that many PR genes, such as PR1, β-1, 3-glucanase (PR2) are targets of the ethylene signal transduction pathway and are regulated by ERF transcription factors (Ohme-Takagi and Shinshi 1995). Similarly, this report suggests that StDREB1 may activate the expression of stress-inducible PR2 genes in transgenic potato plants.
Further investigation should be performed to further confirm the involvement of StDREB1 transcription factor in the regulation of potato biotic stress response. The search for other StDREB target genes is also necessary.
Abbreviations
- CBF/DREB:
-
c-Repeat binding factor/Dehydration-responsive element-binding
- DRE:
-
Dehydration-responsive element
- PR:
-
Pathogenesis related
- ERF:
-
Ethylene-responsive factor
- LEA:
-
Late embryogenesis abundant
- AP2:
-
Apetala2
- bZIP:
-
Basic region leucine zipper
- RAV:
-
Related to ABI3/VP1
References
Agarwal P, Agarwal PK, Joshi AJ, Sopory SK, Reddy MK (2010) Overexpression of PgDREB2A transcription factor enhances abiotic stress tolerance and activates downstream stress-responsive genes. Mol Biol Report 37:1125–1135
Bouaziz D, Pirrello J, Ben Amor H, Hammami A, Charfeddine M, Dhieb A, Bouzayen M, Gargouri-Bouzid R (2012) Ectopic expression of dehydration-responsive element-binding proteins (StDREB2) confers higher tolerance to salt stress in potato. Plant Physiol Biochem 60:98–108
Bouaziz D, Pirrello J, Charfeddine M, Hammami A, Jbir R, Dhieb A, Bouzayen M, Gargouri-Bouzid R (2013) Overexpression of StDREB1 transcription factor increases tolerance to salt in transgenic potato plants. Mol Biotechnol 54:803–817
Bray EA (1997) Plant responses to water deficit. Trends Plant Sci 2:48–54
Carlos M (1987) la bactériose vasculaire de la pomme de terre “pseudomonas Solanaceanum”. Bulletin d’Information Technique 1 à 19. Centre de la pomme de terre (CIP) 83–88
Choi DW, Rodriguez EM, Close TJ (2002) Barley Cbf3 gene identification expression pattern and map location. Plant Physiol 129:1781–1787
Collinge M, Boller T (2001) Differential induction of two potato genes Stprx2 and StNAC in response to infection by Phytophthora infestans and to wounding. Plant Mol Biol 46:521–529
Cong L, Chai TY, Zhang YX (2008) Characterization of the novel gene BjDREB1B encoding a DRE-binding transcription factor from Brassica juncea L. Biochem Biophys Res Commun 371:702–706. doi:10.1016/jbbrc.2008.04.126
D’Ippólito S, Martín ML, Salcedo MF, Atencio HM, Casalongué CA, Godoy AV, Fiol DF (2010) Transcriptome profiling of Fusarium solani f sp Eumartii-infected potato tubers provides evidence of an inducible defense response. Physiol Mol Plant Pathol 75:3–12
Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice Oryza sativa L encode transcription activators that function in drought high salt and cold responsive gene expression. Plant J 33:751–763
Ebel J, Cosio EG (1994) Elicitors of plant defense responses. Int Rev Cytol 148:1–36
Fry WE, Goodwin SB (1997) Re-emergence of potato and tomato late blight in the United States. Plant Dis 81:349–357
Gao MJ, Allard G, Byass L, Flanagan AM, Singh J (2002) Regulation and characterization of four CBF transcription factors from Brassica napus. Plant Mol Biol 49:459–471
Gutha LR, Reddy AR (2008) Rice DREB1B promoter shows distinct stress-specific responses and the overexpression of cDNA in tobacco confers improved abiotic and biotic stress tolerance. Plant Mol Biol 68:533–555
Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M et al (2006) Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol 47:141–153. doi:10.1093/pcp/pci230
Li YC, Bi Y, Ge YH, Sun XJ, Wang Y (2009) Antifungal activity of sodium silicate on Fusarium sulphureum and its effect on dry rot of potato tubers. J Food Sci 74:213–218
Li J, Sima W, Ouyang B, Wang T, Ziaf K, Luo Z, Liu L, Li H, Chen M, Huang Y, Feng Y, Hao Y, Ye Z (2012) Tomato SlDREB gene restricts leaf expansion and internode elongation by down regulating key genes for gibberellins biosynthesis. J Exp Bot 63:6407–6420
Liu Y, Zhao TJ, Liu JM, Liu WQ, Liu Q, Yan YB (2006) The conserved Ala37 in the ERF/AP2 domain is essential for binding with the DRE element and the GCC box. FEBS Lett 580:1303–1308
Mauch F, Mauch-Mani B, Boller T (1988) Antifungal hydrolases in pea tissuesII Inhibition of fungal growth by combinations of chitinase and b-13-glucanase. Plant Physiol 88:936–942
Mizoi J, Ohori T, Moriwaki T, Kidokoro S, Todaka D, Maruyama K, Kusakabe K, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K (2013) GmDREB2A;2 a Canonical dehydration-responsive element-binding protein2 Type Transcription Factor in Soybean is Post translationally Regulated and Mediates Dehydration-Responsive Element-Dependent Gene Expression. Plant Physiol 161:346–361
Morel G, Wetmore RH (1951) Fern callus tissue culture. Am J Bot 38:141–143
Murachige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Nakano T, Suzuki K, Fujimura T, Shinshi H (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140:411–432
Nasreen S, Amudha J, Pandey SS (2013) Isolation and characterization of Soybean DREB 3 transcriptional activator. J Applied Biol Biotechnol 1:9–12
Ohme-Takagi M, Shinshi H (1995) Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7:173–182
Onate-Sanchez L, Singh KB (2002) Identification of Arabidopsis ethylene-responsive element binding factors with distinct induction kinetics after pathogen infection. Plant Physiol 128:1313–1322
Ortiz R, Watanabe KN (2004) Genetic contribution to breeding polyploid crops. Recent Res Develop in Genet Breed 1:269–286
Pirrello J, Prasad N, Zhang W, Chen K, Mila I, Zouine M, Latché A, Pech JC, Ohme-Takagi M, Regad F, Bouzayen M (2012) Functional analysis and binding affinity of tomato ethylene response factors provide insight on the molecular bases of plant differential responses to ethylene. BMC Plant Biol 12–90
Qin F, Sakuma Y, Li J, Liu Q, Li YQ, Shinozaki K, Yamaguchi-Shinozaki K (2004) Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in Zea mays L. Plant Cell Physiol 45:1042–1052
Sabater-Jara AB, Almagro L, Belchí-Navarro S, Ferrer MA, Ros-Barceló A, Pedreno MA (2010) Induction of sesquiterpenes phytoesterols and extracellular pathogenesis related proteins in elicited cell cultures of Capsicum annuum. J Plant Physiol 167:1273–1281
Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Comm 290:998–999
Sambrook J, Fritish EF, Maniatis T (1989) Molecular cloning A Molecular Manuel second edition Cold Spring Harbor
Shen YG, Zhang WK, He SJ, Zhang JS, Liu Q, Chen SY (2003) An EREBP/AP2-type protein in Triticum aestivum was a DRE binding transcription factor induced by cold dehydration and ABA stress. Theor Appl Genet 106:923–930
Shinozaki K, Yamaguchi-Shinozaki K (1997) Gene expression and signal transduction in water-stress response. Plant Physiol 115:327–334
Simmons CR, Litts C, Huang N, Rodriguez RL (1992) Structure of a rice β-glucanase gene regulated by ethylene cytokinin wounding salicylic acid and fungal elicitors. Plant Mol Biol 18:33–45
Sun S, Yu JP, Chen F, Zhao TJ, Fang XH, Li YQ, Sui SF (2008) TINY a dehydration-responsive element (DRE)-binding protein-like transcription factor connecting the DRE and ethylene-responsive element-mediated signaling pathways in Arabidopsis. J Biol Chem 283:6261–6271
Tournier B, Sanchez-Ballesta MT, Jones B, Pesquet E, Regad F, Latche A, Pech JC, Bouzayen M (2003) New members of the tomato ERF family show specific expression pattern and diverse DNAbinding capacity to the GCC box element. FEBS Lett 550:149–154
Tsutsui T, Kato W, Asada Y, Sako K, Sato T, Sonoda Y, Kidokoro S, Yamaguchi-Shinozaki K, Tamaoki M, Arakawa K, Ichikawa T, Nakazawa M, Seki M, Shinozaki K, Matsui M, Ikeda A, Yamaguchi J (2009) DEAR1 a transcriptional repressor of DREB protein that mediates plant defense and freezing stress responses in Arabidopsis. J Plant Res 122:633–643
Van Loon LC, Van Strien EA (1999) The families of pathogenesis-related proteins their activities and comparative analysis of PR-1 proteins. Physiol Mol Plant Pathol 55:85–97
Van Loon LC, Pierpoint WS, Boller T, Conejero V (1994) Recommendations for naming plant pathogenesis-related proteins. Plant Mol Biol Rep 12:245–264
Van Loon LC, Rep M, Pieterse CM (2006) Significance of inducible defense-related proteins in infected plants. Ann Rev Phytopathol 44:135–162
Verwoerd TC, Dekker BM, Hoekema A (1989) A small scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 25:23–62
Welinder KG (1992) Structure and evolution of peroxidases. In: Welider KG Rasmussen SK Penel C Greppin H (eds) Plant Peroxidases: Biochemistry and Physiology University of Geneva Switzerland, pp 35–42
Wu K, Tian L, Hollingworth J, Brown DC, Miki B (2002) Functional analysis of tomato Pti4 in Arabidopsis. Plant Physiol 128:30–37
Xu ZS, Chen M, Li LC, Ma YZ (2011) Functions and Application of the AP2/ERF Transcription Factor Family in Crop Improvement. J Integr Plant Biol 53:570–585
Zhou ML, Ma JT, Pang JF, Zhang ZL, Tang YX, Wu YM (2010) Regulation of plant stress response by dehydration responsive element binding (DREB) transcription factors. Af J Biotechnol 9:255–279
Acknowledgments
This work was financed by the Tunisian Ministry of High Education and Scientific Research. Authors are grateful to Dr. Anne-Lise Haenni from Institute Jacques Monod (France) for her kind help with the English language and to Mofida Bouaziz-Kannoun from the “Institut Supérieur d’Administration des Affaires de Sfax” (Tunisia) for her kind help with the English language.
Author information
Authors and Affiliations
Corresponding author
Additional information
M. Charfeddine and D. Bouaziz have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig S1: RT-PCR analyses of StDREB1 mRNA expression using 25 cycles in potato plants inoculated with F. solani at different treatment periods (a) StDREB1 gene transcription 7 dpi with F. solani (b) log2 (StDREB1 expression level in inoculated plants/control plants) (c) log2 (StDREB1 expression level in transgenic plants/NT ones) (d).
Fig S2: Evaluation of Log2 (stem elongation of transgenic plants/stem elongation of NT) under control conditions and 7dpi with F. solani
Fig S3: RT-PCR analyses of the expression of: a PR2 b PR3 c PR9 d ef1α in Nicola cultivar after 7 dpi with F. solani. The 25 cycles PCR amplification products are presented for Controls: non-infected plants in addition to F. solani infected ones.
Fig S4: RT-PCR analyses using 25 cycles of the expression of the stress-induced PR2 gene (a) and ef1α (b) in transgenic lines (BF1 BF2 BF3 BF4 and BF5) and in NT (BF) plants under standard conditions and 7 dpi with F. solani.
Rights and permissions
About this article
Cite this article
Charfeddine, M., Bouaziz, D., Charfeddine, S. et al. Overexpression of dehydration-responsive element-binding 1 protein (DREB1) in transgenic Solanum tuberosum enhances tolerance to biotic stress. Plant Biotechnol Rep 9, 79–88 (2015). https://doi.org/10.1007/s11816-015-0345-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-015-0345-8