Abstract
A previous study revealed that IbEF1 from sweet potato promoted early flowering in transgenic tobacco and acted as a novel regulator involved in flowering time control. Two putative ortholog genes were found in N. tabacum: NtEF1 and NtEF2. Subcellular localization of NtEF1 and NtEF2 with an in vivo targeting approach revealed that NtEF1 and NtEF2 were correctly targeted to the cytosol in protoplasts when expressed as a green fluorescent fusion protein (GFP). NtEF1 and NtEF2 are not thought to act as transcription factors. In this study, the expression of NtEF2 was compared with that of NtEF1 in the apical buds of N. tabacum of various ages. NtEF2 was only detected in open flowers that were 100 days old, whereas NtEF1 was expressed in apical buds of 60-, 70-, 80-, 90- and 100-day-old plants. NtEF1 was expressed in all floral organs, although its level of expression varied in sepals, petals, stamens and carpels. In contrast, NtEF2 mRNA expression was only detected in the sepals. Transgenic tobacco plants overexpressing NtEF1 and NtEF2 were generated to elucidate the physiological function of NtEF1 and NtEF2. The 35S::NtEF1 and 35S::NtEF2 transgenic lines showed early-flowering phenotypes. Moreover, the constitutive expression of NtEF1 and NtEF2 occasionally engendered an abnormal morphological alteration of anthers. It is assumed that there are only a few differential flowering regulations occurring in tobacco compared to Arabidopsis, indicating that NtEF1 and NtEF2 interfere with the flowering time and floral development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The transition to flowering is a very important developmental step in higher plants. Flowering in plants consists of two different phases: an initial vegetative phase and a subsequent reproductive phase. Responses to environmental and internal signals trigger flowering initiation through a process controlled by several pathways (Jack 2001; Mouradov et al. 2002; Simson and Dean 2002; Fornara et al. 2010). Extensive genetic and physiological studies in the plant model Arabidopsis thaliana have enabled the identification of many genes involved in the regulation of flowering, approximately 180 of which have been shown to be related to flowering time control (Fornara et al. 2010). Floral induction occurs when floral meristem identity genes, such as APETALA1 (AP1), FRUITFULL (FUL), and LEAFY (LFY), are activated by floral pathway integrators, such as FLOWERING LOCUS T (FT), AGAMOUS-LIKE20 (AGL20) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) in Arabidopsis (Jack 2001; Simson and Dean 2002). LFY, which stimulates diverse floral homeotic genes with other floral regulators, directly activates AP1, which is associated with the floral meristem specification and acts as a floral organ identity gene that belongs to class A of the ABC model (Weigel and Meyerowitz 1994; Mandel et al. 1992; Wagner et al. 1999). When the floral organ identity genes, which comprise the proposed ABC model to explain floral organ development, are up-regulated by the floral meristem identity genes, flowering is completed (Jack 2001; Glover 2007). However, the flowering mechanism is very complex and moreover, there are many unknown genes which are regarded as regulators that interact with other floral-related genes in a flowering pathway.
Constitutive expression of many genes involved in flowering has shown early-flowering phenotype and morphological alteration of the flower. The ectopic overexpression of Triticum aestivum VRN1 (TaVRN1), a member of the AP1 subfamily, represents the early-flowering phenotype and causes various levels of defects in the morphology of Arabidopsis (Adam et al. 2007). AGL24, which is closely related to SOC1, one of the floral integrators, stimulates flowering in Arabidopsis. The constitutive expression of AGL24 results in diverse forms of abnormal flowers containing bract-like sepals and ectopic inflorescence formation (Michaels et al. 2003; Yu et al. 2004). The overexpression of AGL6 related to AP1/FUL-like genes and SEPALLATA (SEP) promoted flowering and homeotic conversion (Reinheimer and Kellogg 2009; Koo et al. 2010). The genes involved in flowering participate in various processes, such as transition to the reproductive stage and floral organ development, in the complicated interaction with many other flowering genes.
In a previous study, a novel gene, IbEF1 (GenBank ID: KF286541), was isolated from a sweet potato at levels equivalent to the most abundant mRNA present in root tissue under salt stress. IbEF1 was considered to be a novel regulator involved in the control of flowering because transgenic tobacco (Nicotiana tabacum L. ‘Xanthi’) containing IbEF1 showed an early-flowering phenotype (Kim et al. 2011). IbEF1 was initially derived from expressed sequence tags (ESTs) of dehydration-treated white fibrous roots of sweet potato and its sequence information was assigned to pathogen-related gene by alignment with the translated sequences of GenBank nucleotide sequence database (Kim et al. 2009). However, this annotation was merely classified as putative characteristic of genes by bioinformatics analysis, and other genes such as pathogen-related protein-like (NCBI accession number: XP_002285489), conserved hypothetical protein (GenBank ID: EEF33891), and predicted protein (GenBank ID: EEF11635) which show relatively high identity of deduced amino acid sequence processed by BLASTX through NCBI database were also not identified clearly. Kim et al. (2011) revealed characteristic of the gene and proved that overexpression of IbEF1 gene from sweet potato confers early flowering in tobacco. So, it was devised that some homologue genes which have same function with IbEF1 would exist in Nicotiana tabacum.
In this study, putative homologues of IbEF1, NtEF1 and NtEF2 were screened for in Nicotiana tabacum. A comparison of both 35S::NtEF1 and 35S::NtEF2 transgenic tobacco to wild-type tobacco, based on genetic and molecular analysis of floral-related genes such as NtFUL, NFL1, and NtMADS5, demonstrated that the constitutive expression of NtEF1 and NtEF2 promotes earlier floral-related gene accumulation. Although there is no difference between the expression patterns of 35S::NtEF1 and 35S::NtEF2, NtEF1 and NtEF2 are certainly involved in the flowering time because all transgenic lines showed the accumulation of NtMADS5, an orhologue of AP1, and a molecular indicator of transition to the reproductive phase, earlier than the wild-type. In addition, the results demonstrated that a constitutive expression of these genes promotes flowering and causes the formation of abnormal flowers under the control of the cauliflower mosaic virus 35S (CaMV 35S) promoter in tobacco.
Materials and methods
Plant materials
Tobacco plants (Nicotiana tabacum L. ‘Xanthi’, photoperiod independent) were used in this study. The plants were grown in a climate chamber at 25 ± 3 °C under a 16 h light/8 h dark photoperiod. Transgenic T2 plants were grown under both 460 nm blue and 630 nm red LED light (Dynebio, Korea).
Cloning of full-length NtEF1 and NtEF2 cDNA
Total RNA was isolated from the roots of N. tabacum exposed to 150 mM NaCl using the modified cetyl trimethylammonium (CTAB) method (Kim and Hamada, 2005). cDNA synthesis was conducted using the RACE (Rapid Amplification of cDNA Ends) system (Clontech, Palo Alto, CA, USA) for further finding of 5′ and 3′-UTR regions, according to the manufacturer’s instructions. 3′-RACE cDNA was used as template to search IbEF1 (Kim et al., 2011) orthologue from tobacco by polymerase chain reaction (PCR) using two primers (EF-gene-search-F; 5′-TTTGAAGAAGGCAGAACTAA-3′ and EF-gene-search-R; 5′-ATCCATGTAACCCCAATGCCT-3′) which were designed on the basis of similar sequence through BLAST search results of IbEF1 gene (GenBank ID: KF286541). After obtaining NtEF1 and NtEF2 genes from tobacco, 5′-RACE-PCR and 3′-RACE-PCR was performed to find 5′- and 3′-UTR regions using a SMART RACE cDNA Amplification Kit (Clontech, Palo Alto, CA, USA) in accordance with the manufacturer’s instructions. The amplified fragments were cloned into T&A cloning vector (Real Biotech, Taiwan) and subsequently sequenced. Moreover, detailed comparison and alignment of sequences were performed using the CLC sequence viewer program (CLC Bio, Denmark) and the BLAST protein search program in the NCBI database.
Structural analysis of the NtEF1 and NtEF2 genes
Specific primers for the open reading frame (ORF) were used in the PCR analysis. PCR amplification was conducted using PCR reaction mixtures with a total volume of 50 μL, and with 30 ng of DNA as the template, 5 μL of 10× buffer, 4 μL of 2.5 mM dNTPs, 1 μL of 10 μM primers, and 2.5 U of ExTaq DNA polymerase (Takara Bio, Japan). The PCR reaction mixtures were heated at 94 °C for 5 min, after which they were subjected to 35 cycles of denaturation for 30 s at 94 °C, annealing for 30 s at 60 °C, and extension for 1 min at 72 °C, followed by a 5 min final extension at 72 °C. The amplified PCR products were assayed using 1 % agarose gel electrophoresis; then the purified PCR fragments were cloned into a T&A cloning vector (Real Biotech, Taiwan). Sequence alignment and comparison were executed using the CLC Sequence Viewer program (CLC Bio, Denmark).
Plant transformation and molecular analysis
Tobacco leaves from in vitro grown plants were used for Agrobacterium tumefaciens-mediated tobacco transformation. Each coding region of NtEF1 and NtEF2 was fused to the cauliflower mosaic virus (CaMV) 35S promoter at the KpnI and BamHI sites of the pCAMLA vector (pCAMBIA 1300 + P35S-Tnos cassette) (Lee et al., 2005) (Fig. 5a). Further, the pCAMLA vector was transformed into A. tumefaciens strain EHA 105 via the freeze–thaw method (An 1987). Tobacco leaf discs inoculated with bacterial suspension at OD600 = 0.6–1.0 for 5 min were washed with sterile water and dried on a sterile filter paper. The inoculated leaf discs were incubated in dark for 2 days and then transferred to an MS medium supplemented with 0.1 mg/L 1-naphthaleneacetic acid (NAA), 1.0 mg/L benzylaminopurine (BAP), 50 mg/L hygromycin and 500 mg/L cefotaxime for a shoot induction. Regenerated shoots were then rooted on a half-strength MS medium containing 50 mg/L hygromycin and 500 mg/L cefotaxime. Transgenic lines that were selected on the basis of hygromycin resistance were further identified by hygromycin-resistance gene-specific PCR and Southern hybridization analysis. For the Southern hybridization analysis, genomic DNA was isolated from the transgenic tobacco leaves using a modified CTAB method (Kim and Hamada, 2005). Briefly, more than 27 µg of each DNA sample were digested overnight with restriction enzymes. The genomic DNA isolated from the transgenic tobacco was then digested with HindIII, after which the products were purified and separated by 0.8 % agarose gel electrophoresis. The gel was subsequently agitated in a tray containing 0.25 N HCl for 15 min and then rinsed briefly with dH2O, after which it was agitated again in a tray, including 0.4 N NaOH for 30 min. The DNA was then transferred onto a positively charged nylon membrane (Roche Molecular Biochemicals, Germany) utilizing the capillary transfer method (Sambrook and Russell 2001). The transferred membrane was rinsed in 2x SSC for 5 min, after which it was allowed to dry in a dry oven set to 80 °C. Next, a hygromycin (HPT)-resistance gene fragment was labeled as a probe by two primers (forward; 5′-CCACTATCGGCGAGTACTTCYAC-3′ and reverse; 5′-CTCTCGATGAGCTGATGCTTTG-3′) using a PCR DIG probe synthesis kit (Roche Molecular Biochemicals, Germany). Hybridization, washing and detection were conducted according to the guidelines established for the DIG system (Roche Molecular Biochemicals, Germany). Finally, verified transgenic T2 generation seedlings were transplanted into pots and grown in the growth room for further experiment.
Reverse transcription (RT)-PCR analysis
Total RNA was isolated from tobacco apical shoots of NtEF1 and NtEF2 T2 lines containing apical leaves ≤2 cm in length according to the modified CTAB method (Kim and Hamada 2005). In addition, total RNA was isolated from the apical portion of wild-type tobacco harvested by day. Further, total RNA was also isolated from the leaves, stems and roots of 90-day-old plants that had floral buds or the sepals, petals, stamens and carpels of open flowers. Each total RNA sample was treated with DNase to remove the contaminated genomic DNA using DNase I (Takara Bio, Japan). One microgram of total RNA was then synthesized to the first-strand cDNA using a PrimeScript first-strand cDNA Synthesis Kit (Takara Bio, Japan) and oligo dT primer. The RT reaction mixture was then diluted with 100 μL distilled water, after which the 1 μL RT reaction mixture was used as a template in a 50 μL PCR amplification reaction mixture composed of 5 μL of 10× buffer, 4 μL of 2.5 mM dNTPs, 1 μL of 10 μM primers and 2.5 U of ExTaq DNA polymerase (Takara Bio, Japan).
PCR consisted of 35 cycles of 30 s at 94 °C, 30 s at 56 °C and 1 min at 72 °C, with a 5 min final extension at 72 °C. For calibration of the transcript level, a primer set (forward; 5′-TGGACTCTGGTGATGGTGTC-3′ and reverse; 5′-CCTCCAATCCAAACACTGTA-3′) based on the Nicotiana tabacum actin gene sequence was used in the experiment with tobacco (Kim et al. 2011). To confirm the expression level of NtEF1 and NtEF2, the following five specific primer sets were used (Fig. 8): NtEF1, 5′-TTCGCGTGGGAAGTGATAAACG-3′ and 5′-TGGTATGGGCAGTGTTGAGTGG-3′; NtEF2, 5′-CATGTTTATTCAGGACCACCAGA-3′ and 5′-GTCTACACTTTTGGAGCCTGTT-3′; NtFUL, 5′-GGGAAGCATATCAGAGTAC-3′ and 5′-CAAGGCTGATAAAGATCAG-3′; NFL1, 5′-AAGGAGCGAGGAGAGAATGTTG-3′ and 5′-AGGAGGTAGATATAGGGGCAAGC-3′; NtMADS5, 5′-GCATCTTTCCTCCTACAACCAC-3′ and 5′-GGGCATAACATTGTATTGAAC-3′. The hygromycin (hpt)-resistance gene was also amplified in wild-type tobacco and transgenic T2 lines. The primer set of HPT (forward; 5′-CCCATTCGGACCGCAAGGAA-3′ and reverse; 5′-CCGCTGTTATGCGGCCATTG-3′) was designed based on the hygromycin B phosphotransferase (HPT) gene sequence used in the selection marker of the plant transformation vector pCAMLA. The amplified PCR products were visualized by electrophoresis in 1 % agarose gel.
Subcellular localization of green fluorescent protein (GFP) fusion constructs
To construct the NtEF1:sGFP and NtEF2:sGFP, each coding region of NtEF1 and NtEF2, without the natural termination codon, was amplified by PCR using two specific primers (NtEF1, 5′-CTCGAGATGGAAGGTGAAAAGGAAAGGAT-3′ and 5′-CTCGAGTGTTCTGGTATGGGCAGTGTTGA-3′; NtEF2, 5′-CTCGAGATGGCAAGTTTAGTAGGAGGGG-3′ and 5′-CTCGAGATTTCATGAAAGGACATTTTGAAGC-3′). NtEF1 and NtEF2 were each ligated in frame to the N terminus of the synthetic (s) GFP (Fig. 4a). To construct smGFP:NtEF1 and smGFP:NtEF2, a full length of each NtEF1 and NtEF2 was prepared by PCR amplification using two specific primers (NtEF1; 5′-CTCGAGATGGAAGGTGAAAAGGAAAGGAT-3′ and 5′-CTCGAGTTAGTTCTGGTATGGGCA-3′, NtEF2; 5′-CTCGAGATGGCAAGTTTAGTAGGAGGGG-3′ and 5′-CTCGAGTTATTTCATGAAAGGACATTTTGAAG-3′) and then fused to the C terminus of the soluble-modified (sm) GFP without the terminator codon (Fig. 4a). All chimeric GFP fusion constructs were placed under control of the CaMV 35S promoter in a pUC vector for expression in protoplasts. Plasmids were purified using Qiagen (Valencia, CA) columns according to the manufacturer’s protocol. The fusion constructs were introduced into Arabidopsis protoplasts prepared from whole seedlings via polyethylene glycol-mediated transformation (Kang et al. 1998). The expression of the fusion constructs was monitored at various times after transformation, and images were captured with a cooled charge-coupled device camera using a Zeiss Axioplan fluorescence microscope (Jena, Germany). The filter sets used included XF116 (exciter, 474AF20; dichroic, 500DRLP; emitter, 510AF23) and XF137 (exciter, 540AF30; dichroic, 570DRLP; emitter, 585ALP) (Omega Optical, Brattleboro, VT) for green and red fluorescent proteins, respectively. The images were then processed using Adobe Photoshop (Mountain View, CA, USA).
Scanning electron microscopy
The surface of the anthers of tobacco T2 plants was observed using the scanning electron microscopy, as previously described (Kim 2008). Briefly, selected anthers were immersed in modified Karnovsky’s fixative (Karnovsky 1965) consisting of 2 % (v/v) glutaraldehyde and 2 % (v/v) paraformaldehyde in 0.05 M sodium cacodylate buffer (pH 7.2) overnight at 4 °C, after which they were washed with the same buffer three times for 10 min each. The specimens were then fixed in a second fixative, including 1 % (w/v) osmium tetroxide in the same buffer at 4 °C for 2 h. Next, the samples were washed briefly with distilled water two times and then dehydrated by submersion in 30, 50, 70, 80, 95, and 2× 100 % ethanol at room temperature for 10 min each. The specimens were subsequently treated with isoamyl acetate two times for 10 min each, then dried in a critical point drier (CPD 030; BAL-TEC Inc., Balzers, Liechtenstein). Finally, the specimens were mounted on metal stubs, sputter-coated with gold and observed using the scanning electron microscopy.
Results and discussion
Sequence analysis of NtEF1 and NtEF2
The sequence analysis indicated the full lengths of NtEF1 and NtEF2. Using reconstructed sequences, the cDNA derived from leaf RNAs of tobacco (Nicotiana tabacum L. ‘Xanthi’) was amplified, and the NtEF1 and NtEF2 cDNA was cloned in suitable vectors and sequenced (Fig. 1a, b). The two pairs of specific primers were designed in accordance with the coding regions of these two mRNA sequences and then used to amplify the genomic DNA fragments as well as the entire NtEF1 and NtEF2 genes. The fragments were sequenced, thereby enabling a complete reconstruction of the NtEF1 and NtEF2 genes with the annotation of introns and exons (Fig. 1c). The sequences were deposited in GenBank with accession numbers KF286542 (NtEF1) and KF286543 (NtEF2). The comparison of the two genes (Fig. 1c) demonstrated a strong conservation of the gene structure. The sequenced regions of the two genes were 2,757 and 1,419 bp long, respectively, for NtEF1 and NtEF2. The two genes contained four exons and three introns. The coding sequences were 750 bp for NtEF1 and 729 bp for NtEF2. Although exons 1, 2 and 3 have a similar length, the lengths of all introns differed between the genes. The lengths of the first, second and third introns were 271, 1215 and 521 bp in NtEF1, and 350 bp, 262 bp and 78 bp in NtEF2, respectively. Moreover, the length of the four exons differed between the genes because they coded for regions in which the two proteins differed. The predicted protein sequences were 250 and 243 amino acids for NtEF1 and NtEF1, respectively, and the alignment of the two protein sequences showed a 47 % residue identity (Fig. 2a). In addition, each deduced amino acid sequence of NtEF1 and NtEF2 displayed a high identity with IbEF1 of Ipomoea batatas (KF286541; 71 and 75 %), the pathogen-related protein of Vitis vinifera (NCBI accession number: XP_002285489; 77 and 55 %), predicted protein of Populus trichocarpa (GenBank ID: EEF11635; 71 and 50 %), conserved hypothetical protein of Ricinus communis (GenBank ID: EEF33891; 75 and 54 %) and pathogenesis-related protein of Arabidopsis thaliana (GenBank ID: AAM66077; 71 and 50 %) (Fig. 2a, b). However, the NtEF1 and NtEF2 proteins had no predicted conserved domain based on the NCBI conserved protein domain database.
Structure and sequence analysis of the NtEF1 and NtEF2 genes. a, b Nucleotide and deduced amino acid sequence of the NtEF1 cDNA (a) and NtEF2 cDNA (b). Numbers on the right represent nucleotide and amino acid positions. The deduced amino acid sequence is shown in the single-letter code above the nucleotide sequence. The start codon is indicated by a box and the termination codon is marked with an asterisk. The underlined nucleotide sequences are 5′-UTR and 3′-UTR, respectively. c The structure of the NtEF1 and NtEF2 genes. NtEF1 and NtEF2 consist of four exons and three introns. Filled boxes indicate the ORF and lines between boxes indicate introns. The numbers represent nucleotides. The table below represents structure of NtEF1 and NtEF2 genes
Comparison of the amino acid sequences of NtEF1, NtEF2 and other homologous proteins. a Alignment of deduced amino acid sequences of NtEF1 and NtEF2 with highly homologous sequences, including IbEF1 of Ipomoea batatas (KF286541), pathogen-related protein of Vitis vinifera (XP_002285489), predicted protein of Populus trichocarpa (EEF11635), conserved hypothetical protein of Ricinus communis (EEF33891), and pathogenesis-related protein of Arabidopsis thaliana (AAM66077). Identical amino acid residues in all seven sequences are indicated by white letters with black backgrounds. b Phylogenetic analysis of NtEF1, NtEF2 and homologous sequences via UPGMA
Expression patterns of NtEF1 and NtEF2
To monitor the temporal expression patterns of NtEF1 and NtEF2, the level of the corresponding mRNA in the apical shoots containing apical leaves from 60-, 70- and 80-day-old tobacco, in floral buds from 90-day-old tobacco, and in open flowers from 100-day-old tobacco was monitored by RT-PCR analysis. To confirm the spatial expression patterns of the NtEF1 and NtEF2 gene from tobacco, the level of the mRNA expression in different organs was examined by the RT-PCR analysis. The total RNA samples were isolated from the leaves, stems and roots of 90-day-old tobacco with floral buds as well as from the sepals, petals, stamens and carpels of open flowers.
In the temporal expression patterns, NtEF1 was expressed in the apical part, floral buds and open flower at all stages from 60 to 100 days, while NtEF2 was only detected in open flowers of the 100-day-old plant (Fig. 3a). Before tobacco had a floral bud, the level of NtEF1 expression in the apical parts was rather high at 60, 70 and 80 days (Fig. 3a).
RT-PCR analysis of wild-type tobacco. a Expression patterns of NtEF1, NtEF2, and flowering-related genes in apical shoots with young apical leaves of 60-, 70-, and 80-day-old tobacco, in floral buds of 90-day-old plants, and in open flowers of 100-day-old plants. The specific primer sets of nine genes, NtEF1 (KF286542), NtEF2 (KF286543), NtFUL (DQ534202), NFL1 (U15798), and NtMADS5 (AF068724), were used in the analysis. Actin was amplified as an internal control. b Expression of NtEF1 and NtEF2 in leaves (L), stems (St) and roots (R) of wild-type 90-day-old plants. L5: fifth leaf from flower bud, L10: tenth leaf from flower bud, L15: fifteenth leaf from flower bud. Actin was amplified as an internal control. c Expression of NtEF1 and NtEF2 in sepals (Se), petals (Pe), stamens (St), and carpels (Ca) of open flowers from 100-day-old plants. Actin was amplified as an internal control
In the spatial expression patterns of NtEF1 and NtEF2 from the 90-day-old plant with floral buds, the NtEF1 mRNA expression level in the fifth, tenth and fifteenth leaf apart from the top was similar to that in the stem; however, the level in the roots was slightly higher (Fig. 3b). However, the NtEF2 mRNA expression became more detectable as the location of the leaf was lower and the NtEF2 mRNA level in the roots was comparatively higher (Fig. 3b). Further, the NtEF1 mRNA expression levels differed among the sepals, petals, stamens and carpels; yet, it was expressed in all floral organs (Fig. 3c). In contrast, the NtEF2 mRNA expression was only detected in the sepals (Fig. 3c).
Subcellular localization analysis
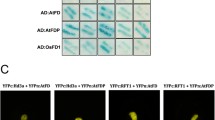
Cellular localization of NtEF1 and NtEF2 was examined by constructing a fusion protein with a GFP (sGFP or smGFP). The GFP was fused to the N or C terminus of NtEF1 or NtEF2 (Fig. 4a) to determine whether some amino acid regions of NtEF1 or NtEF2 had a function or not, even though NtEF1 and NtEF2 have no putative conserved domain based on the NCBI conserved protein domain database. Each fusion construct was introduced into protoplasts prepared from Arabidopsis tissues using the polyethylene glycol transformation method (Kang et al. 1998). After the transient expression of the fusion constructs, an individual protoplast was examined under a fluorescence microscope. As shown in Fig. 4b, the expression patterns of NtEF1:sGFP and smGFP:NtEF1 introduced into the protoplasts were the same, as were those of NtEF2:sGFP and smGFP:NtEF2. These findings indicate that the amino acid residues in the N or C terminus of NtEF1 and NtEF2 have no effect on the expression. The fluorescence of NtEF1 and NtEF2 protein fused to GFP was associated with cytosol, indicating that they were primarily targeted to the cytosol (Fig. 4b). These findings suggest that NtEF1 and NtEF2 do not function as transcription factors.
a Scheme showing the various fusion constructs used in subcellular localization analysis. NtEF1:sGFP, NtEF1 coding region without the natural termination codon was ligated in frame to N-terminus sGFP under control of the CaMV 35S promoter. smGFP:NtEF1, NtEF1 coding region was ligated with C-terminus smGFP removing terminator codon under control of the CaMV 35S promoter. NtEF2:sGFP, NtEF2 coding region removing the natural termination codon was ligated to N-terminus sGFP under control of the CaMV 35S promoter. smGFP:NtEF2, NtEF2 coding region was ligated with C-terminus smGFP without the terminator codon under control of the CaMV 35S promoter. b Subcellular localization of the GFP fusion proteins in protoplasts. The fusion constructs were introduced into Arabidopsis protoplasts. The pictures in the first row are bright-field live images (live). The pictures in the second row represent the overlap of green, red, and blue fluorescent signals of GFP, RFP, and chlorophyll, respectively (merged). The photos in the third row show each GFP fused to NtEF1 or NtEF2 in the transformed protoplasts (GFP fusion). The pictures in the fourth row show cytosolic marker RFP in transformed protoplasts (RFP). The photos in the fifth row indicate the auto-fluorescent signal of chlorophyll colored blue (CH)
Phenotype of NtEF1 and NtEF2 transgenic plants
The NtEF1 and NtEF2 genes under the control of the CaMV 35S promoter were introduced into N. tabacum. Several overexpression lines were obtained. Specifically, 35S::NtEF1 transgenic tobacco plants included ten independent lines, whereas 35S::NtEF2 transgenic plants contained six independent lines. The insertion of T-DNA containing NtEF1 and NtEF2 genes (Fig. 5a) in the transgenic lines for constitutive expression was confirmed by the Southern blot analysis (Fig. 5b). The insertion was detected by probes based on a hygromycin resistance gene sequence that includes T-DNA as a selection marker because NtEF1 and NtEF2 are endogenous genes in tobacco. Lines 3, 6 and 19 of 35S::NtEF1 contain one copy of the T-DNA, while the other lines have multiple copies (Fig. 5b). Lines 4 and 7 of 35S::NtEF2 contain one copy, but the other lines contain multiple copies (Fig. 5b).
Southern blot and RT-PCR analysis of the NtEF1 and NtEF2. a. Schematic diagrams of the expression vectors. Top T-DNA region of pCAMLA containing the NtEF1 coding region. Bottom T-DNA region of pCAMLA containing the NtEF2 coding region. b, c Southern blot analysis of genomic DNA from transgenic T2 tobacco plants. Genomic DNA was digested with HindIII. Hybridizations were carried out with a hygromycin-specific probe. b Southern blot of genomic DNA from transgenic tobacco overexpressing NtEF1. M: DIG-labeled DNA molecular weight marker. Each lane represents an individual line of the transgenic tobacco including NtEF1 and NtEF2. M: DIG-labeled DNA molecular weight marker. Each lane represents an individual line of the transgenic tobacco including NtEF2. c, d RT-PCR analysis in 70-day-old wild-type tobacco and transgenic T2 tobacco lines for analysis of flowering time. Actin was amplified as an internal control. c Expression patterns of the hygromycin B phosphotransferase (HPT) gene and NtEF1 gene in 70-day-old wild-type and 35S::NtEF1 lines. d Expression patterns of the HPT gene and NtEF2 gene in 70-day-old wild-type and 35S::NtEF2 lines
There was no phenotypic difference between the wild type and transgenic lines in the juvenile plant stage, except for early flowering in transgenic plants. After inflorescence, most of the 35S::NtEF1 and 35S::NtEF2 transgenic lines had normal morphology relative to that of wild-type tobacco. A few plants of several overexpressing T1 lines conveyed an abnormal morphological alteration of the stamen (Fig. 6a) and therefore could not undergo the self-pollination problem, although the phenotype frequency was very low. Specifically, only one or two of the eight plants in a single transgenic line showed an alteration of the stamen shape; some lines showed no plants with morphological alteration (data not shown). The morphological alteration of the stamen containing petals appeared somewhat more frequently in 35S::NtEF1 than in 35S::NtEF2 (Fig. 6a).
Phenotypic alterations of the transgenic tobacco plants overexpressing NtEF1 and NtEF2. a Unexpected phenotypic alteration of transgenic tobacco T1 plants overexpressing NtEF1 and NtEF2. A few plants of each transgenic line showed abnormal morphological alteration of the stamen. b, c Unusual morphological alteration of transgenic tobacco T2 plants overexpressing NtEF1 and NtEF2. b Longitudinal sections or open flowers that showed different shape anthers of transgenic lines. c Scanning electron microscopy analysis of surface cell morphology in wild-type tobacco anther and transgenic tobacco anther showing unusual shape
The T2 progeny of the 35S::NtEF1 and 35S::NtEF2 lines was obtained via self-pollination. Four lines of 35S::NtEF1 and four lines of 35S::NtEF2 were selected for the flowering time analysis; however, these lines had fewer copies of T-DNA or abnormal flower morphology in the T1 generation. In addition, the transgenic T2 plants with wild-type tobacco were grown in a climate chamber at 25 ± 3 °C under a 16 h light/8 h dark photoperiod. The transgenic lines were obtained by hygromycin selection and Southern blot analysis. Moreover, the overexpression of NtEF1 and NtEF2 was confirmed by checking the mRNA expression levels of HPT, NtEF1 and NtEF2 (Fig. 5c, d). As shown in Fig. 5c, d, the HPT mRNA expression only appeared in the 35S::NtEF1 and 35S::NtEF2 transgenic lines. Each gene under the control of the CaMV 35S promoter contained the HPT gene positioned in the T-DNA for a constitutive expression of NtEF1 and NtEF2 in transgenic tobacco (Fig. 5a). NtEF1 and NtEF2 were confirmed to be overexpressed in each transgenic line because their mRNA expression levels were higher in transgenic plants than in wild-type tobacco (Fig. 5c, d).
In transgenic T2 plants, the abnormal alteration of the stamens with petals was only observed in 35S::NtEF1-20 plants (Fig. 6b). The abnormal stamen in the 35S::NtEF1-20 showed a normal-shaped filament and an unusual shaped anther covered with petals (Fig. 6b). Some transgenic plants had slim-shaped anthers, a reduced amount of pollen or shorter filaments compared to the round-shaped anthers and length of filament of wild-type tobacco stamens (Fig. 6b). The unusual shaped anthers of the 35S::NtEF1 and 35S::NtEF2 plants were identified by the naked eye, after which the surfaces were observed by a scanning electron microscopy (SEM). There was not a very distinct difference between 35S::NtEF1-3 and wild-type tobacco anthers, except for the volume and length (Fig. 6c). The shape of 35S::NtEF2-7 anthers was twisted and showed an abnormal structure relative to the control (Fig. 6c). The 35S::NtEF1-20 aborted anther covered with petals appeared to have a problem with the differentiation of pollen sacs (Fig. 6c). Self-pollination problems in a few transgenic plants were considered to result from the morphological alteration of anthers. This phenotype corresponded to gai-induced male-sterile phenotype in transgenic tobacco (Huang et al. 2003; Al-Ahmad and Gressel 2005). Although NtEF1 and NtEF2 are not directly involved in gibberellin acid, the constitutive expression of NtEF1 and NtEF2 may affect floral organ development. However, these phenotypic frequencies were also very low in transgenic T2 plants. It is assumed that these unexpected phenotypes are involved with the environmental stimuli, T-DNA insertion copy number or a different expression level of NtEF1 and NtEF2 because the floral development pathway is very complex.
Kim et al. (2011) found that the IbEF1 sweet potato gene promotes rapid flowering through the ectopic constitutive expression in tobacco. To confirm if the NtEF1 and NtEF2, searched as homologue genes of IbEF1 in tobacco, is related to early flowering, the flowering time of 35S::NtEF1 and 35S::NtEF2 transgenic tobacco T2 grown in a climate chamber at 25 ± 3 °C under a 16 h light/8 h dark photoperiod was analyzed. When compared to wild-type tobacco, the 35S::NtEF1 and 35S::NtEF2 lines showed earlier bolting followed by the formation of the floral bud. No wild-type tobacco plants showed floral buds, whereas transgenic 35S::NtEF1 and 35S::NtEF2 plants formed floral buds early; further, the inflorescence of some transgenic plants was completely finished within 100 days of planting (Fig. 7; Table 1). The flowering time was assessed based on node numbers and days at floral bud formation. On average, wild-type tobacco plants first formed floral buds approximately 30 days later than the overexpression lines of 35S::NtEF1 and 35S::NtEF2 (Table 1). Due to the early transition to flowering occurring in the transgenic lines, there was no distinct difference in the number of nodes between wild type and transgenic lines; however, the stem height of transgenic lines was slightly shorter than that of the wild type after inflorescence. These early-flowering phenotypes of 35S::NtEF1 and 35S::NtEF2 were confirmed at the molecular level by RT-PCR, which were conducted to determine if the mRNA expression of floral-related genes occurred earlier in transgenic lines than in wild type.
The flowering pathway is very complex, and plants initiate flowering after the transition from the vegetative to the reproductive stage (Mandel et al. 1992; Araki 2001; Mouradov et al. 2002). Floral induction related to the coordination of many endogenous factors and environmental stimuli is needed during this process; moreover, the floral induction process has been shown to be a genetic and molecular mechanism by diverse studies of Arabidopsis (Mandel et al. 1992; Araki 2001; Mouradov et al. 2002; Fornara et al. 2010). During a floral transition, the meristem identity is changed, and the shoot apical meristem (SAM) is converted into an inflorescence meristem (IM) upon floral induction. The floral transition is controlled by key regulators, including the transcription factors LFY, FUL, and AP1, which specify the floral meristem identity (Benlloch et al., 2007; Sablowsky 2007; Litt and Irish 2003). LFY is a master regulator of flowering because lfy mutation induces a delayed transition to the reproductive phase and development of leafy shoots instead of flowers, while the constitutive expression of LFY confers early flowering and the conversion of shoots into flowers aberrantly in Arabidopsis (Schultz and Haughn 1991; Huala and Sussex 1992; Weigel et al. 1992; Weigel and Nilsson 1995). LFY is believed to occur downstream in all identified flowering pathways, to be controlled by diverse input signals and to regulate many other genes involved in flowering time and floral meristem identity (William et al. 2004; Fornara et al. 2010; Srikanth and Schmid 2011). FUL also functions as a transcription factor associated with the transition to the reproductive phase and the MI transition together with other genes. Specifically, the loss-of-function phenotype in Arabidopsis engenders a change of transition to flowering, whereas a constitutive expression of FUL promotes flowering (Ferrandiz et al. 2000a, b; Teper-Bamnolker and Samach 2005). FUL is believed to occur downstream in the photoperiod pathway, where FT and FD participate as regulators; however, SQUAMOS PROMOTER BINDING LIKEs (SPLs) transcription factors related to the aging pathway directly activate LFY and AP1 in addition to FUL (Abe et al. 2005; Teper-Bamnolker and Samach 2005; Yamaguchi et al. 2009; Srikanth and Schmid 2011). Moreover, AP1 plays an important role in the specification of floral meristem identity and the floral homeotic gene (Bowman et al. 1993; Schultz and Haughn 1993). Accordingly, a suitable regulation of LFY, FUL and AP1 accumulation plays key roles in the initiation of flower formation, in that increased levels of LFY, FUL or AP1 promote MI transition and floral development (Ferrandiz et al. 2000a; Mandel and Yanofsky 1995; Weigel and Nilsson 1995; Yamaguchi et al. 2009). As shown in Fig. 8, the floral-related genes analyzed by RT-PCR are NtFUL, NFL1 and NtMADS5, which are the orthologues of FUL, LFY and AP1, respectively (Smykal et al. 2007; Ahearn et al. 2001; Jang et al. 2002). mRNA expression of NtMADS5, the AP1 orthologue, generally does not occur in apical buds during all vegetative stages; however, NtMADS5 mRNA can be detected when tobacco is transformed from the vegetative to the floral stage (Smykal et al. 2007). NtMADS5 accumulation can be a molecular indicator of the transition to the reproductive phase during the life cycle of tobacco. The transgenic lines of 35S::NtEF1 and 35S::NtEF2 undergo the transition to the reproductive stage earlier than wild type at 80, 90 and 100 days because NtMADS5 mRNA expression appears more rapidly in transgenic lines (Fig. 8a, b). These results correlate with the early-flowering phenotypes in transgenic lines, in which NtEF1 and NtEF2 are overexpressed. When NtMADS5 mRNA expression is not detectable, the tobacco plant is in the vegetative stage. Transcripts of NtFUL and NFL1, or upstream genes of NtMADS5, were visible during the vegetative phase because NtMADS5 mRNA expression was not detected during this phase (Fig. 8a, b). These expression patterns correspond to the results reported in previous studies. FUL and LFY are both expressed in Arabidopsis during the vegetative stage, and their orthologue genes, NtFUL and NFL1, also accumulate in tobacco during the vegetative phase; however, they are all drastically up-regulated in the apical buds after transition to the floral stage (Kelly et al. 1995; Mandel et al. 1995; Blazquez et al. 1997; Borner et al. 2000; Smykal et al. 2007). NFL1 is crucial to the control of the initiation of leaves and inflorescence branches in tobacco shoots, while also acting as a regulator of floral organ development (Ahearn et al. 2001). Although NFL1 is an orthologue of LFY, the function of NFL1 and LFY differs because the ectopic NFL1 expression did not show severe early-flowering phenotype when compared to the tobacco and Arabidopsis with ectopic LFY expression (Kelly et al. 1995; Ahearn et al. 2001). Molecular and genetic studies of flowering time regulation in tobacco are still limited; however, the results of the present study suggest that a slightly different flowering regulation system exists more in tobacco than Arabidopsis.
Expression of the floral-related genes in apical shoots including young apical leaves of 35S::NtEF1 and 35S::NtEF2 transgenic plants at different stages was determined by RT-PCR using the primers shown in the table below. a Total RNA isolated from 80-, 90-, and 100-day-old wild-type tobacco and 35S::NtEF1 transgenic plants was used as a template. Actin was amplified as an internal control. b Total RNA isolated from 80-, 90-, and 100-day-old wild-type tobacco and 35S::NtEF2 transgenic plants was used as a template. Actin was amplified as an internal control
Although the temporal and spatial mRNA expression patterns of the two genes are different and the identity of their two proteins is not high, NtEF1 and NtEF2 may regulate flowering because a constitutive expression of NtEF1 and NtEF2 showed early flowering in tobacco. Further, the phenotype was confirmed by the mRNA expression of floral regulator genes, such as NtMADS5. However, it is not certain as to whether NtEF1 and NtEF2 are transcription factors that directly upregulate or suppress floral-related genes in tobacco. Further studies are needed to demonstrate how NtEF1 and NtEF2 participate in the flowering pathway of tobacco and interact with other floral-related genes, as well as if there is a different action between the NtEF1 and NtEF2 genes.
Above all, this study revealed that constitutive expression of NtEF1 and NtEF2 genes, respectively, promotes early flowering. These days worldwide many countries produce biodiesel which is common name representing an alternative fuel made from crop oils or animal fats (Avinash et al. 2014). Biodiesel has been magnified that it is main substitute and attractive energy source compared to fossil fuel because the biodiesel is not limitative source that could be continuously produced and it has lower emission of combustion gas causing global warming (Ahmad et al. 2011). Most biodiesel is produced from bio-fuel crops such as soybean, sunflower, rubber seed, canola, castor, and rapeseed in various countries (Avinash et al. 2014). Since these oil seed crops have high potential for biodiesel production, promoting early flowering to produce seeds is very important thing besides selection crop cultivar, land use, and modification of oil-extraction methods to increase production efficiency of biodiesel. Thus, it is suggested that NtEF1 and NtEF2 to cause early flowering could be used in gene modification of the oil seed crops, and NtEF1 and NtEF2 genes have industrial utility value on the perspective of biotechnological application.
Abbreviations
- AP1:
-
APETALA1
- FUL:
-
FRUITFULL
- LFY:
-
LEAFY
- FT:
-
FLOWERING LOCUS T
- AGL20:
-
AGAMOUS-LIKE20
- SOC1:
-
SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1
- SEP:
-
SEPALLATA
- CaMV35S:
-
Cauliflower Mosaic Virus 35S
- CTAB:
-
Cetyl trimethylammonium
- RACE:
-
Rapid amplification of cDNA ends
- PCR:
-
Polymerase chain reaction
- ORF:
-
Open reading frame
- NAA:
-
1-Naphthaleneacetic acid
- BAP:
-
Benzylaminopurine
- HPT:
-
Hygromycin B phosphotransferase
- GFP:
-
Green fluorescent protein
- sGFP:
-
Synthetic green fluorescent protein
- smGFP:
-
Soluble-modified green fluorescent protein
- SAM:
-
Shoot apical meristem
- IM:
-
Inflorescence meristem
- SPLs:
-
SQUAMOSA PROMOTER BINDING LIKEs
References
Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309:1052–1056
Adam H, Ouellet F, Kane NA, Agharbaoui Z, Major G, Tominaga Y, Sarhan F (2007) Overexpression of TaVRN1 in Arabidopsis promotes early flowering and alters development. Plant Cell Physiol 8:1192–1206
Ahearn KP, Jonhnson HA, Weigel D, Wagner DR (2001) NFL1, a Nicotiana tabacum LEAFY-like gene, controls meristem initiation and floral structure. Plant Cell Physiol 42(10):1130–1139
Ahmad AL, Mat Yasin NH, Derek CJC, Lim JK (2011) Microalgae as a sustainable energy source for biodiesel production: a review. Renew Sust Energy Rev 15:584–593
Al-Ahmad H, Gressel J (2005) Transgene containment using cytokinin-reversible male sterility in constitutive, gibberellic acid-insensitive (Δgai) transgenic tobacco. J Plant Growth Regul 24:19–27
An G (1987) Binary Ti vectors for plant transformation and promoter analysis. Methods Enzymol 153:292–305
Araki T (2001) Transition from vegetative to reproductive phase. Curr Opin Plant Biol 4:63–68
Avinish A, Subramaniam D, Murugesan A (2014) Bio-diesel-A global scenario. Renew Sust Energy Rev 29:517–527
Benlloch R, Berbel A, Serrano-Mislata A, Madueno F (2007) Floral initiation and inflorescence architecture: a comparative view. Ann. Bot. (Lond.) 100:659–676
Blazquez M, Soowal L, Lee I, Weigel D (1997) LEAFY expression and flower initiation in Arabidopsis. Development 124:3835–3844
Borner R, Kampmann G, Chandler J, Gleißner R, Wisman E, Apel K, Melzer S (2000) A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J 24:591–599
Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119:721–743
Ferrandiz C, Gu Q, Martienssen R, Yanofsky MF (2000a) Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127:725–734
Ferrandiz C, Liljegren SJ, Yanofsky MF (2000b) Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 289:436–438
Fornara F, de Montaigu A, Coupland G (2010) SnapShot: control of flowering in Arabidopsis. Cell 141(550–550):e2
Glover B (2007) Understanding Flowers and Flowering: An integrated Approach, first ed. Oxford University Press, Great Clarendon Street, Oxford OX2 6DP, pp.83-94
Huala E, Sussex I (1992) LEAFY interacts with floral homeotic genes to regulate Arabidopsis floral development. Plant Cell 4:901–913
Huang S, Cerny RE, Qi Y, Bhat D, Aydt CM, Hanson DD, Malloy KP, Ness LA (2003) Transgenic studies on the invlovement of cytokinin and gibberellin in male development. Plant Physiol 131:1270–1282
Jack T (2001) Plant development going MADS. Plant Mol Biol 46:515–520
Jang S, An K, Lee S, An G (2002) Characterization of tobacco MADS-box genes involved in floral initiation. Plant Cell Physiol 42:230–238
Kang SG, Jin JB, Piao HL, Pih KT, Jang HJ, Lim JH, Hwang I (1998) Molecular cloning of an Arabidopsis cDNA encoding a dynamin-like protein that is localized to plastids. Plant Mol Biol 38:437–447
Karnovsky MJ (1965) A formaldehyde-glutaraldehyde fixative of high osmolarity for use in electron microscopy. J Cell Biol 27:137A
Kelly A, Zagotta MT, White RA, Chang C, Meeks-Wagner DR (1995) NFL, the tobacco homolog of Floricaula and Leafy, is transcriptionally expressed in both vegetative and floral meristems. Plant Cell 2:1201–1224
Kim KW (2008) Visualization of micromorphology of leaf epicuticular waxes of the rubber tree Ficus elastica by electron microscopy. Micron 39:976–984
Kim SH, Hamada T (2005) Rapid and reliable method of extracting DNA and RNA from sweetpotato, Ipomoea batatas (L). Lam Biotech Lett 27:1841–1845
Kim SH, Song WK, Kim YH, Kwon SY, Lee HS, Lee IC, Kwak SS (2009) Characterization of full-length enriched expressed sequence tags of dehydration-treated white fibrous roots of sweet potato. BMB reports 42(5):271–276
Kim JS, Seo SG, Jun BK, Lee Y, Jeon SB, Choe J, Kim JB, Kim ST, Kim SH (2011) An IbEF1 from sweet potato promotes flowering in transgenic tobacco. Gens Genom 33:335–341
Koo SC, Bracko O, Park MS, Schwab R, Chun HJ, Park KM, Seo JS, Grbic V, Balasubramanian S, Schmid M, Godard F, Yun DJ, Lee SY, Cho MJ, Weigel D, Kim MC (2010) Control of lateral organ development and flowering time by the Arabidopsis thaliana MADS-box Gene AGAMOUS-LIKE6. Plant J 62:807–816
Lee JH, Kim SH, Jung YH, Kim JA, Lee MO, Choi PG, Choi W, Kim KN, Jwa NS (2005) Molecular cloning and functional analysis of rice (Oryza sativa L.) OsNDR1 on defense signaling pathway. Plant Pathol J 21:149–157
Litt A, Irish VF (2003) Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: implications for the evolution of floral development. Genetics 165:821–833
Mandel MA, Yanofsky MF (1995) A gene triggering flower formation in Arabidopsis. Nature 377:522–524
Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky M (1992) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360:273–277
Mandel T, Fleming AJ, Kra¨henbu¨hl R, Kuhlemeier C (1995) Definition of constitutive gene expression in plants: the translation initiation factor 4A gene as a model. Plant Mol Biol 29:995–1004
Michaels SD, Ditta G, Gustafson-Brwon C, Pelaz S, Yanofsky M, Amasino R (2003) AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J 33:867–874
Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14(Suppl. S):S111–S130
Reinheimer R, Kellogg E (2009) Evolution of AGL6-like MADS box genes in grasses (Poaceae): ovule expression is ancient and palea expression is new. Plant Cell 21:2591–2605
Sablowsky R (2007) Flowering and determinacy in Arabidopsis. J Exp Bot 58(5):899–907
Sambrook J, Russell DW (2001) Molecular cloning: A laboratory manual, third ed. Cold Spring Harbor Press, Cold Spring Harbor, New York, pp. 6.39-6.46
Schultz EA, Haughn GW (1991) LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis. Plant Cell 3:771–781
Schultz EA, Haughn GW (1993) Genetic analysis of the floral initiation process (FLIP) in Arabidopsis. Development 119:745–765
Simson GG, Dean C (2002) Arabidopsis, the Rosetta Stone of Flowering Time? Science 296:285–289
Smykal P, Gennen J, Bodt SD, Ranganath V, Melzer S (2007) Flowering of strict photoperiodic Nicotiana varieties in non-induction conditions by transgenic approaches. Plant Mol Biol 65:233–242
Srikanth A, Schmid M (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68:2013–2037
Teper-Bamnolker P, Samach A (2005) The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17:2661–2675
Wagner D, Sablowski RW, Meyerowitz EM (1999) Transcriptional activation of APETALA1 by LEAFY. Science 285:582–584
Weigel D, Meyerowitz EM (1994) The ABCs of floral homeotic genes. Cell 78:203–209
Weigel D, Nilsson O (1995) A developmental switch sufficient for flower initiation in diverse plants. Nature 377:495–500
Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69:843–859
William DA, Su Y, Smith MR, Lu M, Baldwin DA, Wagner D (2004) Genomic identification of direct target genes of LEAFY. Proc Natl Acad Sci USA 101:1775–1780
Yamaguchi A, Wu MF, Yang L, Wu G, Poethig RS, Wagner D (2009) The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev Cell 17:268–278
Yu H, Ito T, Wellmer F, Meyerowitz EM (2004) Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat Genet 36:157
Acknowledgments
This work was supported by Korea Institute of Planning and Evaluation for Technology (IPET) funded by Ministry for Food, Agriculture, Forestry and Fisheries (311048-05).
Author information
Authors and Affiliations
Corresponding author
Additional information
J. M. Shin and S.-G. Seo are the first authors and contributed equally to this work.
Rights and permissions
About this article
Cite this article
Shin, J.M., Seo, SG., Kim, JS. et al. Ectopic expression of NtEF1 and NtEF2 promotes flowering and alters floral organ identity in Nicotiana tabacum . Plant Biotechnol Rep 9, 11–26 (2015). https://doi.org/10.1007/s11816-014-0336-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-014-0336-1