Abstract
The novel coronavirus disease (COVID-19) has spread throughout the globe, affecting millions of people. The World Health Organization (WHO) has declared this infectious disease a pandemic. At present, several clinical trials are going on to identify possible drugs for treating this infection. SARS-CoV-2 Mpro is one of the most critical drug targets for the blockage of viral replication. The aim of this study was to identify potential natural anthraquinones that could bind to the active site of SARS-CoV-2 main protease and stop the viral replication. Blind molecular docking studies of 13 anthraquinones and one control drug (Boceprevir) with SARS-CoV-2 Mpro were carried out using the SwissDOCK server, and alterporriol-Q that showed the highest binding affinity towards Mpro were subjected to molecular dynamics simulation studies. This study indicated that several antiviral anthraquinones could prove to be effective inhibitors for SARS-CoV-2 Mpro of COVID-19 as they bind near the active site having the catalytic dyad, HIS41 and CYS145 through non-covalent forces. The anthraquinones showed less inhibitory potential as compared to the FDA-approved drug, boceprevir. Among the anthraquinones studied, alterporriol-Q was found to be the most potent inhibitor of SARS-CoV-2 Mpro. Further, MD simulation studies for Mpro- alterporriol-Q system suggested that alterporriol-Q does not alter the structure of Mpro to a significant extent. Considering the impact of COVID-19, identification of alternate compounds like alterporriol-Q that could inhibit the viral infection will help in accelerating the process of drug discovery.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
December 2019 saw the emergence of an array of severe pneumonia cases caused by a coronavirus (CoV) in Wuhan, China (Huang et al. 2020). CoV is an enveloped positive-stranded RNA virus, portrayed by club-like spikes on their surface and belongs to the group Cornonaviridae of the Nidovirales order (Fehr and Perlman 2015). The genomic sequencing of the 2019 CoV showed that it 96.2% alike to a bat coronavirus and shares 79.5% sequence similarity to SARS-CoV (Dai et al. 2020). Hence the International Committee on Taxonomy of Viruses named this novel coronavirus as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and the associated pneumonia was named as COVID-19 on 11th of February, 2020 by the World Health Organization (WHO). The pandemic has spread to more than 210 countries affecting a cumulative population of 162 million and leading to the death of over 3.3 million people as on 18th May, 2021 (https://www.who.int/publications/).Footnote 1 As COVID-19 is new to the immune system of humans, people throughout the globe are at risk of becoming sick on exposure to SARS-CoV-2 (Rahimi and Talebi Bezmin Abadi 2020). Therefore, considering the global threat due to this viral infection, several clinically effective vaccine have been made to treat COVID-19, and altogether more than 300 vaccine projects are going on (Forni et al. 2021). Prophylactic vaccines are the only way to successfully contain and eradicate pandemic viruses, but the development of vaccines is time consuming as compared to the conventional medicines (Calina et al. 2020). Moreover, risk factors of COVID-19 vaccine efficacy particular to the elder people and children cannot be ignored (Calina et al. 2020; Müller et al. 2021; Ryzhikov et al. 2021). Antivirals have the potential to minimise the morbidity and mortality associated with viral infections, thus global preparedness programmes that enhance our ability to produce or repurpose existing antivirals are critical. Several drugs such as hydroxychloroquine (Gautret et al. 2020), remdesivir, chloroquine (Wang et al. 2020), favipiravir (Cai et al. 2020; Khambholja and Asudani 2020), ivermectin (Caly et al. 2020), and several other small molecules (Fu et al. 2020; Ghosh et al. 2021) have been found to inhibit the viral disease. SARS-CoV-2 contains the 3C-like protease (3CLpro), also known as the main protease (Mpro), which consists of a catalytic domain (highly conserved) from the SARS virus and is essential for controlling several CoV functions (Hall and Ji 2020). One vital function is the replication of the virus, making it one of the best-characterized target for drug development (Adeola Falade et al. 2021), hence targeting the Mpro would prevent the virus from building its proteins (Baildya et al. 2020).
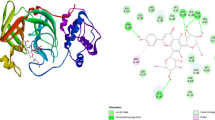
SARS-CoV-2 Mpro (Fig. 1) is a three-domain (I to III) cysteine protease and is a homodimer. The domain I (8-101) and II (102-184) consists of β-barrels mostly, and III (201-306) is made up of mainly of α-helices (Khan et al. 2020). The structure consists of a conserved non-canonical HIS41-CYS145 dyad located within the cleft between domains I and II (Dai et al. 2020). Moreover, it has been reported that [LEU-GLN↓ (SER, ALA, GLY)] (↓: cleavage site) is the cleavage site in Mpro (Borkotoky et al. 2021). No human protease has been found to have such cleavage selectivity, and these functional characteristics make Mpro an appealing target for drug development investigations (Ullrich and Nitsche 2020). Targeting the substrate binding site of Mpro that harbors the catalytic dyad formed by HIS41 and CYS145 has been the protocol for screening of molecules for the inhibition of SARS-CoV-2 (Gil et al. 2020; Borkotoky et al. 2021). The physicochemical properties of SARS-CoV-2 Mpro as obtained from the ExPASy ProtParam tool (https://web.expasy.org/protparam/), listed in Table S1 indicate that Mpro has a molecular weight of 33,796.64 Da, with a GRAVY score of −0.019 and instability index of 27.65, suggesting that the protease is stable. Furthermore, the total number of positively (ARG + LYS) and negatively (ASP + GLU) charged amino acid residues were observed to be 22 and 26, respectively, which indicates that it is hydrophilic in nature capable of forming hydrogen bonds (Tahir ul Qamar et al. 2020).
Natural-source compounds are progressively becoming important therapeutic and pharmacological agents in drug discovery and development (Atanasov et al. 2021). They have minimal sid-effects when administered, a unique method of action, and a high level of chemical diversity, which improves their therapeutic interaction with a wide range of biological targets as compared to synthetic medications. Hence, here in this study anthraquinones that occur naturally were screened for the their inhibitory potential towards SARS-CoV-2 infection. Natural anthraquinones are a class of aromatic compounds having low toxicity and high bioactivity (Chien et al. 2015; Malik and Müller 2016). One of the important properties of anthraquinones is based on their antiviral activity (Barnard et al. 1992; Cohen et al. 1996), which is needed in the current context of the COVID-19 pandemic to be analyzed for their inhibitory potential against the SARS-CoV-2 infection. Here, in this report, we have analyzed the inhibitory potentials of 13 naturally occurring anthraquinones such as emodin, aloe emodin, chrysophanic acid, tetrahydroaltersolanol C, aloin A and B, rhein, rubiadin, alterporriol Q, damnacanthal, hypericin, pseudohypericin and isopseudohypericin against SARS-CoV-2 Mpro through blind molecular docking analysis and compared their results with a FDA approved drug, boceprevir, that has been shown to target Mpro and efficaciously inhibit SARS-CoV-2 (Fu et al. 2020). The other antiviral drugs such as remdesivir, favipiravir, and sofosbuvir targets RNA-dependent RNA polymerase (RdRp) of SARS-CoV-2 (Allen et al. 2020; Jácome et al. 2020; Kokic et al. 2021). Hence, boceprevir was chosen as the control since it targets Mpro which falls in line with our objective of Mpro targeting anthraquionones. Boceprevir is a drug used to treat the Hepatitis C virus (HCV) (Behmard and Barzegari 2020). It is highly selective against the HCV serine protease and has shown potential in the blockage of viral replication, thus inhibiting the life cycle of HCV in vitro and in vivo (Ascione 2012).
The antiviral activities of the above mentioned natural anthraquinones along with their sources are listed in Table S2. It is to be noted that isopseudohypericin has not been reported for its antiviral property till date, but as Hypericum perforatum extract has antiviral effects (Chen et al. 2019), and isopseudohypericin is isolated from H. perforatum; therefore, it might have an antiviral effect; hence we studied its binding efficacy with SARS-CoV-2 Mpro. This work concentrates on recognizing natural anthraquinones compounds with a particular objective to accelerate the process of identifying specific/alternate drugs for COVID-19 treatment.
Methodology
Geometry optimization of the compounds
The 3D coordinates of the compounds, boceprevir, emodin, aloe emodin, chrysophanic acid, tetrahydroaltersolanol C, aloin A and B, rhein, rubiadin, alterporriol Q, damnacanthal, hypericin, pseudohypericin and isopseudohypericin were downloaded as a .mol file from ChemSpider (www.chemspider.com) and geometry optimized further using the Parametric Method 3 (PM3) in ArgusLab (Bitencourt-Ferreira and de Azevedo 2019; Das et al. 2020b). The optimized structures of boceprevir and the natural anthraquinones are depicted in Fig. S1. The ChemSpider ID of the compounds is listed in Table S2. The logP values were obtained from SwissADME analysis (Daina et al. 2017).
Molecular docking analyses and visualization
The blind molecular docking method has become an increasingly essential technique for drug discovery and understanding protein-ligand interactions. The blind docking procedures carry out an unbiased search over the entire surface of the protein/enzyme to identify binding sites. Hence, blind molecular docking studies of several natural antiviral anthraquinones were carried out with SARS-CoV-2 Mpro, and their results were compared with that of boceprevir.
The 3D crystal structure of SARS-CoV-2 MPro (PDB ID. 6Y84) was downloaded from Protein Data Bank (PDB) (Owen et al. 2020). A molecular docking study on a single chain was carried out by removing the water molecules from the PBD using PyMOL (Yuan et al. 2017). The final PDB file of Mpro and optimized ligands using ArgusLab were directly fed into an online docking server, SwissDock (http://www.swissdock.ch/docking). SwissDock incorporates an automated in silico molecular docking procedure based on EADock DSS docking algorithm, which utilizes the CHARMM (Chemistry at HARvard Macromolecular Mechanics) forcefield (Grosdidier et al. 2011). According to SwissDock, the minimum energy docked conformers are ranked in terms of their fullfitness score. According to a report on EADock (Grosdidier et al. 2007), ‘the fullfitness of a cluster is calculated by averaging the 30% most favorable effective energies of its elements, in order to limit the risk of a few complexes penalizing the whole cluster’. The docked pose that has the least fullfitness score is used for further analysis. The molecular visualization were carried out using UCFS Chimera (Pettersen et al. 2004), PyMOL, and the 2D interaction plots were created using Discovery Studio Visualizer.Footnote 2 The online server available at http://cib.cf.ocha.ac.jp/bitool/ASA/ was used to calculate the changes in the accessible surface area (∆ASA) of the Mpro protease on interactions with the compounds.
Molecular dynamics simulation
GROMACS 2018.2 was used for the molecular dynamics study of the following three complex system: (i) alterporriol Q with Mpro, (ii) only Mpro, and (iii) only alterporriol Q. Each complex was prepared in an aqueous medium. Charm 36–mar2019 force field was used in alterporriol Q with Mpro and only alterporriol Q system. At the same time, OPLS-AA force field was used for the Mpro protein system. For the generation of ligand topology, we have used the CGenFF server (Vanommeslaeghe et al. 2010), and for only alterporriol Q system SwissParam (Zoete et al. 2011) server was used. To neutralize the system, 4 Na+ were added. All three systems were equilibrated at the same condition where we had done the energy minimization, then followed with the NVT and NPT equilibration process. During the NVT equilibration, systems were assigned isothermal-isobaric, and in NPT, the system was assembled into the isothermal-isobaric condition. All the system was equilibrated for 100 ps in NVT and NPT equilibration process. The temperature and pressure of the system were controlled through the Nosé-Hoover thermostat (Hoover 1985) and Parrinello-Rahman barostat (Parrinello and Rahman 1981), respectively. Finally, molecular dynamics simulations were run for 100 ns, and all trajectories were saved at each 10 ps under periodic boundary condition (PBC). The Binding free energy of the complex was calculated through the g_mmpbsa (Kumari et al. 2014) module in each 1 ns of formed trajectories after the molecular dynamic simulation of the complex.
Results and discussion
Toxicity prediction of the natural anthraquinones
Drug discovery and drug development is a risky and time-consuming process. Paul et al. (Paul et al. 2010) estimated that the rate of drug attrition is about 96%, while the average cost during the 2000s -early 2010s to develop a new drug amounts to 2.6 billion U.S. dollars (http://phrma-docs.phrma.org/sites/default/files/pdf/biopharmaceutical-industry-profile.pdf). The high attrition rate is attributed to drug safety, which corresponds to about 30% of the drug failures (Giri and Bader 2015). The first step of drug safety assessment involves only the knowledge about the toxicity of a compound (Yang et al. 2018). Therefore, the toxicity prediction and drug likeliness of the natural anthraquinones using DataWarrior (Sander et al. 2015) and SwissADME are listed in Table 1. It could be observed that the control drug, boceprevir has tumourigenic and irritation effects, whereas most of the anthraquinones have comparatively lesser toxic effects. Among the 13 anthraquinones, tetrahydroaltersolanol C, aloin A, and aloin B have none of the four effects, whereas rhein, rubiadin, alterporriol Q and damnacanthal is predicted to have only irritant effect.
Molecular docking analysis
The fullfitness score, the free energy of binding, and the logP values of the control drug, boceprevir, and that of the 13 natural anthraquinones have been listed in Table 2. The binding affinity of each anthraquinone towards SARS-CoV-2 MPro was discussed based on the estimated ∆G value (Ulloa-Guerrero et al. 2018; Kumar et al. 2020). From this Table 2, it could be observed that the estimated ∆G is higher for boceprevir (−8.66 kcal/mol) as compared to all the anthraquinones, which means that the inhibitory potency of these anthraquinones is lesser than that of the control drug. But alterporriol Q (−8.48 kcal/mol) has a very close ∆G value to that of boceprevir, while others have energies between 6.64-7.75 kcal/mol. The logP values listed in Table 2 measures the molecular hydrophobicity or lipophilicity of a particular compound. High logP values show poor absorption or low permeability, whereas low logP values indicate increased absorption and permeability. A logP value greater than 5 indicates a high hydrophobic character of a compound (Ditzinger et al. 2019). Here, the logP values of most of the anthraquinones are less than 5, excluding only hypericin. The logP values are essential for the understanding of how the compounds may penetrate cell membranes. Unfortunately, a relationship between the estimated binding energy and the logP values could not be obtained here.
The minimum energy docked poses and the 2D interaction plots of the compounds within the substrate-binding site of SARS-CoV-2 Mpro are depicted in Figs. 2, 3, 4 and 5. The nearby residues interacting with the compounds through non-covalent forces, except the van der Waals forces of attraction (shown in the 2D interaction plots) are also listed in Table 3. It could be observed from Figs. 2, 3, 4 and 5 that boceprevir and all the natural anthraquinones could bind to the active site of SARS-CoV-2 Mpro, which is lined up by residues such as THR25, THR26, HIS41, MET49, GLY143, CYS145, GLU166, PRO168, etc. The significance of the two catalytic residues HIS41 and CYS145, and other residues like GLY143, CYS145, HIS163, HIS164, GLU166, PRO168, and GLN189 was indicated in a recent study by Zhang et al. (2020) (Zhang et al. 2020) for the design of α-ketoamide inhibitors for SARS-CoV-2 Mpro. Similarly, the significance of these residues for the design and synthesis of antiviral compounds as inhibitors of SARS-CoV-2 Mpro was also demonstrated by Dai et al. (Dai et al. 2020). Therefore, the natural anthraquinones studied here could inhibit the viral disease by binding to the active site of Mpro.
The compounds stabilize within the active site of Mpro by different non-covalent forces such as hydrogen-bonding, π-alkyl, π-sigma, π-π stacked interactions, and others as shown in the 2D interaction plots of Figs. 2, 3, 4 and 5. The stability of ligand within the binding site of a macromolecule is related mainly to the hydrogen bonding interactions formed between the two counterparts (Chen et al. 2016; Szefler 2019). Boceprevir forms two hydrogen bonds (H-bonds) with SER46 (3.62 Å) and GLU166 (4.34 Å), and is surrounded by residues such as HIS41, CYS145, HIS164, and GLU166 (Fig. 2a), which is similar to that reported in the crystal structure of Mpro complexed with boceprevir (Fu et al. 2020).
Among the anthraquinones from Rhubarb, emodin (Fig. 2b) forms H-bonds with THR25 (3.81 Å) and GLY143 (3.63 Å), rhein forms H-bonds with THR25 (3.65 Å), GLY143 (3.73 Å) and GLU166 (3.97 Å) (Fig. 2c), while chrysophanic acid interacts with GLY143 through H-bonds at a distance of 3.61 Å (Fig. 2d). For the anthraquinones from aloe, aloe emodin forms H-bonds with PHE140 and GLY143 (3.57 Å) as depicted in Fig. 3a, aloin A forms two H-bonds with GLN189 at a distance of 3.89 and 3.76 Å, one each with CYS145 (4.95 Å) and GLU166 (5.05 Å) as shown in Fig. 3b, while aloin B forms two H-bonds with ASN142 (Fig. 3c).
Rubiadin, an anthraquinone from Rubia Cordifolia, forms H-bonds with HIS41 (3.14 Å) and SER46 (3.69 Å) as can be seen from Fig. 3d. Anthraquinones from Alternaria sp. fungus, tetrahydroaltersolanol C (Fig. 4a) interacts with CYS44 (4.09 Å) and CYS145 (3.74 Å) through H-bonds, while alterporriol Q (Fig. 4b) forms H-bonds with ASN142 (3.66 Å), GLY143 (4.22 Å) and GLU166 (4.08 Å). Damnacanthal, an anthraquinone of Noni, forms H-bond with GLY143 (3.77 Å), as seen from Fig. 4c.
The anthraquinones (Fig. 5a–c) of H. perforatum, hypericin forms two H-bonds with GLU166 at a distance of 4.87 and 4.83 Å, pseudohypericin interacts with GLY143 (3.69 Å) and GLN189 (3.55 Å) residues through H-bonds, while isopseudohypericin forms two H-bonds with GLU166 at a distance of 4.13 and 4.24 Å. Besides the H-bonds, the conformational energy of the interactions are minimized through other non-covalent forces such as π-sigma, π-π stacked, amide-π, π-alkyl, π-sulphur and van der Waals forces as shown in the 2D interactions plots of the docked poses of Figs. 2, 3, 4 and 5 (Arthur and Uzairu 2019).
Herein, the blind molecular docking studies of the natural anthraquinones with Mpro indicated that they possess inhibitory potential towards SARS-CoV-2, as they can bind to the substrate-binding site of SARS-CoV-2 Mpro, which is essential for inhibiting the viral replication (Zhang et al. 2020). This substrate binding site of Mpro is lined up by residues such as HIS41, MET49, GLY143, CYS145, HIS163, HIS164, GLU166, PRO168, and GLN189. As HIS41 and CYS145 are the two important catalytic residues, therefore the distance of the compounds from these two residues, along with the change in accessible area of the residues, are listed in Table 4. In terms of the estimated free energy of binding (∆G) values, the control (boceprevir) has the highest affinity (−8.66 kcal/mol) to function as a potential inhibitor for SARS-CoV-2 Mpro, which indeed supports the promising role of boceprevir as a potential anti- COVID-19 drug. Although, the none of the anthraquinones could cross that of the boceprevir inhibitory potential, but among the anthraquinones the inhibitory potential follows the following order alterporriol Q (−8.48 kcal/mol) > aloin A (−7.75 kcal/mol) > aloin B (−7.64 kcal/mol) > tetrahydroaltersolanol C (−7.38 kcal/mol) > pseudohypericin (−7.26 kcal/mol) > isopseudohypericin (−7.23 kcal/mol) > hypericin (−7.18 kcal/mol) > damnacanthal (−7.16 kcal/mol) > aloe emodin (−7.12 kcal/mol) > rhein (−7.00 kcal/ mol) > emodin (−6.90 kcal/mol) > chrysophanic acid (−6.83 kcal/mol) > rubiadin (−6.64 kcal/mol).
Molecular dynamics (MD) simulation studies
From the docking results, alterporriol Q was found to possess the best binding affinity towards the Mpro. Hence, the complex of alterporriol Q with Mpro (6y84) was subjected to 100 ns of MD simulation. The dynamics and stability of the complex was assessed through root mean square deviation (RMSD), root mean square fluctuation (RMSF), and radius of gyration (Rg) and compared with the native Mpro and the native ligand (Das et al. 2019).
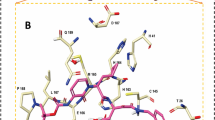
The dynamics of alterporriol Q (at different time intervals) within the 3D matrix of Mpro could be observed from Fig. 6a, which suggests that the ligand optimizes its position and resides approximately within the active site as the simulation period increases. This could be further substantiated from Fig. 6b, which shows the participation of different amino acid residues within the alterporriol Q binding site, common among them being THR25, HIS41, SER46, MET49, CYS145, GLN189, and others. The different kinds of non-covalent interactions between alterporriol Q and the active site residues of SARS-CoV-2 Mpro at 0 ns, 50 ns and 100 ns of simulation time are depicted in Fig. S2.
The RMSD value is a measure of the comprehensive stability of the protein complex in terms of determining the deviation from its initial structure (Das et al. 2020a). Smaller deviations in the RMSD values indicate that the protein is more stable. To gauge the structural and conformational stability of SARS-CoV-2 Mpro and its complex with alterporriol Q, the difference between the backbone atoms of Mpro from its initial to final conformation was monitored through the RMSD analysis. The time evolutions of RMSDs of the complexed Mpro and alterporriol Q are depictesd in Fig. 7a and b along with RMSDs of the native forms of Mpro and alterporriol Q. From Fig. 7a, it could be observed that the RMSD of the native Mpro increases at around 30 ns and then maintains its stability throughout the simulation period. This observation correlates well with that observed for boceprevir binding to Mpro (Borkotoky et al. 2021), indicating that alterporriol Q also has similar effect on the structure of Mpro as does boceprevir.
Similarly, in the presence of alterporriol Q (black curve) the variation in the RMSD value follows a similar trend except for some variations at around 70 ns-80 ns after which it again stabilizes and follows a similar pattern as that of the native Mpro. Hence, it could be inferred that the ligand does not alter the stability of the SARS-CoV-2 Mpro significantly. Figure 7b depicts the variation in the RMSD of alterporriol Q in the free (red curve) and Mpro bound (black curve) states throughout the simulation period. It is apparent that the RMSD of alterporriol Q increases in the complex form in comparison to the native form, but the variations are smooth and remain steady throughout the simulation time.
The RMSF is a measure of the deviations of a particle from its original position and serves as a crucial structural parameter for identifying the flexible regions within the protein structure, which in turn provides an idea about the conformational flexibility of the protein residues. From Fig. 8, for free Mpro, fluctuations were observed in all three domains, the highest being in domain III (201-303). It is evident that the Mpro complexed with alterporriol Q (black line) showed fluctuations in most regions as compared to the free Mpro, but are in minimal order which is suggestive of no major alteration between the initial and final structures. Moreover, the catalytic dyad consisting of HIS41 and CYS145 remained stable during the course of simulation. The reported RMSF analysis of boceprevir binding of Mpro follows a similar trend (Borkotoky et al. 2021).
Next, to acquire a comparable structural envelope, the variation in the radius of gyration (Rg) was monitored for 100 ns of simulation. The radius of gyration (Rg) offers an idea about the level of compactness in the structure of macromolecules upon interaction with ligands (Das et al. 2018). Loosely packed or unfolded protein is characterized by a relatively high value, whereas a more compact and folded protein has a lower Rg value. From Fig. 9a, it could be observed that Mpro has a Rg value of around 2.22 nm, which upon complexation with alterporriol Q shows a slight/ negligible increase which indicates that the ligand does not alter the structure of the protein significantly. Hence, it could be inferred that alterporriol Q maintained the compactness of Mpro throughout the simulation time, and the system is well converged. The variation in the Rg of alterporriol Q in the free and Mpro bound state is also shown in Fig. 9b, it could be seen that the Rg value of alterporriol Q decreases significantly when bound to Mpro, suggesting the increase in the compactness of the ligand in the protease environment.
The free energy calculations for the binding of alterporriol Q with SARS-CoV-2 Mpro using Molecular Mechanics-Poisson–Boltzmann Solvent-Accessible surface area (MMPBSA) method (Kollman et al. 2000). The calculated binding energy for the complex system was found out to be −46.76 kJ mol−1, which indicates that the alterporriol Q-Mpro complex was stable. Further from Table 5, it is evident that the major favorable contributors towards binding energy was van der Waals energy followed by electrostatic energy, while the polar solvation energy contributed unfavorably towards the binding of alterporriol Q with Mpro. To further delve into key residues involved in the complexation process, the binding free energy was split into energy contribution from each individual residue (Fig. 10).
From the plot it could be observed that the binding site residues such as LEU27, HIS41, SER46, MET49, GLY143, CYS145, HIS164, MET165 and GLN189 participated actively in the complexation process. The energy contribution per residue profile indicated that most of the residues have negative binding energy (these residues played a major role in stabilizing the complex), whereas few residues had a positive affinity. The residues like MET49 with −3.53 kJ mol−1 and LEU27 with −1.76 kJ mol−1 energy had the highest binding energy among all the residues. Boceprevir-Mpro system also has similar energy contribution from HIS41, MET49, CY145 and GLN189, therefore the alterporriol Q-Mpro system correlates well with the control drug boceprevir (Borkotoky et al. 2021).
Since 1972 onwards, the world has seen the emergence of more than 50 new viruses, which been recognized as etiologic agents of human diseases (Bryan-Marrugo et al. 2015). Antiviral drug development is a complex and time-consuming phenomenon. Thus the evolution of new viruses calls for the development and usage of efficient strategies to synthesize or identify already known antiviral drugs that limit the spread or treat the virus. Over the years, since the discovery of idoxuridine (IDU) in 1959 (Bryan-Marrugo et al. 2015), several antivirals that affect the viruses life cycle have been determined, which lead to a number of antiviral protocols being proposed that includes targeting intracellular signal transduction pathways or inhibiting the viral replication (Bryan-Marrugo et al. 2015). While a variety of antivirals have been reported, when subjected to selective antiviral therapy, only very few molecules have proved to be safe and effective. In the current context, the repurposing of licensed FDA drugs or the use of compounds from natural sources is a key concept due to its economic viability and ease of availability in terms of research and development of new drugs, particularly at this junction where the COVID-19 pandemic is posing as a global threat. In addition to the FDA-approved drug, boceprevir here we observed that the anthraquinones, particularly alterporriol Q possess significant inhibitory potential towards SARS-CoV-2 Mpro. Naturally occurring anthraquinones have low toxicity and different biological activities (Chien et al. 2015; Islam et al. 2015). Therefore, these observations indicate promising potential for the use of natural anthraquinones for the treatment of COVID-19.
Conclusion
Blind molecular docking has been used for studying the inhibitory potentials of natural anthraquinones against SARS-CoV-2 Mpro of COVID-19. Around 13 hit natural anthraquinones reported here to have potential inhibitory effects against the SARS-CoV-2 main protease. Out of the 13 anthraquinones, the compound having the highest binding affinity was further subjected to MD simulation studies. This study provides a foundation for computational drug discovery of new natural compounds to treat and reduce the transmission COVID-19. It was observed that the anthraquinones could bind to the substrate-binding site of SARS-CoV-2 Mpro through different non-covalent forces. Although the inhibitory potential of the natural anthraquinones was found to be lesser than that of boceprevir in terms of the estimated ∆G value, alterporriol Q could be the most potent inhibitor of SARS-CoV-2 Mpro among the natural anthraquinones studied here, as its ∆G value differed from that of boceprevir by 0.18 kcal/ mol. Then, the MD simulations studies for the interaction of alterporriol Q with SARS-CoV-2 Mpro depicted that the ligand had no significant impact on the structure of Mpro, and it remains within the active site throughout the simulation period. Further, the contribution energy per residue suggested that MET49 and LEU27 played a major role in stabilizing the complex formed. This study leads to the possibility of natural anthraquinones being used as a treatment for COVID-19, but as this study has been carried out using a computational method, detailed in vivo and in vitro experiments are required to be carried out to gauge the applicability and toxicity of these anthraquinones.
Notes
World Health Organization (2021) onward (continuously updated). WHO Coronavirus Disease (COVID-19) Dashboard [online]. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19%2D%2D-18-may-2021 [accessed 18 May 2021]
Dassault Systèmes BIOVIA, Discovery Studio Visualizer, Release 2019, San Diego: Dassault Systèmes, 2019.
Abbreviations
- CoV:
-
Coronavirus
- COVID-19:
-
Coronavirus Disease 2019
- SARS-CoV-2:
-
Severe Acute Respiratory Syndrome Coronavirus 2
- Mpro :
-
Main Protease
- MD:
-
Molecular Dynamics
- RMSD:
-
Root Mean Square Deviation
- RMSF:
-
Root Mean Square Fluctuation
- Rg:
-
Radius of Gyration
References
Adeola Falade V, Isaac AT, Olaide AI, Abdul-Hammed M, Alabi LT, Alabi AS, Silico PI (2021) In silico investigation of saponins and tannins as potential inhibitors of SARS-CoV-2 main protease (Mpro). Silico Pharmacol 9:1–15. https://doi.org/10.1007/s40203-020-00071-w
Allen CNS, Arjona SP, Santerre M, Sawaya BE (2020) Potential use of RNA-dependent RNA polymerase (RdRp) inhibitors against SARS-CoV2 infection. 13:608–614. https://doi.org/10.1080/26895293.2020.1835741
Arthur DE, Uzairu A (2019) Molecular docking studies on the interaction of NCI anticancer analogues with human phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit. J King Saud Univ - Sci 31:1151–1166. https://doi.org/10.1016/j.jksus.2019.01.011
Ascione A (2012) Boceprevir in chronic hepatitis C infection: a perspective review. Ther Adv Chronic Dis 3:113–121. https://doi.org/10.1177/2040622312441496
Atanasov AG, Zotchev SB, Dirsch VM, Orhan IE, Banach M, Rollinger JM, Barreca D, Weckwerth W, Bauer R, Bayer EA, Majeed M, Bishayee A, Bochkov V, Bonn GK, Braidy N, Bucar F, Cifuentes A, D’Onofrio G, Bodkin M et al (2021) Natural products in drug discovery: advances and opportunities. Nat Rev Drug Discov 20:200–216. https://doi.org/10.1038/s41573-020-00114-z
Baildya N, Ghosh NN, Chattopadhyay AP (2020) Inhibitory activity of hydroxychloroquine on COVID-19 main protease: an insight from MD-simulation studies. J Mol Struct 1219:128595. https://doi.org/10.1016/j.molstruc.2020.128595
Barnard DL, Huffman JH, Morris JLB, Wood SG, Hughes BG, Sidwell RW (1992) Evaluation of the antiviral activity of anthraquinones, anthrones and anthraquinone derivatives against human cytomegalovirus. Antivir Res 17:63–77. https://doi.org/10.1016/0166-3542(92)90091-I
Behmard E, Barzegari E (2020) Insights into resistance mechanism of hepatitis C virus nonstructural 3/4A protease mutant to boceprevir using umbrella sampling simulation study. J Biomol Struct Dyn 38:1938–1945. https://doi.org/10.1080/07391102.2019.1621212
Bitencourt-Ferreira G, de Azevedo WF (2019) Molecular docking simulations with ArgusLab. Pp. 203–220. Methods in molecular biology, Humana press Inc. https://doi.org/10.1007/978-1-4939-9752-7_13
Borkotoky S, Banerjee M, Modi GP, Dubey VK (2021) Identification of high affinity and low molecular alternatives of boceprevir against SARS-CoV-2 main protease: a virtual screening approach. Chem Phys Lett 770:138446. https://doi.org/10.1016/j.cplett.2021.138446
Bryan-Marrugo OL, Ramos-Jiménez J, Barrera-Saldaña H, Rojas-Martínez A, Vidaltamayo R, Rivas-Estilla AM (2015) History and progress of antiviral drugs: from acyclovir to direct-acting antiviral agents (DAAs) for hepatitis C. Med Univ 17:165–174. https://doi.org/10.1016/j.rmu.2015.05.003
Cai Q, Yang M, Liu D, Chen J, Shu D, Xia J, Liao X, Gu Y, Cai Q, Yang Y, Shen C, Li X, Peng L, Huang D, Zhang J, Zhang S, Wang F, Liu J, Chen L et al (2020) Experimental treatment with Favipiravir for COVID-19: an open-label control study. Engineering. 6:1192–1198. https://doi.org/10.1016/j.eng.2020.03.007
Calina D, Docea AO, Petrakis D, Egorov AM, Ishmukhametov AA, Gabibov AG, Shtilman MI, Kostoff R, Carvalho F, Vinceti M, Spandidos DA, Tsatsakis A (2020) Towards effective COVID-19 vaccines: updates, perspectives and challenges (review). Int J Mol Med 46:3–16. https://doi.org/10.3892/ijmm.2020.4596
Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM (2020) The FDA-approved drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir Res 178:104787. https://doi.org/10.1016/j.antiviral.2020.104787
Chen D, Oezguen N, Urvil P, Ferguson C, Dann SM, Savidge TC (2016) Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci Adv 2:e1501240. https://doi.org/10.1126/sciadv.1501240
Chen H, Muhammad I, Zhang Y, Ren Y, Zhang R, Huang X, Diao L, Liu H, Li X, Sun X, Abbas G, Li G (2019) Antiviral activity against infectious bronchitis virus and bioactive components of Hypericum perforatum L. Front Pharmacol 10:1272. https://doi.org/10.3389/fphar.2019.01272
Chien SC, Wu YC, Chen ZW, Yang WC (2015) Naturally occurring anthraquinones: chemistry and therapeutic potential in autoimmune diabetes. Evidence-based Complement Altern Med 2015:357357. https://doi.org/10.1155/2015/357357
Cohen PA, Hudson JB, Towers GHN (1996) Antiviral activities of anthraquinones, bianthrones and hypericin derivatives from lichens. Experientia. 52:180–183. https://doi.org/10.1007/BF01923366
Dai W, Zhang B, Jiang XM, Su H, Li J, Zhao Y, Xie X, Jin Z, Peng J, Liu F, Li C, Li Y, Bai F, Wang H, Cheng X, Cen X, Hu S, Yang X, Wang J et al (2020) Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science (80-) 368:1331–1335. https://doi.org/10.1126/science.abb4489
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:42717. https://doi.org/10.1038/srep42717
Das S, Bora N, Rohman MA, Sharma R, Jha AN, Singha Roy A (2018) Molecular recognition of bio-active flavonoids quercetin and rutin by bovine hemoglobin: an overview of the binding mechanism, thermodynamics and structural aspects through multi-spectroscopic and molecular dynamics simulation studies. Phys Chem Chem Phys 20:21668–21684. https://doi.org/10.1039/C8CP02760A
Das S, Santra S, Rohman MA, Ray M, Jana M, Singha Roy A (2019) An insight into the binding of 6-hydroxyflavone with hen egg white lysozyme: a combined approach of multi-spectroscopic and computational studies. J Biomol Struct Dyn 37:4019–4034. https://doi.org/10.1080/07391102.2018.1535451
Das S, Sarmah S, Hazarika Z, Rohman MA, Sarkhel P, Jha AN, Singha Roy A (2020a) Targeting the heme protein hemoglobin by (−)-epigallocatechin gallate and the study of polyphenol–protein association using multi-spectroscopic and computational methods. Phys Chem Chem Phys 22:2212–2228. https://doi.org/10.1039/C9CP05301H
Das S, Sarmah S, Lyndem S, Singha Roy A (2020b) An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. J Biomol Struct Dyn 39:3347–3357. https://doi.org/10.1080/07391102.2020.1763201
Ditzinger F, Price DJ, Ilie A-R, Köhl NJ, Jankovic S, Tsakiridou G, Aleandri S, Kalantzi L, Holm R, Nair A, Saal C, Griffin B, Kuentz M (2019) Lipophilicity and hydrophobicity considerations in bio-enabling oral formulations approaches - a PEARRL review. J Pharm Pharmacol 71:464–482. https://doi.org/10.1111/jphp.12984
Fehr AR, Perlman S (2015) Coronaviruses: an overview of their replication and pathogenesis. pp. 1–23. Coronaviruses: methods and protocols, Springer New York. https://doi.org/10.1007/978-1-4939-2438-7_1
Forni G, Mantovani A, Forni G, Mantovani A, Moretta L, Rappuoli R, Rezza G, Bagnasco A, Barsacchi G, Bussolati G, Cacciari M, Cappuccinelli P, Cheli E, Guarini R, Bacci ML, Mancini M, Marcuzzo C, Morrone MC, Parisi G et al (2021) COVID-19 vaccines: where we stand and challenges ahead. Cell Death Differ 28:626–639. https://doi.org/10.1038/s41418-020-00720-9
Fu L, Ye F, Feng Y, Yu F, Wang Q, Wu Y, Zhao C, Sun H, Huang B, Niu P, Song H, Shi Y, Li X, Tan W, Qi J, Gao GF (2020) Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat Commun 11:4417. https://doi.org/10.1038/s41467-020-18233-x
Gautret P, Lagier JC, Parola P, Hoang VT, Meddeb L, Mailhe M, Doudier B, Courjon J, Giordanengo V, Vieira VE, Tissot DH, Honoré S, Colson P, Chabrière E, La Scola B, Rolain JM, Brouqui P, Raoult D (2020) Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents 56:105949. https://doi.org/10.1016/j.ijantimicag.2020.105949
Ghosh K, Amin SA, Gayen S, Jha T (2021) Chemical-informatics approach to COVID-19 drug discovery: exploration of important fragments and data mining based prediction of some hits from natural origins as main protease (Mpro) inhibitors. J Mol Struct 1224:129026. https://doi.org/10.1016/j.molstruc.2020.129026
Gil C, Ginex T, Maestro I, Nozal V, Barrado-Gil L, Cuesta-Geijo MÁ, Urquiza J, Ramírez D, Alonso C, Campillo NE, Martinez A (2020) COVID-19: drug targets and potential treatments. J Med Chem 63:12359–12386. https://doi.org/10.1021/acs.jmedchem.0c00606
Giri S, Bader A (2015) A low-cost, high-quality new drug discovery process using patient-derived induced pluripotent stem cells. Drug Discov Today 20:37–49. https://doi.org/10.1016/j.drudis.2014.10.011
Grosdidier A, Zoete V, Michielin O (2007) EADock: docking of small molecules into protein active sites with a multiobjective evolutionary optimization. Proteins Struct Funct Bioinforma 67:1010–1025. https://doi.org/10.1002/prot.21367
Grosdidier A, Zoete V, Michielin O (2011) SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res 39:W270–W277. https://doi.org/10.1093/nar/gkr366
Hall DC, Ji HF (2020) A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease. Travel Med Infect Dis 35:101646. https://doi.org/10.1016/j.tmaid.2020.101646
Hoover WG (1985) Canonical dynamics: equilibrium phase-space distributions. Phys Rev A 31:1695–1697. https://doi.org/10.1103/PhysRevA.31.1695
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M et al (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China Lancet 395:497–506. https://doi.org/10.1016/S0140-6736(20)30183-5
Islam R, Mamat Y, Ismayil I, Yan M, Kadir M, Abdugheny A, Rapkat H, Niyaz M, Ali Y, Abay S (2015) Toxicity of anthraquinones: differential effects of Rumex seed extracts on rat organ weights and biochemical and haematological parameters. Phyther Res 29:777–784. https://doi.org/10.1002/ptr.5317
Jácome R, Campillo-Balderas JA, Ponce de León S, Becerra A, Lazcano A (2020) Sofosbuvir as a potential alternative to treat the SARS-CoV-2 epidemic. Sci Reports 10:1–5. https://doi.org/10.1038/s41598-020-66440-9
Khambholja K, Asudani D (2020) Potential repurposing of Favipiravir in COVID-19 outbreak based on current evidence. Travel Med Infect Dis 35:101710. https://doi.org/10.1016/j.tmaid.2020.101710
Khan SA, Zia K, Ashraf S, Uddin R, Ul-Haq Z (2020) Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. J Biomol Struct Dyn 39:2607–2616. https://doi.org/10.1080/07391102.2020.1751298
Kokic G, Hillen HS, Tegunov D, Dienemann C, Seitz F, Schmitzova J, Farnung L, Siewert A, Höbartner C, Cramer P (2021) Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat Commun 12:1–7. https://doi.org/10.1038/s41467-020-20542-0
Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, Cheatham TE (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 33:889–897. https://doi.org/10.1021/ar000033j
Kumar N, Awasthi A, Kumari A, Sood D, Jain P, Singh T, Sharma N, Grover A, Chandra R (2020) Antitussive noscapine and antiviral drug conjugates as arsenal against COVID-19: a comprehensive chemoinformatics analysis. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2020.1808072
Kumari R, Kumar R, Lynn A (2014) G-mmpbsa -a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54:1951–1962. https://doi.org/10.1021/ci500020m
Malik EM, Müller CE (2016) Anthraquinones as pharmacological tools and drugs. Med Res Rev 36:705–748. https://doi.org/10.1002/med.21391
Müller L, Andrée M, Moskorz W, Drexler I, Walotka L, Grothmann R, Ptok J, Hillebrandt J, Ritchie A, Rabl D, Ostermann PN, Robitzsch R, Hauka S, Walker A, Menne C, Grutza R, Timm J, Adams O, Schaal H (2021) Age-dependent immune response to the Biontech/Pfizer BNT162b2 coronavirus disease 2019 vaccination. Clin Infect Dis ciab381. https://doi.org/10.1101/2021.03.03.21251066
Owen CD, Lukacik P, Strain-Damerell CM, Douangamath A, Powell AJ, Fearon D, Brandao-Neto J, Crawshaw AD, Aragao D, Williams M, Flaig R, Hall DR, McAuley KE, Mazzorana M, Stuart DI, von Delft F, Walsh MA (2020) COVID-19 main protease with unliganded active site (2019-nCoV, coronavirus disease 2019, SARS-CoV-2). DOI. https://doi.org/10.2210/pdb6Y84/pdb
Parrinello M, Rahman A (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys 52:7182–7190. https://doi.org/10.1063/1.328693
Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, Schacht AL (2010) How to improve RD productivity: the pharmaceutical industry’s grand challenge. Nat Rev Drug Discov 9:203–214. https://doi.org/10.1038/nrd3078
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Rahimi F, Talebi Bezmin Abadi A (2020) Challenges of managing the asymptomatic carriers of SARS-CoV-2. Travel Med Infect Dis 37:101677. https://doi.org/10.1016/j.tmaid.2020.101677
Ryzhikov AB, Ryzhikov E, Bogryantseva MP, Usova SV, Danilenko ED, Nechaeva EA, Pyankov OV, Pyankova OG, Gudymo AS, Bodnev SA, Onkhonova GS, Sleptsova ES, Kuzubov VI, Ryndyuk NN, Ginko ZI, Petrov VN, Moiseeva AA, Torzhkova PY, Pyankov SA et al (2021) A single blind, placebo-controlled randomized study of the safety, reactogenicity and immunogenicity of the “EpiVacCorona” vaccine for the prevention of COVID-19, in volunteers aged 18–60 years (phase I–II). Russ J Infect Immun 11:283–296. https://doi.org/10.15789/2220-7619-ASB-1699
Sander T, Freyss J, Von Korff M, Rufener C (2015) DataWarrior: an open-source program for chemistry aware data visualization and analysis. J Chem Inf Model 55:460–473. https://doi.org/10.1021/ci500588j
Szefler (2019) Docking linear ligands to glucose oxidase. Symmetry (Basel) 11:901. https://doi.org/10.3390/sym11070901
Tahir ul Qamar M, Alqahtani SM, Alamri MA, Chen LL (2020) Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J Pharm Anal 10:313–319. https://doi.org/10.1016/j.jpha.2020.03.009
Ulloa-Guerrero CP, Delgado MDP, Jaramillo CA (2018) Structural analysis of variability and interaction of the N-terminal of the oncogenic effector CagA of helicobacter pylori with phosphatidylserine. Int J Mol Sci 19. https://doi.org/10.3390/ijms19103273
Ullrich S, Nitsche C (2020) The SARS-CoV-2 main protease as drug target. Bioorg Med Chem Lett 30:127377. https://doi.org/10.1016/j.bmcl.2020.127377
Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD (2010) CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 31:671–690. https://doi.org/10.1002/jcc.21367
Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M, Shi Z, Hu Z, Zhong W, Xiao G (2020) Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 30:269–271. https://doi.org/10.1038/s41422-020-0282-0
Yang H, Sun L, Li W, Liu G, Tang Y (2018) In Silico prediction of chemical toxicity for drug design using machine learning methods and structural alerts. Front Chem 6. https://doi.org/10.3389/fchem.2018.00030
Yuan S, Chan HCS, Hu Z (2017) Using PyMOL as a platform for computational drug design. Wiley Interdiscip Rev Comput Mol Sci 7:e1298. https://doi.org/10.1002/wcms.1298
Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R (2020) Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science (80) 368:409–412. https://doi.org/10.1126/science.abb3405
Zoete V, Cuendet MA, Grosdidier A, Michielin O (2011) SwissParam: a fast force field generation tool for small organic molecules. J Comput Chem 32:2359–2368. https://doi.org/10.1002/jcc.21816
Acknowledgments
ASR and SD are indebted to NIT Meghalaya for providing research platform. SD is grateful to the TEQIP III, NIT Meghalaya for fellowship. The study is not supported by any funding agency. AS and SKS are thankful to Indian Institute of Information Technology, Allahabad for providing high-speed central computation facility (CCF). Authors acknowledge Mr. Viswajit Mulpuru and Mr. Vishal from Indian Institute of Information Technology, Allahabad for their helpful suggestions. The authors thank the reviewers for their valuable comments and suggestions for enhancing the scientific quality of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declared that no competing conflict of interest exists. All authors have read and approved this version of the article.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 1135 kb)
Rights and permissions
About this article
Cite this article
Das, S., Singh, A., Samanta, S.K. et al. Naturally occurring anthraquinones as potential inhibitors of SARS-CoV-2 main protease: an integrated computational study. Biologia 77, 1121–1134 (2022). https://doi.org/10.1007/s11756-021-01004-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11756-021-01004-4