Abstract
Globally, Brassica juncea is one of the significant oilseed crops, which is commercially important and cultivated under diverse agro-climatic region preferably for its high oil content as well as superior oil quality. Biotic stress is one of the dominant factors which hinders plant growth and yield globally. Though substantial progress has been made in breeding and genetic manipulation of plants for imposing resistance, the task remains as a challenge even today. Here, in this study, two major defense genes of plant origin, lectin from lentil and protease inhibitors from chickpea, which have insecticidal activity against phytophagous insect pests have been isolated and stacked into one ORF by overlapping extension PCR and a fusion gene construct is prepared. For tissue specific expression of this fusion gene, phloem specific promoter rolC was utilized and the construct was mobilized into Agrobacterium tumefaciens strain GV3101. Further, genetic transformation of B. juncea cv. Varuna was performed with fusion gene using Agrobacterium-mediated genetic transformation. The regeneration of the transformed putative plants was screened with hygromycin selection. Fusion gene integration in randomly selected transformed plants gives 17% overall transformation efficiency by PCR and Southern hybridization. A quantitative real-time PCR analysis showed that the transcript level of the fusion gene was within 1.8–2.8-fold. The expression of the fusion gene was significantly higher in the third line (2.8-fold). The aphid resistance test demonstrated that inhibition of larval survivability is about 40%, and the extent of leaf damage measured was also diminished in the transgenic plants compared to non transformed control plants. This study provides insight the multi-toxin approach developed through overlap extension PCR in B. juncea and may facilitate genetic engineering for the potential improvement of resistance against aphids.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Brassica juncea (L.) is one of the significant oilseed crop which contributes 82.7% to total national hectarage and 86.9% to production (Kumar and Shauhan 2005). One of the major biotic factors which decrease crop productivity is predatory insect pests. Plant diseases can reduce crop yield dramatically and the major impact of disease outbreaks is particularly acute in tropical and subtropical nations. One of the major phytophagous insect pests of mustard crop is an aphid species, Lipaphis erysimi, which is a sap-sucking phytophagous insect-pest and acts as a mediator for many diseases causing various viruses and other pathogens (Kumar 1999; Patel et al. 2004). The nymph and adult stages of this insect suck the plant sap during flowering and seed formation which prevents the nutrient flow and hence affects the plant growth and crop productivity. The breeding of insect pests were considerably controlled by utilization of toxic chemicals for crop protection and to enhance crop yield over the past few decades, but the indiscriminate use of these chemical pesticides resulted in environmental pollution and adverse effect on human health (Jouanin et al. 1998). Not only this, these chemicals have also led to reduction in beneficial insect population. The traditional breeding programme used for aphid resistance has been proved insignificant due to non-availability of aphid resistance genes in the available collection of germplasms (Rogers et al. 1993; Pradhan et al. 1993; Yadav and Singh 1999).
Advancements in plant molecular biology provide approaches of plant genetic engineering for introducing one or multiple insecticidal molecules in crop plants to develop transgenic plants for improved and durable insect resistance. In the last few decades, several insecticidal genes have been incorporated into different plants to control the infestation of insect pests. One of the well-documented microbial genes introduced in various plants is Bacillus thuringiensis (Bt), which has proved effective against numerous insect pests as lepidopteran and coleopteran which is chewing type of insect pests. Plant-based carbohydrate-binding plant lectins have also been well documented to have insecticidal activity when expressed in transgenic plants against sap-sucking insects including aphids, with no effect on human metabolism (Hilder et al. 1995a, b; Gatehouse et al. 1996; Dutta et al. 2005). Lectins such as ASAL (Allium sativum leaf agglutinin) are mannose-binding lectin and their expression provides resistance against aphid attack in mustards (Bandyopadhyay et al. 2001; Dutta et al. 2005). Galanthus nivalis (snowdrop) bulb lectin agglutinin commonly known as GNA has found to be effective against homopteran (Hilder et al. 1995a, b; Gatehouse et al. 1996; Rao et al. 1998). Wheat germ agglutinin (WGA) has also been expressed into many plants to provide insect resistance in various crop plants; they mainly bind to glycoprotein located in the mid gut of mustard aphids which inhibits the absorption of nutrients and hence leads to the destruction and mortality of the insect pests (Majumder et al. 2004; Zhu-Salzman et al. 1998).
Numerous literatures have also proclaimed the potency of plant-based protease inhibitor genes as effective anti-digestive compounds to defend crop plants from pathogenic infestation (Broadway 2000; Haq et al. 2004). Potential candidate to boost up insect-resistant transgenic crops, in particular, is serine protease inhibitors (Daun et al. 1996). Mostly, the protease inhibitors of plants origin are proteinaceous in nature and basically competitive inhibitors, acting as pseudo-substrates which bind to the active site of respective proteases, and the targeted proteases cannot cleave the peptide bonds which adversely cause a disruption in assimilation of dietary protein in herbivorous insect pests primarily delays the significant growth and development (Birk 2003; Haq et al. 2004). Increased level of protease inhibitor is found to be inducible and there accumulations in transgene expression have been explored as possible avenues to confer resistance against aphids (Habib and Fazili 2007). Recently, the insecticidal effects of protease inhibitors in plants have been developed and in-planta expression of the protease inhibitors showed the inhibition of constitutive-targeted proteases in native herbivores.
For tissue-specific expression, rolC isolated from Agrobacterium rhizogenes strain A4 was utilized for phloem specific expression. This study has been effectively utilized for developing phloem-specific expression of fusion lectin-protease inhibitors genes to protect plants against aphid attack. The mannose-binding lectin has been isolated from lentil (LL) and protease inhibitor gene from chickpea (CPPI) and both are isolated from edible crops, hence are expected to be safe for consumers.
In the recent years, studies have shown that the defense-related proteins when used in combinations may accomplish enhanced toxicity and durability against majority of predatory insect pests. Fusion proteins strategy such as gene stacking by integrating two or more lectin genes (Hossain et al. 2006; Bharathi et al. 2011), pyramiding of two different protease inhibitor genes (Senthil Kumar et al. 2010) as well as stacking of lectin gene with protease inhibitor have been designed to increase the efficacy and broaden the insecticidal action (Zhu-Salzman et al. 1998, Maqbool et al. 2001). In this study, the multi-toxin approach has been used and for this fusion product of both lentil lectin and chickpea protease inhibitor gene has been developed through overlap extension PCR approach which was further utilized for Agrobacterium-mediated transformation of B. juncea for the potential improvement of resistance against aphids.
Materials and methods
Vector construction
Preparation of fusion gene
Overlapping extension PCR is one of the precise procedures for the assembly of two or more multiple genes using chimeric primers. To assemble two insecticidal genes, lentil lectin (LL) and chickpea protease inhibitor (CPPI), four chimeric primers were used. Chimeric primers were designed in a manner that the forward primer is same as first gene and reverse primer of first gene (LL) is reverse complementary having flanking sequences of second gene (CPPI). Similarly, primers for the second gene were designed where forward primer having sequences same as second gene sequence (CPPI) but having flanking sequences same as first gene (LL) for overlapping during hybridization and reverse primer same as the second gene (CPPI). The overlapping regions of the four chimeric primers are shown clearly (in bold)
LL-Kp-F- 5′ GCGGGTACC ATGGCTTCTCTTCAA 3′
Fu-LLPI-R- 5′TACAATGGATTTCATTGCATCTGCAGCTTG3′
Fu-LLPI-F- 5′CAAGCTGCAGATGCA ATGAAATCCATTGTA3′
PI-Xb-R- 5′GCGTCTAGACTAAACTGACGCATC3′
PCR amplification of LL and CPPI gene using chimeric primers
To prepare fusion gene, the LL gene was PCR amplified from earlier cloned in pGEMT-Easy vector with gene-specific primers having KpnI restriction site incorporated in the chimeric forward (LL-Kp-F) and reverse primer (Fu-LLPI-R). PCR was performed for 30 cycles with annealing at temperature of 60 °C and final extension of 72 °C for 1 min using Pfu DNA polymerase (Thermo, USA). The amplification products were analyzed on 1% (w/v) agarose gel and remaining product was used for hybridization step.
Similarly, the CPPI gene was also PCR amplified from earlier cloned in pCR2.1 TOPO vector with gene specific primers having chimeric forward primer (Fu-LLPI-F) and reverse primer (PI-Xb-R) of CPPI gene having XbaI restriction site. PCR was performed for 30 cycles with annealing temperature of 60 °C using Pfu DNA polymerase. The amplification products were run on 1% (w/v) agarose gel and utilized for further fusion gene preparation.
Assembling of fusion gene construct
Fusion reaction step A The Equi-molar aliquots of both the gel-eluted gene fragments (LL and CPPI) having overlapping sequences were taken in a PCR tube and added the remaining components of PCR mix except primers in the reaction volume of 50 µl each in five different PCR tubes kept at five different annealing temperatures ranging between 50 and 55 °C. Fragments carrying overlaps sequence were fused at different annealing temperatures programmed with an initial denaturation at 94 °C for 2 min, subsequent steps 94 °C for 15 s, annealing at 50–55 °C for 20 s and extension at 72 °C for 2 min, for 10 cycles. The hybridization product was utilized for the PCR amplification of whole fusion product of both the genes in step B.
Fusion reaction step B The unpurified PCR fusion product (5–10 µl) of step A was used as template and remaining components of PCR was added with forward primer of LL gene (LL-Kp-F) and reverse primer of CPPI gene (PI-Xb-R). The PCR reaction was performed in an programmed thermal cycler with an preliminary DNA denaturation at 94 °C for 2 min, subsequent steps 94 °C for 15 s, annealing at 62 °C for 20 s, extension at 72 °C for 2 min and additional extension of 72 °C for 10 min. Resulting PCR product of 5 µl was run and analyzed on 1% agarose gel and remaining product was purified using Qiagen PCR cleanup kit (Qiagen, Germany) and send for sequencing before cloning.
Cloning of fusion gene construct in pCAMBIA1300 (nosT) binary vector
The binary vector pCAMBIA1300 was used to create the gene construct. The eluted fusion PCR product and plasmid pCAMBIA1300 was separately digested with KpnI and XbaI and further ligated to generate the construct and transformed into E. coli XL1-Blue. Colony PCR was performed with transformed colonies and the recombinants were also confirmed by restriction digestion for the presence of fusion gene construct.
Cloning of phloem-specific promoter (rolC) into pCAMBIA1300 binary vector construct
Cloning of phloem-specific promoter (rolC), plasmid DNA of rolC from pGEMT-Easy vector was amplified with forward primer having EcoRI restriction site RF- 5′ GCGAATTCTTAGCGAAAGGATGTCA 3′ and reverse primer having KpnI restriction site RR-5′ CCCGGTACCATGGTAACAAAGTAGGA 3′. PCR amplification product was run on 1% agarose gel and the desired band was eluted and restricted with both the restriction enzymes. The restricted rolC was purified and ligated to linearized pCAMBIA1300 having fusion gene and transform into E. coli XL1-Blue. Specific primers of rolC were used to performed colony PCR and restriction analysis to confirm the presence of rolC promoter in pCAMBIA1300 vector construct.
Finally, the whole construct was confirmed by restriction digestion of the recombinants with EcoRI and SalI which included rolC + fusion gene + nosT and backbone of binary vector pCAMBIA1300. Fusion gene construct in the pCAMBIA1300 binary vector were mobilized to Agrobacterium strain GV3101 through heat and thaw method. The binary vector pCAMBIA1300 is competent enough to replicate in E.coli and Agrobacterium and having unique restriction sites as well as hptII marker for plant selection between its disarmed borders. Fusion gene-specific primers were used for PCR amplification to confirm the recombinant plasmid in A. tumefaciens.
Agrobacterium-mediated transformation of B. juncea
Plant material and conditions
HgCl2 0.1% and SDS 0.1% were used for surface sterilization of B. juncea cv. Varuna seeds for 5–8 min and washed with sterilized water 7–8 times and dried on sterile Whatman filter paper No. 4 (Whatman International, UK) and inoculated on ½ MS (Murashige and Skoog (1962) [MS medium with 1.5% sucrose] for 2 days in dark condition and then transferred to 16-h light/8-h dark cycle and maintained at 25 ± 2 °C.
Co-cultivation of explants
Cotyledons as well as hypocotyls were excised from 4- to 5-day-old seedling, hypocotyls were cut into 0.5–0.8 cm and both the explants were cultured on pre-culture medium (PCM) [MS medium with 2 mg l−1 BAP and 0.5 mg l−1 NAA with 0.8% agar (w/v)] (Himedia, India) at 25 °C under 16-h light/8-h dark cycle. Agrobacterium harboring fusion gene construct was grown in 10 ml of YEM media (Primary culture) with 50 mg l−1 of rifampicin and 50 mg l−1 of kanamycin at 28 °C until the A600 reaches ~0.6–0.8. From the primary grown culture, 100 µl aliquot was added to 10 ml of liquid MS medium and grown at 28 °C till the A600 reaches ~0.3. The pre-cultured explants (cotyledons and hypocotyls) were incubated in A. tumefaciens suspension for 10–15 min and dried on Whatman filter paper No. 4 for 20–25 min and incubated on co-cultivation medium (CCM) (MS medium with 3% sucrose, 2 mg l−1 BAP, 0.5 mg l−1 NAA and acetosyringone 200 µM) for 48 h in dark.
Regeneration and selection
Explants after co-cultivation were washed twice with sterile distilled water as well as with cefotaxime of 250 mg l−1 in liquid MS (without sucrose) and dried on sterile Whatman filter paper No. 4 properly; and cotyledons were cultured on shoot regeneration and growth medium (SRM) (MS medium with 3% sucrose, 1 mg l−1 of BAP, 0.5 mg l−1 of NAA), and hypocotyls on the SRM with 3 mg l−1 of AgNO3 at 25 ± 2 °C. After 3 days, cotyledons were transferred to selection culture medium (SM) [MS medium with 3% sucrose, 1 mg l−1 of BAP, 0.5 mg l−1 of NAA, 250 mg l−1 of cefotaxime, 25 mg l−1 of hygromycin], whereas hypocotyls on the similar medium supplemented with 3 mg l−1 of AgNO3 at 25 ± 2 °C. Green shoots growing on selection culture medium were repeatedly sub-cultured after every 10–15 days on same medium, and explants having well-developed shoots were excised. Small plantlets from both the explants were transferred to shoot elongation and growth medium (SEM) [MS medium with 0.05 mg l−1 of IAA, 0.5 mg l−1 of BAP, 250 mg l−1 of cefotaxime, 25 mg l−1 of hygromycin] at 25 ± 2 °C for elongation. The multiplied shoots of 2–4 cm of length were excised and kept for rooting on rooting medium (RM) [MS medium with 1 mg l−1 of IBA, 250 mg l−1 of cefotaxime, 25 mg l−1 of hygromycin] at 25 ± 2 °C. Explant with well-developed roots on RM after 15–20 days were transplanted into small pots (4 inch dm) containing sterile soil, vermiculite and sand (2:1:1). The plantlets were maintained in plant growth chamber under controlled conditions at 80–90% relative humidity and 16-h light/8-h dark photoperiod at 25 ± 2 °C for 25–30 days. Thereafter, the plantlets which grown well were transferred to larger pots (8 inch dm) containing the mixture of soil, vermiculite and sand in 2:1:1 ratio and shifted to phytotron green house maintaining 65% relative humidity and 14-h light/10-h dark photoperiod at 25 ± 2 °C for complete acclimatization.
Genomic DNA extraction, PCR and Southern blot analysis
To confirm the transgene integration, molecular analysis was carried out and total gDNA was extracted, following CTAB method from the leaf tissues of putative transformed and non-transformed B. juncea plants (Murrey and Thompson 1980). To amplify fusion gene and hptII gene from gDNA of putative transformants plants, gene-specific primers were used. Plasmid pCAMBIA1300 having fusion gene was used as a positive control and non-transformed tissue culture plant (NTC) was used as a negative control. The PCR amplified products were run and analyzed on 1% (w/v) agarose gel.
Southern blot hybridization was carried out according to the method described in Sambrook and Russell (2001). Genomic DNA was extracted from PCR positive-transformed B. juncea leaves. Approximately 5–10 µg of total gDNA was completely digested with EcoRI and electrophoreses on 0.8% (w/v) agarose gel and transferred onto a nylon membrane Biodyne (Hybond N+, Amershan Pharmecia Biotech, UK). Fusion gene in pCAMBIA1300 was restricted with EcoRI and SalI and used as probe after labelling using (α32P) dCTP-labeled DNA labelling kit from Fermentas as manufacturer’s instructions.
Total RNA isolation and RT-PCR analysis
Fusion gene expression was analyzed through RT-PCR in selected transformants which were confirmed positive by PCR. Total RNA was isolated from the young leaves of PCR positive transformed and NTC plants by Tri-reagent method. The integrity of total RNA was evaluated through electrophoresis of denatured RNA on agarose gel. Approximately 100 ng of total RNA was utilized and cDNA was prepared (Thermo, USA) for the transgenic analysis according to manufacturer’s instruction. Amplified product for the fusion gene was size fractioned separately on 1% agarose gel.
Expression analysis of the transgene
To synthesize cDNA, 100 ng of total RNA was used and amplification of the target gene fragment primer pair are used as LL-FP-5′ATGGCTTCTCTTCAAAC 3′ and CPPI-RP-5′ATGGCTTCTCTTCAAAC 3′ and internal control actin used with primer pairs FP-5′TGGGTTTGCTGGTGACGAT3′ and RP-5′TGCCTAGGACGACCAACAATACT 3′ and electrophoreses in 1% (w/v) agarose gel. qRT-PCR analysis was performed for fusion gene expression with relative quantification against actin gene using Stratagene Mx3000P. Different transformed plants lines and non-transformed control plant expression analysis were conducted using SuperScript® III Platinum® SYBR® Green One-Step qRT-PCR Kit (Invitrogen, USA). For a single reaction, 50 µl of mixture contained 100 ng of cDNA, 25 pM of each of the forward and reverse primers, and 25 µl of PCR master mix were used. qRT-PCR conditions consisted of an primarily denaturation at 95 °C for 4 min followed by 35 cycles of 95 °C for 30 s, 60 °C for 30 s, 95 °C for 30 s with a final incubation at 95 °C for 10 min. To avoid incorrect normalization by the reference gene, fusion gene and actin gene were measured in different wells with precise pipetting and performed in triplicates. MxPro QPCR Software used for data analyses.
Insect bioassay by detached leaf of transgenic
Aphid bioassay was performed to detect the efficacy of fusion gene transformants. Leaves of putative transformants as well as control NTC were collected by cutting from growing plants and the detached ends were immediately immersed in 3% agar in Petri plates. Around 20 mustard aphids (third instars larvae) were maintained on ventral surface of leaves in triplicates, of which three leaves are from each transgenic plant as well as from control plants. The survival of aphids was monitored at regular intervals for a period of 0–9 days. Experiment was carried out under controlled conditions at 16-h light/8-h dark intervals with 75% relative humidity at 25 °C temperature. The effect of fusion gene transgenic on survivability of aphids was assessed by the total number of insects survived on the leaves of transgenic plants during the entire bioassay period. The mean data was expressed as the percentage of total surviving aphids per plant basis and the extent of leaf damage was measured using Leaf Area Meter of all the triplicates.
Agronomic performance of transgenic plants
For the evaluation of the agronomic performance of transgenic (T), tissue-cultured generated non-transgenic (NTC) and wild type plants (WT), different parameters like plant height, seed/pod, number of branches/plant, yield/plant (gram) were observed. From Southern positive transgenic lines, five plants were picked and evaluated for studying each parameter.
Statistical analysis
To find the significance of difference between means of transgenic and control (NTC) lines of each treatment group, data were subjected to analysis of variance (ANOVA) and expressed as mean with standard error (±SE). Each assay was carried out in three replicates. Statistical significance was determined using a Student’s t test, at the p = 0.05 significance level.
Results and discussion
The availability of diverse genes from different plant species makes it a possibility to use one or more genes in combination whose products are targeted at different biochemical and physiological processes within the insects. Plants with two pyramided distinct toxin genes deployed either sequentially or in mosaics, exhibited delayed expansion of insect resistance more efficiently in comparison to the plants having single-toxin genes (Roush 1998; Zhao et al. 2003). Fusion gene cloning and its expression would be a potential candidate and might be promising for crop plants engineering against major insect pests. Keeping this in mind, we prepared chimeric primers of genes with overlapping regions of other gene and prepare fusion of two genes by overlapping extension PCR method.
For pyramiding of LL and CPPI gene, we prepared overlapping chimeric primers and fusion of two genes by two-step overlapping extension PCR (Fig. 1). Plasmid DNA of LL gene was isolated, restricted with EcoRI as well as PCR amplified with gene specific primers and sequenced. Nucleotide sequence of LL gene revealed ~826 bp sequence without any stop codon. Another gene CPPI was also isolated, restricted with KpnI and XbaI as well as PCR amplified with gene specific primers and sequenced. Nucleotide sequence analysis of CPPI gene has revealed ~669 bp ORF (open reading frame).
For stacking of LL with CPPI gene, both amplified gene products with overlapping region were mixed in equi-molar ratio and hybridization was performed for 30 min and final PCR was performed using both end primers having KpnI in forward and XbaI in reverse primer. The whole fusion gene product of ~1494 bp was generated in one ORF which was analyzed on agarose gel, and the sequencing results also showed no variation in nucleotide assembly (Fig. 2). Further, the fusion gene product was cloned in binary vector pCAMBIA1300 having nosT by restricting KpnI and XbaI. Promoter rolC was also cloned for phloem-specific expression of fusion gene product using EcoRI and KpnI restriction site and the intact cassette was confirmed in recombinants by restriction digestion using EcoRI and SalI gives ~2.6 kb fragment which included rolC promoter, fusion gene, nosT terminator and backbone of binary vector pCAMBIA1300. Different strategies, like gene constructs nature and their expression profiles, multiple transgenes in different combinations, domains of toxin and non-toxin chains fusion constructs to discourage the expansion of resistant insect populations adoption of a range of deployment strategy populations have been proposed to ensure the durable effectiveness of potential insecticidal genes expressing in transgenic crops (Maqbool et al. 2001; Mehlo et al. 2005; Manyangarirwa et al. 2006).
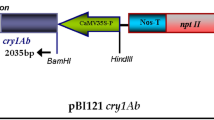
Explants such as hypocotyls have been utilized in majority for Agrobacterium-mediated transformation of B. juncea (Dutta et al. 2005; Sharma et al. 2004). Only a few reports are available for the development of transgenic using stem segments (Singh et al. 2010; Pental et al. 1993) or cotyledons (Bhuiyan et al. 2011). The shoot regeneration in B. juncea was achieved with cotyledons and hypocotyls where cotyledons explants responded better with higher transformation efficiency (Bhuiyan et al. 2011). Here, in this study, the fusion gene construct (Fig. 3) was mobilized into A. tumefaciens strain GV3101 by freeze and thaw method and genetic transformation of B. juncea using cotyledons and hypocotyls as explants. To ascertain the transformation parameters, inoculation period of explants in Agrobacterium culture and co-cultivation period was optimized in subsequent transformation purpose, as co-cultivation time was critical for high transformation efficiency. After co-cultivation, explants were kept in dark for 24–48 h for Agrobacterium infection on co-cultivation culture medium with 3 mg l−1 of AgNO3. To remove excess Agrobacterium growth, the co-cultivated explants were washed 2–3 times with sterile distilled water and liquid MS supplemented with 250 mg l−1 of cefotaxime. Explants were blotted dry on sterile Whatman filter paper No. 4 and inoculated on delayed regeneration medium at 25 ± 2 °C for 3–5 days (Fig. 3a, b). Further, explants were sub-cultured on regeneration medium and hypocotyls on the regeneration growth culture medium supplemented with 3 mg l−1 of AgNO3. The positive effect of delayed application of the selective agent in potato has suggested that the cell division of transformed cells were proliferated by delayed selection, which has ability to provide greater protection against selection pressure (Visser et al. 1989). This provides enhanced transformation frequency in cases where there was steady transcription of selection marker. Greenish shoots started differentiating into green multiple shoots on selection culture medium, and explants were further sub-cultured to fresh culture medium after every 10–15 days (Fig. 3c). Shoots which survived on selection culture medium were transferred to elongation medium and plantlets having length of about 3–4 cm were excised and culture onto rooting medium (Fig. 3d). Regenerated plantlets with well-developed roots were washed carefully with sterile water and transferred into pots for growth and maturation in environmentally controlled growth chamber (Fig. 3e). We observed that the transformed plants harboring fusion gene showed normal morphology and flowering characteristics at maturation stage (Fig. 3f).
T-DNA region of the binary vector pCAMBIA1300. LB left border, RB right boarder, PolyA, terminator, HYG hygromycin selectable marker gene, MCS multiple cloning sites having fusion gene. A modified B. juncea transformation method using hygromycin as the selection agent. Explants were prepared from 4- to 5-day-old seedlings of Varuna. a 4–5-day-old seedling for explant preparation. b Hypocotyl explants on SRM. c Cotyledon explants. d Shoot buds from cotyledons. e Elongated plantlets on SEM. f Elongated plantlets on rooting medium. g Rooted plantlets were hardened in sterilized pot mix in the tissue culture facility. h Growing plantlets were shifted to greenhouse for acclimatized and reached maturity. Scale bar 0.8 cm (a, h); 1.0 cm (b, c, f); 1.2 cm (d, e, g)
Reports have shown 6–7% of transformation efficiency in B. juncea transformation by Sharma et al. (2004) as well as Bhuiyan et al. (2011) and Singh et al. (2010) have shown increased transformation efficiency in cotyledons as 16.2%. However, in our study with cotyledons as explants, 16.4–19.7% transformation efficiency was achieved from six independent batches with fusion gene construct (Table 1).
The putative transformed plants were screened for the existence of fusion gene product of LL and CPPI in the transformed plants by PCR. Genomic DNA of putative transformants was extracted by CTAB method and analyzed by PCR using fusion and hptII gene-specific primers showed expected band size of ~1496 bp and ~1 kb, respectively (Fig. 4a, b). Based on PCR analysis, plants were randomly selected to confirm the transgene integration and copy number in transformed B. juncea and analyzed by Southern hybridization. The gDNA was restricted with EcoRI and hybridized with α32P dCTP labeled fusion gene probe. In putative transformants, a distinctive hybridizing signal was observed whereas the lane with untransformed (NTC) plant gDNA showed the absence of any hybridization. On autoradiogram, the number of bands obtained indicates the corresponded transgene integration and also the transgene loci numbers varied for different events (Fig. 4c).
Molecular analysis of the putative transformed B. juncea. a PCR analysis with fusion gene specific primers. Lane M, shows 1 kb Ladder; Lane NTC, shows negative control; Lane P, shows positive control; Line 1–Line 5, shows transformed plant lines. b PCR analysis with HptII gene specific primers. Lane M shows 1 kb Ladder, Lane NTC shows negative control; Lane P shows positive control; Line 1–Line 5, shows transformed plant lines. c Southern blot hybridization analyses of putative transformed plants. Genomic DNA was extracted from PCR positive plants and digested with EcoRI and the restricted product of fusion gene construct was used as a probe to hybridized and produce fragments for integrated T-DNA representing the transgene; Lane P shows positive control, Lane NTC shows negative control, Line 1–Line 6, shows independent transgenic lines
For confirmation of the presence of gene transcript, total RNA was extracted from the young leaves of transformants and NTC plants by Tri-reagent method. The cDNA was prepared from the isolated RNA by two-step approach. Molecular analysis through semi-quantitative RT-PCR showed the presence of fusion gene in transformants (Fig. 5). It appears that the single copy nature of transgenes and low sequence homology between lectin and protease inhibitor genes have contributed to the prevention of gene silencing in pyramided transgenic plants under the control of phloem-specific rolC promoter (Dutta et al. 2005; Nagadhara et al. 2003).
Fusion gene expression in putative transformed B. juncea plants. a RT-PCR amplification of fusion gene, Lane M shows 1 kb DNA ladder; Lane NTC shows control plants; Line 1–Line 6 shows, fusion gene expression in transformed plants. b RT-PCR amplification of actin gene, Lane M shows 1 kb DNA ladder; Lane NTC shows control plants; Line 1–Line 6 shows, actin expression in transformed plants
In the six transgenic events, up-regulation of normalized fold expression in transcript level of fusion gene varied. However, in the third transgenic line the maximum 2.8-fold induction was observed, which revealed up-regulation of both transgenes distinctly (Fig. 6). Whereas the line 1 and line 6 was showed minimal fold up-regulation of transgenes, i.e., 1.8 and 2.0-fold. A resemblance in the expression pattern was also observed in semi-quantitative RT-PCR which validated the transcript expression data. The transgenic lines observed with differentially over-expressed transgenes were propagated further for subsequent analyses in glass house.
The detached leaf bioassay was also performed to study the multi-toxin activity of fusion gene expressed in transgenic lines. The aphid’s survival as well as leaf damage cause was monitored at regular interval of 24 h on fusion transformed plants under phloem-specific rolC and control NTC plants. To avoid the redundant expression, phloem-specific rolC promoter was used which is highly desirable and reduced the expose probability of non-target organism directly to fusion protein. The mean percentage of aphid survival declined slowly to approximately 45% on fusion gene transformants as compared to control plants where the aphid survival was around 80% as shown in Fig. 7a. Similar type of findings was observed by Hilder et al. (1995a, b) who reported the reduction in aphid survival on GNA expressing rice transgenic plants by 8.3% compared to control plants. The damage caused by the attack of aphids on the leaves of transformed as well as non-transformed plants was measured by Leaf Area Meter. The extent of leaf damage was much more (~30%) in control plant on 9 days as compared to transgenic plants (line 1, line 2 and line 3) as shown in Fig. 7b (Each bar represents mean data ± standard error). There are reports which show that GNA when expressed under RSs1 promoter (rice sucrose synthase gene) in wheat led to decrement of grain aphids by 30% as compared to control plant (Stoger et al. 1999). Bonning et al. (2014) fused two genes (CP-P and Hva1) and fusion protein product was isolated from rice transgenic plants, it was found toxic to four homopteran species. In the near future, we expect the additional fusion genes will be expressed under phloem-specific rolC promoter which will be evaluated for their exploit in various other crops, which will greatly speed up in providing new insights in multi-defense genes. The pyramided transgenic plants with durable resistance emerged more promising in the cultivation area, which is highly prone for the sucking pest.
Aphids bioassay of fusion gene transformed B. juncea plants and NTC plant. a The graph shows the % survival of mustard aphids on control and transformed plants. b The graph depicts the leaf area recorded at regular intervals of 0, 3, 6 and 9 days. Data represent mean ± SD of three biological replicates (P value <0.01)
The transformed lines were compared with the non-transformed control plants (NTC) as well as wild types (WT) for their agronomic performance; and different phenotypical and morphological personas were well-thought-out and evaluated. However, all the transformed lines and NTC control plants showed significantly parallel morphologies at all the stages under study in terms of plant height, seed/pod, number of branches/plant, yield/plant (gram) and no significant (P < 0.05) variation was observed in the mentioned parameters (Table 2).
From this study, we conclude that the pyramiding of two different plant-based genes lectin and protease inhibitor into B. juncea is promising as it has increased resistance against major phytophagous sap-sucking insect pests and might serve as novel genetic resource. We also conclude that the reported protocol in the manuscript with transformation efficiency ranging from 16.4 to 19.7% can be utilized for genetic transformation of B. juncea. The development of such transgenic with plant insecticidal genes which are otherwise difficult to transfer through conventional breeding, will be very helpful in minimizing the losses by the sap-sucking insects and enhancing the crop productivity.
Abbreviations
- LL:
-
Lentil lectin gene
- CPPI:
-
Chickpea protease inhibitor gene
- ORF:
-
Open reading frame
- CCM:
-
Co-cultivation medium
- PCM:
-
Pre-cultivation medium
- SRM:
-
Shoot regeneration and growth medium
- SEM:
-
Shoot elongation and growth medium
- SM:
-
Selection culture medium
- RM:
-
Rooting medium
- MS:
-
Murashige and Skoog culture medium
- NTC:
-
Non-transgenic control
- WT:
-
Wild type
References
Bandyopadhyay S, Roy A, Das S (2001) Binding of garlic (Allium sativum) leaf lectin to the gut receptors of homopteran pest is correlated to its insecticidal activity. Plant Sci 161:1025–1033. doi:10.1016/S0168-9452(01)00507-6
Bharathi Y, Vijaya KS, Pasalu IC, Balachandran SM, Reddy VD, Rao KV (2011) Pyramided rice lines harbouring Allium sativum (ASAL) and Galanthus nivalis (GNA) lectin genes impart enhanced resistance against major sap-sucking pests. J Biotechnol 152:63–71. doi:10.1016/j.jbiotec.2011.01.021
Bhuiyan MS, Min SR, Jeong WJ, Sultana S, Choi KS, Lim YP, Song WY, Lee Y, Liu JR (2011) An improved method for Agrobacterium-mediated genetic transformation from cotyledon explants of Brassica juncea. Plant Biotechnology. 28:17–23. doi:10.5511/plantbiotechnology.10.0921a
Birk Y (2003) Plant protease inhibitors: significance in nutrition, plant protection, cancer prevention and genetic engineering. Springer Science and Business Media, Germany
Bonning BC, Pal N, Liu S, Wang Z, Sivakumar S, Dixon PM, King GF, Miller WA (2014) Toxin delivery by the coat protein of an aphid-vectored plant virus provides plant resistance to aphids. Nat Biotechnol 32:102–105. doi:10.1038/nbt.2753
Broadway RM (2000) The adaptation of insects to protease inhibitors. In: Michaud D (ed) Recombinant Protease Inhibitors in Plants. Landes Bioscience, Georgetown, pp 80–88
Daun X, Li X, Xue Q, Abo-El-Saad M, Xu D, Wu R (1996) Transgenic rice plants harbouring an introduced potato proteinase inhibitor II gene are insect resistant. Nat Biotechnol 14:494–498. doi:10.1038/nbt0496-494
Dutta I, Majumder P, Saha P, Ray K, Das S (2005) Constitutive and phloem specific expression of Allium sativum leaf agglutinin (ASAL) to engineer aphid (Lipaphiserysimi) resistance in transgenic Indian mustard (Brassica juncea). Plant Sci 169:996–1007. doi:10.1016/j.plantsci.2005.05.016
Gatehouse AMR, Down RE, Powell KS, Sauvion N, Rahbe Y, Newell CA, Merryweather A, Hamilton WD, Gatehouse JA (1996) Transgenic potato plants with resistance to the peach potato aphids Myzuspersicae. Entomol Exp Appl 79:295–307. doi:10.1111/j.1570-7458.1996.tb00837.x
Habib H, Fazili KM (2007) Plant protease inhibitors: a defense strategy in plants. Biotechnology and Molecular Biology Reviews 2:068–085
Haq SK, Atif SM, Khan RH (2004) Protein proteinase inhibitor genes in combat against insects, pests, and pathogens: natural engineered phytoprotection. Arch Biochem Biophys 431:145–159. doi:10.1016/j.abb.2004.07.022
Hilder VA, Powell KS, Gatehouse A, Gatehouse J, Gatehouse LN, Shi Y, Hamilton W, Merry Whether A, Newell CA, Timens JC (1995a) Expression of snowdrop lectin in transgenic tobacco plants results in added protection against aphids. Transgenic Res 4:18–25. doi:10.1007/BF01976497
Hilder VA, Powell KS, Gatehouse AM, Gatehouse JA, Gatehouse LN, Shi Y, Hamilton WD, Newell CA, Merryweather A, Timans JC (1995b) Expression of snowdrop lectin in transgenic tobacco plants result in added protection against aphids. Transgenic Res 4:18–25. doi:10.1007/BF01976497
Hossain MA, Maiti MK, Basu A, Sen S, Ghosh AK, Sen SK (2006) Transgenic Expression of Onion leaf lectin gene in Indian mustard offers protection against colonization. Cro Sci 46:2022–2032. doi:10.2135/cropsci2005.11.0418
Jouanin L, Bonade-Bottino M, Girard C, Marrot G, Giband M (1998) Transgenic plants for insect resistance. Plant Sci 131:1–11. doi:10.1016/S0168-9452(97)00239-2
Kumar PR (1999) Rapeseed mustard research in India: 21st century strategies. 10th International Rapeseed Congress, Canberra, Australia
Kumar A, Shauhan JS (2005) Status and future thrust areas of rapeseed-mustard research in India. Indian Journ Agric Sci 75:621–635
Majumder P, Banerjee S, Das S (2004) Identification of receptors responsible for binding of the mannose specific lectin to the gut epithelial membrane of the target insects. Glycoconjugate J 20:525–530. doi:10.1023/B:GLYC.0000043288.72051.7c
Manyangarirwa W, Turnbull M, McCutcheon GS, Smith JP (2006) Gene pyramiding as a Bt resistance management strategy: how sustainable is this strategy? Afr J Biotech 5(10):781–785. doi:10.4314/ajb.v5i10.42833
Maqbool SB, Riazuddin S, Loc NT, Gatehouse AMR, Gatehouse JA, Christou P (2001) Expression of multiple insecticidal genes confers broad resistance against a range of different rice pests. Mol Breed 7:85–93. doi:10.1023/A:1009644712157
Mehlo L, Gahakwa D, Nghia PT, Loc NT, Capell T, Gatehouse JA, Gatehouse AMR, Christou P (2005) An alternative strategy for sustainable pest resistance in genetically enhanced crops. Proc Natl Acad Sci 102:7812–7816. doi:10.1073/pnas.0502871102
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tobacco cultures. Physiol Plant 15:473–497. doi:10.1111/j.1399-3054.1962.tb08052.x
Murrey MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326. doi:10.1093/nar/8.19.4321
Nagadhara D, Ramesh S, Pasalu IC, Rao YK, Krishnaiah NV, Sarma NP, Bown DP, Gatehouse JA, Reddy VD, Rao KV (2003) Transgenic indica rice resistant to sap-sucking insects. Plant Biotechnol J 1:231–240. doi:10.1046/j.1467-7652.2003.00022.x
Patel SR, Awasthi A, Tomar RKS (2004) Assessment of yield losses in mustard (Brassica juncea L.) due to mustard aphids (Lipaphis erysimi Kalt.) under different thermal environment in eastern central India. App Ecol Environ Res 2:1–5. doi:10.15666/aeer/02001015
Pental D, Pradhan AK, Sodhi YS, Mukhopadhyay A (1993) Variation amongst Brassica juncea cultivars for regeneration from hypocotyl explants and optimization of conditions for Agrobacterium-mediated genetic transformation. Plant Cell Rep 12:462–467. doi:10.1007/BF00234713
Pradhan AK, Sodhi YS, Mukhopadhay A, Pental D (1993) Heterosis breeding in Indian Mustard (Brassica juncea L. Czern and Coss): analysis of components characters contributing to heterosis for yield. Euphytica 69:219–229. doi:10.1007/BF00022368
Rao KV, Rathore KS, Hodges TK, Fu X, Stoger E, Sudhakar D, Williams S, Christou P, Bharathi M, Bown DP, Powell KS, Spence J, Gatehouse AM, Gatehouse JA (1998) Expression of snowdrop lectin (GNA) in transgenic rice plants confers resistance to rice brown plant hopper. Plant J 15:469–477. doi:10.1046/j.1365-313X.1998.00226.x
Rogers BL, Pollock J, Klapper DG, Griffith IJ (1993) Sequence of the proteinase inhibitor cystatin homologue from the pollen of Ambrosia artemisiifolia (short ragweed). Gene 133:219–221. doi:10.1016/0378-1119(93)90641-F
Roush RT (1998) Two-toxin strategies for management of insecticidal transgenic crops: can pyramiding succeed where pesticide mixtures have not? Philos Trans R Soc Lond B Biol Sci 353:1777–1786. doi:10.1098/rstb.1998.0330
Sambrook J, Russell DW (2001) Molecular cloning. A Laboratory Manual. 3rd edn. Cold Spring Harbor, New York
Senthil Kumar R, Cheng CP, Yeh KW (2010) Genetically pyramiding protease-inhibitor genes for dual broad-spectrum resistance against insect and phytopathogens in transgenic tobacco. Plant Biotechnol J 8:65–75. doi:10.1111/j.1467-7652.2009.00466.x
Sharma VK, Hänsch R, Mendel RR, Schulze J (2004) A highly efficient plant regeneration system through multiple shoot differentiation from commercial cultivars of barley (Hordeum vulgare L.) using meristematic shoot segments excised from germinated mature embryos. Plant Cell Rep 23:9–16. doi:10.1007/s00299-004-0800-4
Singh R, Tiwari IM, Kansal R, Saini R, Koundal KR (2010) An efficient Agrobacterium-mediated genetic transformation method of Brassica juncea (L.) Czern & Coss cv. Varuna with a lectin gene of moth bean. J Plant Biol 37:123–130
Stoger E, Williams S, Christou P, Down RE, Gatehouse JA (1999) Expression of the insecticidal lectin from snowdrop (Galanthus nivalis agglutinin; GNA) in transgenic wheat plants: effects on predation by the grain aphid Sitobion avenae. Mol Breed 5:65–73. doi:10.1023/A:1009616413886
Visser RG, Jacobsen E, Hesseling-Meinders A, Schans MJ, Witholt B, Feenstra WJ (1989) Transformation of homozygous diploid potato with an Agrobacterium tumefaciens binary vector system by adventitious shoot regeneration on leaf and stem segments. Plant Mol Biol 12:329–337. doi:10.1007/BF00043210
Yadav JS, Singh NB (1999) In: Proceedings of the 10th international rapeseed congress, Canberra, Australia
Zhao JZ, Cao J, Li Y, Collins HL, Roush R, Earle ED, Shelton AM (2003) Transgenic plants expressing two Bacillus thuringiensis toxins delay insect resistance evolution. Nat Biotechnol 21:1493–1497. doi:10.1038/nbt907
Zhu-Salzman K, Shade RE, Koiwa H, Salzman R, Narasimhan M, Bressan IA, Hasegawa PM, Murdock L (1998) Carbohydrate binding and resistance to proteolysis control activity of Grifinia simplicifolia Lectin II. Proc Natl Acad Sci USA 95:15123–15128
Acknowledgements
We are grateful to Council of Scientific and Industrial Research (CSIR), New Delhi, India, for providing the financial assistance to Prof. K.R. Koundal under Emeritus Scientist scheme to perform this research work. The authors are sincerely thankful to Dr. P. Anand Kumar, former Project Director of National Research Centre on Plant Biotechnology (NRCPB), New Delhi, India for his kind support.
Author information
Authors and Affiliations
Contributions
RK and RCB, conceived and designed the research. SR, performed experiments and wrote the manuscript. AH, analyzed the experimental data and helped in experimental and writing part. VS, contributed to part of conception and finalized the revised manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The author(s) declare that they have no competing interests.
Additional information
Communicated by J.-H. Liu.
Rights and permissions
About this article
Cite this article
Rani, S., Sharma, V., Hada, A. et al. Fusion gene construct preparation with lectin and protease inhibitor genes against aphids and efficient genetic transformation of Brassica juncea using cotyledons explants. Acta Physiol Plant 39, 115 (2017). https://doi.org/10.1007/s11738-017-2415-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-017-2415-8