Abstract
MYB genes are extensively distributed in higher plants and constitute one of the largest transcription factors (TFs) families. These TFs have been proved to be implicated in the regulation of plant growth, development, metabolism, and multiple abiotic stress responses. In the present study, a new soybean MYB gene, denoted GmMYBJ2, was isolated and its function was characterized. The GmMYBJ2 cDNA is 1428 bp in length with an open reading frame (ORF) of 960 bp encoding 319 amino acids. Sequence and yeast one-hybrid analyses showed GmMYBJ2 contains two MYB domains and belongs to R2R3-MYB protein with transactivation activity. Transient expression analysis using the GmMYBJ2-GFP fusion gene in onion epidermal cells showed GmMYBJ2 protein is targeted to the nucleus. GmMYBJ2 was induced by drought, cold, salt, and exogenous abscisic acid (ABA). Arabidopsis overexpressing GmMYBJ2 exhibited a higher seed germination rates (GRs), a notable increase in the soluble sugar content under water-deficit stress, and a lower water loss rate (WLR) when water is sufficient. These results indicated the overexpression of GmMYBJ2 make transgenic Arabidopsis more tolerant to drought stress than wild-type (WT) plants, and GmMYBJ2 may be useful for improving drought stress tolerance in transgenic plant breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants are frequently confronted with adverse environmental conditions, for instance, drought, salt, and extreme temperature, which may seriously affect their growth and development. These adverse conditions have always resulted in decreased crop yields and economic losses. Therefore, cultivating the plant species tolerant to abiotic stresses has become a main objective of many crop breeders. In recent years, overexpression or knockdown of some key genes that mediate plant responses to environmental stress (e.g., drought stress) provides an effective method to enhance tolerance to water-deficit stress (Valliyodan and Nguyen 2006). The regulatory proteins translated from these pivotal genes often act as responders to various signals, resulting in expression modifications of stress-related genes. Moreover, the function of regulatory proteins can also be amplified and strengthened via some signal transduction cascades. Thus, some researchers working on plant stress response have focused on regulatory proteins (Kreps et al. 2002; Ahuja et al. 2010). TFs are pivotal regulatory proteins regulating its downstream target genes, and it has been proven that some of these TFs are implicated in stress responses in plants (Hu et al. 2008). In terms of differences in their DNA-binding domains, these TFs can be divided into diverse families, for instance, MYB, NAC, AP2, and WRKY (Dubouzet et al. 2003; Mare et al. 2004; Song et al. 2011).
In plants, MYB proteins comprise a large, functionally various family of TFs (Riechmann et al. 2000). These proteins have a structurally conserved DNA-binding domain, namely the MYB domain, which is made up of approximately 53 amino acid residues, each of which often forms a helix-turn-helix structure. In terms of the number of imperfect repeats (one, two, three, or four) in their MYB domain, MYB proteins can be classified into different subfamilies, such as MYB-related (with a partial or a single MYB repeat), R2R3-MYB (2R-MYB), R1R2R3-MYB (3R-MYB), and 4R-MYB (Dubos et al. 2010). MYB-related proteins are implicated in the regulation of cellular morphogenesis (Simon et al. 2007; Pesch and Hulskamp 2009) and secondary metabolism (Dubos et al. 2008; Matsui et al. 2008). Genes encoding 3R-MYB proteins have been observed in most eukaryotic genomes and participate in cell cycle control (Ito 2005; Haga et al. 2007). The 4R-MYB group is the smallest class, and in contrast with other groups, very little is known regarding the functions of these proteins in plants. It is worth noting that a majority of plant MYB genes encode 2R-MYB proteins and are confirmed to be concerned in cell fate and identity, regulation of primary and secondary metabolism, developmental processes, and especially responses to various stresses (Du et al. 2009). For instance, AtMYB2 plays a role in drought stress response in an ABA-dependent manner (Abe et al. 2003). Arabidopsis AtMYB41 is induced by drought, ABA and salt, and presents up-regulated expression pattern (Cominelli et al. 2008; Lippold et al. 2009). Overexpressed OsMYB4 has been demonstrated that it could enhance Arabidopsis tolerance to freezing stress (Vannini et al. 2004). TaMYB33, a novel member of MYB TF family from wheat, improves Arabidopsis tolerance to drought and salt stresses (Qin et al. 2012).
Soybean [Glycine max (L.)] is a vital crop that provides humans with abundant proteins and vegetable oil. However, with the rapid development of society and global climate change, its growth and development are increasingly affected by diverse environmental stresses. In soybean, some genes, whose expressions were induced by various stresses, have been isolated and their functions have been discussed (He et al. 2002; Luo et al. 2005; Wang et al. 2005; Liao et al. 2008a, b; Zhai et al. 2012, 2013). Some soybean MYB genes have been reported. For example, the expression of GmMYB76, GmMYB92, and GmMYB177 is induced by NaCl, cold, or drought whereas not by ABA treatment (Liao et al. 2008b). Li et al. (2013) reported that GmMYB12B2 exhibits transcriptional activity and is involved in plant flavonoid biosynthesis. Su et al. (2014) reported that Arabidopsis overexpressing GmMYBJ1 exhibited high resistance to drought and cold stresses. There are 252 MYB-encoding genes in soybean genome, including 244 typical 2R-, six 3R-, and two 4R-like MYB proteins (Du et al. 2012). Nevertheless, in contrast with other plant species, few soybean MYB genes participating in environmental stresses have been characterized. An in silico analysis looking for the Arabidopsis gene homologous to soybean is an effective way to predict gene function, and can provide extensive information regarding the gene which will be cloned. Based on in silico analysis, a soybean R2R3-MYB gene, which is designated GmMYBJ2, was isolated from soybean in this study, and its role in response to drought stress was characterized in Arabidopsis through the generation of transgenic plants overexpressing GmMYBJ2.
Materials and methods
Isolation and sequence analysis of GmMYBJ2 from soybean
Through screening the gene expression profiles of soybean Jilin32, together with the NCBI blast tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi), we found a cDNA clone exhibiting high homology with other plant MYB genes from the differently expressed genes. Total RNA was extracted from soybean Jilin32 and cDNA was synthesized using PrimeScript™ 1st Strand cDNA Synthesis Kit (Takara, China). Subsequently, reverse transcription polymerase chain reaction (RT-PCR) was carried out to obtain this clone. The MYB gene cloned was designated GmMYBJ2 (GenBank accession number: KC759158). The primer pairs used for the isolation of GmMYBJ2 were 5′-GAACCTTGTCTCTGCTTCACCA-3′ (forward) and 5′-CACACACGCACGCATACTCTTT-3′ (reverse). The specific PCR product was inserted into the pMD18-T cloning vector (Takara, China) and sequenced for its accuracy. The conserved MYB domain of the GmMYBJ2 protein was analyzed using online SMART software (Letunic et al. 2006), and phylogenetic analysis was performed using the Neighbor-Joining (NJ) algorithm of MEGA 5.1 (Tamura et al. 2011).
Plant materials and stress treatments
For soybean seedlings treatment, about 24 plants (~8 plants per pot, each pot is considered as an independent repeat) for each treatment were used as starting material. Seedlings of the soybean cultivar Jilin32 were cultivated in Hoagland’s solution under 16-h light/8-h dark cycles at 25 °C and 70 % humidity for 2 weeks (four-leaf stage) and then maintained in same solution with or without 150 mM NaCl, 100 μM ABA and 10 % polyethylene glycol (PEG) 8000. Regarding cold treatment, the seedlings were kept in an incubator at 4 °C. For sample preparation, ~0.2 g plant leaves were randomly collected from each pot (~8 plants) and then pooled as one sample. The collection time were 0, 1, 3, 6, 9, 12, and 24 h (0 h was the non-treated control) from the initiation of the treatments and immediately frozen in liquid nitrogen for total RNA extraction.
For transgenic Arabidopsis treatment, transgenic seeds from Arabidopsis plants were sterilized, planted in MS medium supplemented with different concentrations (treatment) of mannitol and incubated under normal conditions for germination. The germination rates (GRs) were scored on the 6th day after sowing. All of the treatments were repeated three times, and the data were assessed from the results of three independent experiments. For dehydration treatment in soil, the WT and transgenic plants were exposed to stress for 19 days by withholding water, and the phenotype was observed.
Quantitative RT-PCR (qRT-PCR) analysis
Total RNA from soybean leaves was extracted using RNAiso Plus (Takara, China) in accordance with the manufacturer’s instruction, and converted to cDNA using an M-MLV kit (Takara, China). The qRT-PCR system and reaction program were carried out following the protocol described in Su et al. (2014). Each reaction was composed of 10 μL of SYBR Green I, 2 μL of the cDNA (~25 ng/μL) samples, 0.4 μL of ROX reference Dye II, 0.4 μL of 10 μM gene-specific primers, and finally added distilled water to a total volume of 20 μL. The primer pairs 5′-TCGGTTCCCACTAATACAGGTTT-3′ (forward) and 5′-ATAGTTGGTCCATCTGAGTCTGC-3′ (reverse) were used for GmMYBJ2 amplification. β-tubulin (GenBank accession number: GMU12286) was amplified and used as an internal reference to standardize the data. The fold changes were determined using the 2−ΔΔCt method (Livak and Schmittgen 2001). All qRT-PCR reactions were run in triplicate.
Subcellular localization and transactivation assay of GmMYBJ2
The GFP fusion vector was constructed as described in Su et al. (2014) to generate pBI121-GmMYBJ2-GFP construct controlled by CaMV 35S promoter. The recombinant construct was then introduced into A. tumefaciens strain EHA105 and subsequently transformed into onion epidermal cells. After 24 h of incubation at 25 °C in the dark, the GFP fluorescence was detected under a confocal microscope (Olympus, Japan). To explore the transactivation activity, the full-length ORF of GmMYBJ2 was cloned into pGBKT7 vector (Invitrogen), which was in advance digested with the Eco RI and Sal I restriction enzymes. The positive plasmid pGBKT7-GmMYBJ2 was subsequently transformed into yeast AH109 strain, and the transformants were selected on SD medium lacking Trp (SD/-Trp) at 30 °C for 3 days. After 2 days of culture at 30 °C, the yeast colonies were shifted from SD/-Trp medium to SD/-Trp-His-Ade medium, which 10 mM 3-amino-1, 2, 4-triazole (3-AT) was included. Furthermore, the β-galactosidase assay was conducted for examination of transactivation ability within 8 h.
Plasmid construction and Arabidopsis transformation
The GATEWAY clone technology was used to construct a plant expression vector based on the detailed method described by Karimi et al. (2002) and Su et al. (2014). The pCB35SR1R2-GFP-GmMYBJ2 resultant was electroporated into A. tumefaciens EHA105 and the method transforming Arabidopsis Col-0 was in accordance with the description of Clough and Bent (1998). The transformed Arabidopsis plants were selected on MS medium containing 4 mg L−1 glufosinate-ammonium (Sigma). The homozygous T3 progeny was confirmed by PCR and RT-PCR prior to further analysis.
Determination of excised-leaf water loss rate (WLR) and soluble sugar contents
Leaves from WT and overexpressed plants were detached at the rosette stage and weighed immediately at room temperature to obtain the initial weight (~0.5 g). Subsequently, the weight of the excised leaf was measured and recorded at different time points. Three replicates were used for each transgenic line and WT. The total soluble sugar contents were measured following the methods described by Bailey (1958), Song et al. (2011) and Su et al. (2014).
Analysis of genes regulated by GmMYBJ2 using qRT-PCR
Total RNAs from WT and transgenic Arabidopsis were extracted as mentioned above. The gene-specific primers for qRT-PCR are listed in Supplementary Table S1. The Arabidopsis actin gene (GenBank accession number: NM_112764) was chosen as an internal control.
Statistical analysis
Statistical analyses were performed using the SPSS 13.0 program. The differences were considered significant at P < 0.05 or P < 0.01.
Results
Isolation and sequence analysis of GmMYBJ2
Soybean Jilin32 is an improved variety with better tolerance to stress (e.g., drought and salt) and better disease resistance (Fu 1995). Based on an analysis of the gene expression profiles of soybean Jilin32 immature embryos (20, 30, and 50 d), we found six unknown cDNA clones that were homologous to other plant MYB TFs from the differentially expressed genes. These clones contained the full-length ORF, and their predicted translation product contained two conserved MYB domains. qRT-PCR was conducted to determine the expression pattern of these GmMYBs in response to abiotic stresses (not published). One of these showed significant modifications under abiotic stresses, and this gene was designated GmMYBJ2. Based on this finding and the results of phylogenetic analyses, this gene was picked for further functional analysis. The ORF of GmMYBJ2 was 960 bp in length and encodes a protein with 319 amino acids with a calculated molecular mass of 36.09 kDa and a theoretical pI of 5.72. The alignment results between cDNA and soybean GmGDB database (http://www.plantgdb.org/GmGDB/cgi-bin/blastGDB.pl) demonstrated that GmMYBJ2 contains two introns and is located on chromosome 4. The predicted GmMYBJ2 protein contains two conserved MYB domains, as determined by SMART analysis (Fig. 1), and the phylogenetic analysis revealed that it clustered with Arabidopsis AtMYB60, which belongs to the MYB protein family and participates in plant drought tolerance (Cominelli et al. 2005) (Fig. 2).
Alignments of the deduced amino acids of GmMYBJ2 with those of other plant MYBs: TaMYB2 (AAT37168.1), TaMYB73 (AEW23186), AtMYB13 (NP_172108), AtMYB17 (XP_002876601), GmMYB92 (ABH02844.1), AtMYB60 (NP_973795.1), GmMYBJ2 (AGN96216.1), CpMYB10 (AAM43912.1), GmMYB76 (ABH02836.1), AtMYB18 (XP_002869667), and AtMYB101 (AEC08688.1)
GmMYBJ2 is located in the nucleus and has transactivation activity in yeast
The transient expression assay using onion epidermis cells showed that the GmMYBJ2-GFP fusion protein was specifically localized in the nucleus, whereas the GFP fluorescence was visualized in the whole cells transformed with the GFP control vector (Fig. 3). MYB proteins usually play roles as a transcriptional activator or repressor. To verify our presumption, a yeast assay system was used for the analysis. The transactivation assay showed that all of the transformants could grow well on the selective SD/-Trp medium (Fig. 4a). However, only the transformants of pGBKT7 fused with the full-length ORF of GmMYBJ2 could grow normally on the selective SD/-Trp/-Ade/-His medium and exhibited β-galactosidase activity when X-gal was added to the Whatman filter paper (Fig. 4b, c). The results confirmed that GmMYBJ2 is a transcriptional activator that activates the transcription of the reporter genes Ade, His and LacZ.
Transactivation analysis of GmMYBJ2 in the yeast AH109 strain. The fusion protein of the GAL4 DNA-binding domain and GmMYBJ2 were expressed. The empty pGBKT7 vector was expressed as a control. a Growth of the transformants on an SD plate lacking tryptophan (SD/-Trp plus 10 mM 3-AT). b Growth of the transformants on an SD plate lacking tryptophan, histidine and adenine (SD/-Trp-Ade-His plus 10 mM 3-AT). c β-galactosidase activity assay
Expression patterns of GmMYBJ2 under abiotic stress
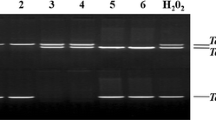
To further elucidate the role of GmMYBJ2 in plants, its expression patterns in soybean seedlings treated with drought, salt, cold stresses, and ABA were monitored by qRT-PCR (Fig. 5), and photographs of the soybean seeds and non-treated seedlings are shown in Supplementary Fig. S1. A clear increase in the GmMYBJ2 transcript was observed at 6 and 9 h after initiation of PEG treatment, and the transcript then rapidly decreased. A similar trend was observed with ABA treatment. For cold and salt treatments, the transcripts showed a gradual increase, and both peaked 24 h after initiation of the treatments. GmMYBJ2 was induced by salt, ABA, cold, and drought, but the transcripts increased more obviously under PEG treatment to levels nearly 16-fold higher compared with those observed in the control.
Monitoring of seed germination and the changes in physiological parameters of GmMYBJ2-overexpressing Arabidopsis under stress conditions
Due to the marked change in the expression of GmMYBJ2 under drought stress, we inferred that this gene may be implicated in drought tolerance in plants. Four transgenic lines, which were positive plants, as verified by PCR and RT-PCR, were used in these experiments. Transgenic seeds harvested from overexpressed GmMYBJ2 and WT plants were sowed on MS medium supplemented with various concentrations of mannitol for germination, and the GRs were measured (Fig. 6). The seeds from all of the transgenic lines displayed higher GRs in contrast with the WT when the mannitol concentration was 200 and 300 mM. For instance, the WT seed GRs were approximately 73.85 and 53.85 % with mannitol concentrations of 200 and 300 mM, respectively, whereas approximately 86 and 78.5 % of the transgenic seeds could germinate at the corresponding mannitol concentrations. The seedlings in normal MS medium were then transferred into soil for normal growth (Fig. 7a). After 7 days, all of the plants were subjected to water withholding for 19 days. The results indicated that the WT plants presented smaller leaves with obvious etiolation and wilting, whereas the transgenic line showed better growth (Fig. 7b).
Comparison of the germination rates of transgenic seeds under drought stress. BJ2-1, BJ2-5, BJ2-6, and BJ2-7 represent the transgenic lines. The data represent the average of three independent experiments ± SD. * and ** indicate significant (P < 0.05) and highly significant differences (P < 0.01) between the transgenic lines and the WT
Effects of drought stress on the transgenic plants overexpressing GmMYBJ2. a Phenotypic comparison of plant growth before treatment. b Phenotypic comparison of plants a after 7 days of normal growth followed by water withholding for 19 days. Col-0 indicates the WT plants, and TL1 and TL2 represent the transgenic lines
The physiological traits of plants are often altered under stress conditions. To explore the physiological mechanism when plants were under stress conditions, the changes in water loss and soluble sugar contents in overexpressed lines and WT were assessed. The fresh weights of the detached leaves from transgenic and WT plants were used for assessment of the water loss at the designated time points. The four transgenic lines displayed lower WLR than the WT plants within 10 h after leaf detachment (Fig. 8). Soluble sugar is an osmotic regulation substance. Alterations in the soluble sugar content are reported to be connected with the plant response to unfavorable conditions, and its accumulation is considered as a very important adaptive mechanism for coping with stress. No significant difference was observed in soluble sugar content between the transgenic and WT plants when plants were normally incubated. However, its content in the transgenic plants displayed obviously higher levels than that in the WT plants after drought stress treatment (Fig. 9).
Soluble sugar content in the GmMYBJ2-overexpressing transgenic plants, and the transgenic lines BJ2-1, BJ2-5, BJ2-6, and BJ2-7 were used for the analysis. The data represent the means from three independent tests ± SD. ** indicates that the differences between the transgenic lines and WT are highly significant (P < 0.01)
GmMYBJ2 altered the expression of abiotic stress-responsive genes
The overexpression of GmMYBJ2 enhanced plant resistance to drought stress. To further elucidate the molecular mechanism, the expression levels of six downstream stress-responsive genes, which were chosen according to Liao et al. (2008a) and Qin et al. (2012), in GmMYBJ2-overexpressed lines grown under non-stressed conditions were examined by qRT-PCR. In contrast with WT plants, ERD10, DREB2A, COR15a, RD29A, and RD29B were shown to be up-regulated to varying degrees in transgenic plants (Fig. 10). Among these genes, the expression level of RD29B increased by approximately 14-fold in the BJ2-1 transgenic line compared with the WT plant and by almost six-fold in the other three lines. However, no obvious expression changes in RD17 were observed.
Expression levels of stress-responsive genes in the WT and GmMYBJ2 transgenic plants under normal growth conditions, and BJ2-1, BJ2-5, BJ2-6, and BJ2-7 represent the transgenic lines. The data represent the means of three replicates. The error bars indicate the SD. * and ** indicate significant (P < 0.05) and highly significant differences (P < 0.01) between the transgenic lines and the WT
Discussion
TFs play a very important role in regulating gene expression when plants suffer from adverse conditions. The plant MYB family is large and these regulatory proteins are frequently exhibits multiple functions (Dubos et al. 2010). Recent studies on MYB genes in rice, wheat, Arabidopsis, and other plants have demonstrated that numerous MYB proteins are implicated in stress response (He et al. 2012; Qin et al. 2012; Yang et al. 2012). In the present study, a new member of the soybean MYB gene family, denoted GmMYBJ2, was isolated, and its function was analyzed.
Sequence analysis using some databases and software packages is a primary and effective way to predict gene function. Analyses of the amino acid sequence and phylogenetic tree of GmMYBJ2 showed that it contains two highly conserved MYB domains and clusters with R2R3-MYB proteins from other plants (Figs. 1, 2), indicating that GmMYBJ2 is an R2R3-MYB TF. A majority of MYB proteins are presumed to be transcriptional activators with activation domains (ADs) in the C-terminal region (Weston 1998). It is well-known that N-terminal region of MYB proteins are conserved, however, the sequences in the C termini are generally not, and not all MYB TFs function as activators in the nucleus. The sub-localization of GmMYBJ2 showed that it is a nucleus-localized protein (Fig. 3), which is in agreement with its role as a TF. β-galactosidase and transactivation assays using a yeast one-hybrid system pointed that it has transactivation activity and may act as an activator in the yeast assay system (Fig. 4). This observation needs to be further examined in a plant system.
In plants, such as Arabidopsis, wheat and rice, many MYB TFs have been depicted to taken part in stress response. AmMYB1, a single-repeat MYB TF, is powerfully induced by diverse stresses, such as salt, light and ABA, and confers NaCl tolerance to tobacco (Ganesan et al. 2012). He et al. (2012) described that TaMYB73 is induced by NaCl, dehydration and several phytohormones and that ectopic expression of TaMYB73 improves plant resistance to salinity. Zhang et al. (2012) reported that TaMYB30-B is a PEG stress-induced gene, and overexpression of the gene uncovered TaMYB30-B protein could improve stress tolerance to drought during the germination and seedling stages. OsMYB2 is induced by cold, salt, drought, and ABA and enhances the tolerance of plants to salinity, chilling, and dehydration in rice (Yang et al. 2012). In soybean, Liao et al. (2008b) reported that GmMYB76, GmMYB92, and GmMYB177 was induced by salt, cold, or drought but not influenced by ABA treatment, and overexpressed Arabidopsis displayed higher resistance to salt and freezing. GmMYBJ1 was remarkably induced by cold and drought stresses (Su et al. 2014). The present study showed the MYB gene, GmMYBJ2, from soybean could be induced by diverse stresses, particularly drought, at the seedling stage (Fig. 5), inferring that GmMYBJ2 may play important roles in multiple signaling transduction pathways responsive to abiotic stress.
To further investigate the role of GmMYBJ2 in plant systems, overexpressed plants were generated. The seed GRs were compared between transgenic and WT seeds (Fig. 6). An increase in the mannitol concentration resulted in decreases in the GRs of WT and transgenic seeds, whereas a markedly lower GR was observed in the WT seeds. After 19 days of withholding water, the transgenic lines displayed better growth in comparison with WT plants (Fig. 7). These findings inferred that GmMYBJ2 is implicated in regulating the plant response to water-deficit stress and may be used for plant improvement. In addition, these results are also partly consistent with the function of AtMYB60, which clusters with GmMYBJ2 (Fig. 2). It has been reported that AtMYB60 exhibits guard-cell-specific expression and participates in the tolerance of plants to drought by regulating stomatal movements. Moreover, ABA usually participates in the process by strongly down-regulating the expression of AtMYB60 (Cominelli et al. 2005). In response to drought stress, stress response hormone ABA was usually accumulated, resulting in rapid closing of the stomata, to restrain water loss by transpiration (Cominelli et al. 2005; Gray 2005). Hamanishi et al. (2012) also found that drought exposure results in decreases in stomatal conductance and an alteration in stomatal development in Populus balsamifera, and some genes that could strengthen drought-responsive changes in stomatal development were found to be altered in poplar. Further studies are required to elucidate whether a similar mechanism occurs with GmMYBJ2, which regulates the stomatal movements of plants and consequently enhances their tolerance to drought. Previous studies have demonstrated that numerous genes responding to drought stress, including the MYB genes, could be induced by the application of exogenous ABA (Nakashima et al. 2009). Zhu et al. (2014) reported that the EsWAX1 gene from E. salsugineum, whose product is a 2R-MYB protein, is induced rapidly by exogenous ABA and drought stress. Overexpression of EsWAX1 improves Arabidopsis tolerance to drought. In the present study, GmMYBJ2 was also found to be induced rapidly by exogenous ABA and drought stress (Fig. 5); thus, we inferred that GmMYBJ2 may be positively correlated with the ABA signaling pathways and may regulate ABA-mediated gene expression when plants undergone environmental stress. More experiments will be required to verify our presumption in the future.
Various physiological responses can be induced in plants when suffering from drought stress (Seki et al. 2007). The WLR and soluble sugar content are considered as important indicators of drought resistance and are widely used in this research field (Dedio 1975; Vinocur and Altman 2005). In our present study, in contrast with WT, the WLR was found to be lower in the detached leaves from overexpressed lines (Fig. 8), which is in concert with the results reported for the soybean ERF, soybean MYB, wheat MYB and WRKY gene family (Zhang et al. 2012; Niu et al. 2012; Zhai et al. 2013; Zhu et al. 2014; Su et al. 2014). The results suggest that the transgenic plants may exhibit better water retention abilities. Soluble sugar is an important osmolyte for facilitating osmoregulation, thereby protecting plants from dehydration due to osmotic stress by reducing the cell water potential. Previous studies have confirmed the correlation between the existence of exceptional soluble sugars and the acquisition of stress tolerance (Kerepesi and Galiba 2000; Cominelli et al. 2005; Song et al. 2011; Zhang et al. 2012; Zhai et al. 2012; Su et al. 2014). In this study, more soluble sugars were accumulated in overexpressed lines compared with WT plants when drought stress was presented (Fig. 9), supporting the hypothesis that a greater accumulation of soluble sugar contributes to plant resistance to stress. The transactivation assay results showed that GmMYBJ2 may be an activator (Fig. 4). Some downstream genes, including AtERD10, AtDREB2A, AtCOR15a, AtRD29B, and AtRD29A, were activated and up-regulated to different levels in the transgenic plants (Fig. 10). It has been confirmed that these genes are implicated in multiple stress responses (Kang et al. 2002; Sakuma 2006). DREB2A, a TF containing AP2 domain, is induced by salt and osmotic stress. Overexpression of DREB2A could induce feeble expression of its downstream genes in the case of unstressed situations (Liu et al. 1998). DRE or related motifs are existed in the promoter regions of RD29A, RD17, and COR15a, and they are induced by cold, salt, and dehydration stresses (Narusaka et al. 2003; Simpson et al. 2003). The RD29B gene could slowly make response to dehydration stress and is probably induced by endogenous ABA produced under dehydration conditions (Yamaguchi-Shinozaki and Shinozaki 1993). Kiyosue et al. (1994) reported that ERD10 could make rapidly response to water-deficit stress in Arabidopsis, and its expression could be induced when ABA was applied. Thus, we reasoned that GmMYBJ2 may confer drought stress tolerance through the up-regulation of downstream genes. Additionally, some research indicated that overexpression of some genes in plant can cause severe growth retardation, a stunted phenotype or a never-produced seed (Dai et al. 2007; Onate-Sanchez et al. 2007). In our study presented here, the GmMYBJ2-overexpressing lines exhibited normal growth under normal conditions (Fig. 7a).
Collectively, GmMYBJ2, an R2R3-MYB gene, was isolated from soybean. GmMYBJ2 encodes a nucleus-localized protein and functions as a transcriptional activator, and its transcript was found to be rapidly induced by ABA and drought stress. Overexpression of GmMYBJ2 in Arabidopsis improved plant tolerance to dehydration stress, and the enhanced tolerance maybe resulted from the activation of stress-responsive genes and alterations in some physiological traits. The results provide insights into the soybean MYB TFs activated in response to environmental stresses and suggest this gene could be a candidate for crop improvement.
Author contribution statement
L. T. Su and Q. Y. Wang designed the research. L. T. Su and Y. Wang performed the majority of the experiments and wrote most of the paper. D.Q. Liu provided assistance during the research study and analyzed the gene expression profile. X. W. Li and Y. Zhai provided guidance and advice on experimental techniques and performed the yeast one-hybrid and subcellular localization assays. All of the statistical analyses were performed by X. Sun and J. W. Li, and these authors performed the sample collection for qPCR and the transformation of Arabidopsis. X. Y. Li and Y.J. Liu contributed technical assistance and insightful discussions. Q.Y. Wang supervised the study. All of the authors have read and approved the final manuscript.
Abbreviations
- TF(s):
-
Transcription factor(s)
- ORF:
-
Open reading frame
- ABA:
-
Abscisic acid
- WT:
-
Wild-type
- PEG:
-
Polyethylene glycol
- β-tubulin:
-
Beta-tubulin
- CaMV35S:
-
Cauliflower mosaic virus 35S
- 3-AT:
-
3-Amino-1, 2, 4-triazole
References
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
Ahuja I, de Vos RC, Bones AM, Hall RD (2010) Plant molecular stress responses face climate change. Trends Plant Sci 15:664–674
Bailey RW (1958) Reaction of pentoses with anthrone. Biochem J 68:669–672
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium -mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C (2005) A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol 15:1196–1200
Cominelli E, Sala T, Calvi D, Gusmaroli G, Tonelli C (2008) Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. Plant J 53:53–64
Dai X, Xu Y, Ma Q, Xu W, Wang T, Xue Y, Chong K (2007) Over-expression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought and salt stress in transgenic Arabidopsis. Plant Physiol 143:1739–1751
Dedio W (1975) Water relations in wheat leaves as screening tests for drought resistance. Can J Plant Sci 55:369–378
Du H, Zhang L, Liu L, Tang XF, Yang WJ, Wu YM, Huang YB, Tang YX (2009) Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry 74:1–11
Du H, Yang SS, Liang Z, Feng BR, Liu L, Huang YB, Tang YX (2012) Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol 12:106
Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55:940–953
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15:573–581
Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33:751–763
Fu J (1995) A breeding report on new soybean cultivar of Jilin32. J Jilin Agric Sci 2:18–20 (in Chinese)
Ganesan G, Sankararamasubramanian HM, Harikrishnan M, Ashwin G, Parida A (2012) A MYB transcription factor from the grey mangrove is induced by stress and confers NaCl tolerance in tobacco. J Exp Bot 63:4549–4561
Gray J (2005) Guard cells: transcription factors regulate stomatal movements. Curr Biol 15:R593–R595
Haga N, Kato K, Murase M, Araki S, Kubo M, Demura T, Suzuki K, Müller I, Voss U, Jürgens G, Ito M (2007) R1R2R3-Myb proteins positively regulate cytokine sis through activation of KNOLLE transcription in Arabidopsis thaliana. Development 134:1101–1110
Hamanishi ET, Thomas BR, Campbell MM (2012) Drought induces alterations in the stomatal development program in Populus. J Exp Bot 63:4959–4971
He CY, Zhang JS, Chen SY (2002) A soybean gene encoding a proline-rich protein is regulated by salicylic acid, an endogenous circadian rhythm and by various stresses. Theor Appl Genet 104:1125–1131
He Y, Li W, Lv J, Jia Y, Wang M, Xia G (2012) Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. J Exp Bot 63:1511–1522
Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L (2008) Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 67:169–181
Ito M (2005) Conservation and diversification of three-repeat Myb transcription factors in plants. J Plant Res 118:61–69
Kang JY, Choi HI, Im MY, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14:343–357
Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7:193–195
Kerepesi I, Galiba G (2000) Osmotic and salt stress-induced alteration in soluble carbohydrate content in wheat seedlings. Crop Sci 40:482–487
Kiyosue T, Yamaaguchi-Shinozaki K, Shinozaki K (1994) Characterization of two cDNAs (ERD10 and ERD14) corresponding to genes that respond rapidly to dehydration stress in Arabidopsis thaliana. Plant Cell Physiol 35:225–231
Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130:2129–2141
Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P (2006) SMART 5: domains in the context of genomes and networks. Nucleic Acids Res 34:D257–D260
Li XW, Li JW, Zhai Y, Zhao Y, Zhao X, Zhang HJ, Su LT, Wang Y, Wang QY (2013) A R2R3-MYB transcription factor, GmMYB12B2, affects the expression levels of flavonoid biosynthesis genes encoding key enzymes in transgenic Arabidopsis plants. Gene 532:72–79
Liao Y, Zou HF, Wei W, Hao YJ, Tian AG, Huang J, Liu YF, Zhang JS, Chen SY (2008a) Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 228:225–240
Liao Y, Zou HF, Wang HW, Zhang WK, Ma B, Zhang JS, Chen SY (2008b) Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res 18:1047–1060
Lippold F, Sanchez DH, Musialak M, Schlereth A, Scheible WR, Hincha DK, Udvardi MK (2009) AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in Arabidopsis. Plant Physiol 149:1761–1772
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10:1391–1406
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Luo GZ, Wang HW, Huang J, Tian AG, Wang YJ, Zhang JS, Chen SY (2005) A putative plasma membrane cation/proton antiporter from soybean confers salt tolerance in Arabidopsis. Plant Mol Biol 59:809–820
Mare C, Mazzucotelli E, Crosatti C, Francia E, Stanca AM, Cattivelli L (2004) Hv-WRKY38: a new transcription factor involved in cold and drought-response in barley. Plant Mol Biol 55:399–416
Matsui K, Umemura Y, Ohme-Takagi M (2008) AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J 55:954–967
Nakashima K, Ito Y, Yamaguchi-Shinozaki K (2009) Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 149:88–95
Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T, Abe H, Narusaka M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J 34:137–148
Niu C, Wei W, Zhou QY, Tian AG, Hao YJ, Zhang WK, Ma B, Lin Q, Zhang ZB, Zhang JS, Chen SY (2012) Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant, Cell Environ 5:1156–1170
Onate-Sanchez Anderson JP, Young J, Singh KB (2007) AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense. Plant Physiol 143(1):400–409
Pesch M, Hulskamp M (2009) One, two, three models for trichome patterning in Arabidopsis? Curr Opin Plant Biol 12:587–592
Qin YX, Wang MC, Tian YC, He WX, Han L, Xia GM (2012) Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis. Mol Biol Rep 39:7183–7192
Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290:2105–2110
Sakuma Y (2006) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18:1292–1309
Seki M, Umezawa T, Urano K, Shinozaki K (2007) Regulatory metabolic networks in drought stress responses. Curr Opin Plant Biol 10:296–302
Simon M, Lee MM, Lin Y, Gish L, Schiefelbein J (2007) Distinct and overlapping roles of single-repeat MYB genes in root epidennal patterning. Dev Biol 311:566–578
Simpson SD, Nakashima K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J 33:259–270
Song SY, Chen Y, Chen J, Dai XY, Zhang WH (2011) Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234:331–345
Su LT, Li JW, Liu DQ, Zhai Y, Zhang HJ, Li XW, Zhang QL, Wang Y, Wang QY (2014) A novel MYB transcription factor, GmMYBJ1, from soybean confers drought and cold tolerance in Arabidopsis thaliana. Gene 538:46–55
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Valliyodan B, Nguyen HT (2006) Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr Opin Plant Biol 9:189–195
Vannini C, Locatelli F, Bracale M, Magnani E, Marsoni M, Osnato M, Mattana M, Baldoni E, Coraggio I (2004) Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J 37:115–127
Vinocur B, Altman A (2005) Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotechnol 16:123–132
Wang YJ, Li YD, Luo GZ, Tian AG, Wang HW, Zhang JS, Chen SY (2005) Cloning and characterization of an HDZip I gene GmHZ1 from soybean. Planta 221:831–843
Weston K (1998) Myb proteins in life, death and differentiation. Curr Opin Genet Dev 8:76–81
Yamaguchi-Shinozaki K, Shinozaki K (1993) Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol Gen Genet 236:331–340
Yang A, Dai XY, Zhang WH (2012) A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot 63:2541–2556
Zhai Y, Wang Y, Li Y, Lei T, Yan F, Su L, Li XW, Zhao Y, Sun X, Li J, Wang QY (2012) Isolation and molecular characterization of GmERF7, a soybean ethylene-response factor that increases salt stress tolerance in tobacco. Gene 513:174–183
Zhai Y, Li JW, Li XW, Lei TT, Yan F, Zhao Y, Li YJ, Su LT, Wang Y, Wang QY (2013) Isolation and characterization of a novel transcriptional repressor GmERF6 from soybean. Biol Plantarum 57:26–32
Zhang LC, Zhao GY, Xia C, Jia JZ, Liu X, Kong XY (2012) A wheat R2R3-MYB gene, TaMYB30-B, improves drought stress tolerance in transgenic Arabidopsis. J Exp Bot 63:5873–5885
Zhu L, Guo JS, Zhu J, Zhou C (2014) Enhanced expression of EsWAX1 improves drought tolerance with increased accumulation of cuticular wax and ascorbic acid in transgenic Arabidopsis. Plant Physiol Biochem 75:24–35
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 30971808) and the Major Science and Technology Sponsored Program for Transgenic Biological Breeding (No. 2013ZX08004003-004).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There are no conflicts of interest.
Additional information
Communicated by O. Ferrarese-Filho.
L. T. Su and Y. Wang contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Su, LT., Wang, Y., Liu, DQ. et al. The soybean gene, GmMYBJ2, encodes a R2R3-type transcription factor involved in drought stress tolerance in Arabidopsis thaliana . Acta Physiol Plant 37, 138 (2015). https://doi.org/10.1007/s11738-015-1889-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-015-1889-5