Abstract
An early light-inducible protein (GbELIP) isolated from immature ginkgo leaves by rapid amplification of cDNA ends method contains a 570 bp open reading frame encoding a 189 amino acid protein, with significant homology to early light-inducible proteins from other plant species. The genomic DNA of GbELIP gene contains three exons and two introns. Southern blotting revealed that GbELIP is a single-copy gene in ginkgo. Light, defense and stress responsive element were found in the promoter region of GbELIP. GbELIP expressional patterns were detected by Real-time PCR at various conditions. ELIP expressions were variable at different time points of 1 day and increasing significantly at the commencement of light illumination then dropping to a basal level gradually. ELIP expression in small size leaves (1–1.5 cm width and 7-day age) was higher then middle (2–3 cm width and 15-day age) and large (4 cm width and 30-day age) size leaves. It revealed that ELIP expression was under the control of developmental regulation. Under temperature and light intensity treatments, GbELIP transcripts accumulation may relate to photoinhibition. The peak expression of GbELIP appeared later by chilling than heating. Meanwhile, GbELIP expression under high light was higher than low light at both two temperatures. GbELIP expression was also increased by wounding and methyl jasmonate (Me-JA) treatment, but not corresponded with increasing malondialdehyde (MDA) concentration. In conclusion, GbELIP appears to be induced by an imbalance between chlorophyll formation and degradation during development or under abiotic stress. These results suggest that GbELIP may function in response to environmental signals, possibly regulating responses to abiotic stresses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants in nature frequently experience extreme environmental conditions for factors including light, temperature, and other abiotic stresses. Through long-term evolution, plants have adapted to various stresses. Chloroplast, the place for photosynthetic reaction, is the most sensitive organ under stress. In former researches, ELIPs may have a role in protecting chloroplast function (Tao et al. 2011). ELIP was initially found as a transiently expressed protein during greening of etiolated pea seedlings, with expression disappearing before chloroplast development was completed (Kolanus et al. 1987; Meyer and Kloppstech 1984). Hence, ELIP was hypothesized to be replaced by light-harvesting chlorophyll a/b binding protein (LHC) in a late step of photosystem assembly in the process of plastid development (Grimm et al. 1989; Meyer and Kloppstech 1984; Tzvetkova-Chevolleau et al. 2007). Subsequently, ELIP transcripts and proteins have been found in the mature or young leave of some plants under photoinhibition. The deduced biological functions of ELIP are serving as a transient pigment binder that form complexes with free chlorophyll and an energy-dissipating antenna activating the xanthophyll cycle (Montané et al. 1999).
There is still an issue about the ELIP (RNA and protein) expressional pattern under some kinds of stresses. It is widely accepted that high light stress increases ELIP (RNA or protein) expression in several plants, e.g. Arabidopsis thaliana (Heddad and Adamska 2000; Hutin et al. 2003), Triticum aestivum (Shimosaka et al. 1999), Hordeum vulgare (Montané et al. 1997, 1999), Spinacia oleracea (Lindahl et al. 1997), Vitis vinifera (Pinto et al. 2011), Rhododendron catawbiense, R. ponticum (Peng et al. 2008; Wang et al. 2009; Wei et al. 2005). For temperature stress, several researches clarified high ELIP expression under photoinhibition by chilling (Casazza et al. 2005; Han and Kim 2013; Montané et al. 1999; Shimosaka et al. 1999; Wang et al. 2009). While, just a few researches focused on ELIP (RNA and protein) expression under heating stress. It is still an issue about effect of heating on ELIP (RNA and protein) accumulation. Among the former researches, ELIP did not increase obviously by heating (Han and Kim 2013; Osipenkova et al. 2010; Pinto et al. 2011; Wang et al. 2009; Zarter et al. 2006), but ELIP in Capsicum annuum was induced evidently by high temperature (Tao et al. 2011). In addition, ELIP RNA or protein could also be induced under wounding (Adamska et al. 1992; Sävenstrand et al. 2004). Meanwhile, phytohormones also affect the expression of ELIP. For example, light stress-induced ELIP expression can be enhanced by Me-JA (Bartels et al. 1992; Tao et al. 2011; Wierstra and Kloppstech 2000).
In this paper, we selected the ancient species—G. biloba originated in the Carboniferous and thrived in the Jurassic period. It is the only extant representative of the Ginkgoaceae and deciduous broad-leaved gymnosperm. Although its current natural range is a small area within southwestern China, it is cultivated throughout the world and exhibits great tolerance to various environmental conditions. Therefore, analyzing expression pattern GbELIP under various stresses will not only be beneficial to deduce the functions of GbELIP during environmental adaptation, but also will reveal the functional divergences during ELIP evolution in different species. In this paper, we try to isolate GbELIP gene and detect its expression under various stresses and provide a foundation for exploring the function and regulation patterns of ELIP—an important stress indicator in different species.
Materials and methods
Plant materials and treatments
Two-year-old grafted ginkgo ramets were growing in the outdoor ginkgo garden of Nanjing Forestry University in Jiangsu Province. Immature and folded leaves with 0.5-1 cm width and an age of 4–5 days were collected for DNA and RNA extraction (April 15th). On April 16th, grafted ramets were transferred to an artificial climate chamber for one week (25 °C, 1,000 μmol m−2 s−1, 60 % relative humidity and 12 h photoperiod). Then, different samples were collected under the following conditions, frozen in liquid nitrogen and stored at −80 °C.
To determine the best time for sampling, we tested ELIP expression at different time points during 1 day. After 7-day acclimation under the above condition, three grafted ramets were processing in the following test. The immature (1–1.5 cm width and 7-day age) leaves on short branches were selected and marked. Three leaves were sampled on each of the three ramets at the end of darkness (the control) and then sampled every 2 h.
To reduce difference from leaf-age in the following stress experiment, the relative quantification of GbELIP was tested in three kinds of leaves. Leaves on three grafted ramets were divided into three groups based on leaf width and age, respectively, small (1–1.5 cm width and 7-day age, S), middle (2–3 cm width and 15-day age, M) and large (more than 4 cm width and 30-day age, L). Different sized leaves on short branches were collected after 4 h illumination. Each sample includes three biological replicates.
Combined with the above experiments, middle (2–3 cm width and 15-day age, M) size leaves were selected and sampled for the following light, temperature, wounding and Me-JA treatments. For temperature and light treatments, nine acclimated grafted ramets were transferred to four different phytotrons with different temperature and light intensity (4 °C, 400 photons μmol m−2 s−1; 4 °C, 1,500 photons μmol m−2 s−1; 45 °C, 400 photons μmol m−2 s−1; 45 °C, 1,500 photons μmol m−2 s−1) at the end of darkness. Three biological replicates were included under the above four conditions. Samples (25 °C, 60 % relative humidity, 12 h photoperiod and 1,000 μmol m−2 s−1) at the end of darkness were collected before transfer as a control for real-time PCR analysis. Samples were collected at four time points, 2, 6, 12 and 24 h under 4 °C and 0.5, 1, 2 and 4 h under 45 °C. All samples were frozen in liquid nitrogen and stored at −80 °C.
To get rid of the difference from sampling time during 1 day and just focus on the effect of Me-JA (100 mM) and wounding, nine grafted ramets were kept for 5 days with 24 h light/0 h dark, 25 °C and 1,000 photons μmol m−2s−1. Leaves were collected from untreated plants at the same sampling time as the relative control of the real-time PCR. Me-JA (100 mM) was sprayed on the leaves surface of the three tested plants in Me-JA treatment. In the wound treatment, the 1 cm edge of leaves was cut and the 1 cm region adjacent to wound edge was collected. Sampling was at 2, 4, 6, 12 and 24 h after the beginning of treatment. Each sample includes three biological replicates.
Cloning and sequence analysis of ELIP from ginkgo
Based on the EST sequence (EX930691) available in the NCBI database, specific primer pairs (ELIP5R and ELIP3R) and nested primer pairs (ELIP5N and ELIP3N) were designed to amplify the 5′ and 3′ end of GbELIP using the Smarter™ RACE cDNA Amplification Kit (Clontech, USA). Primer sequences are listed in Table S1. The nested PCR products were purified using the TaKaRa Agarose Gel DNA Purification Kit Ver.2.0 (Takara, Japan) and cloned into PMD18-T Vector (Takara, Japan) for sequencing (Sangon, China). After comparing and aligning the 5′RACE, 3′RACE and EST sequences, two gene-specific primers, ELIPU and ELIPD (Table S1), were designed to amplify the full-length cDNA and genomic sequence of GbELIP. Sequence analysis and alignment were performed using the software Vector NTI Advance 11 (Life Technologies, USA). Sequences of homologous ELIP proteins present in the NCBI database was retrieved using BLAST network services (http://www.ncbi.nl.nih.gov). The signal peptide of the deduced amino acid sequence was predicted using SignalP 4.1 Server (http://www.cbs.dtu.dk/services/SignalP/). Hydrophobicity of GbELIP was analyzed using ProtScale Server (http://www.expasy.org/cgi-bin/protscale.pl). Transmembrane topology prediction was performed using TMHMM Server Version 2.0 (http://www.cbs.dtu.dk/services/TMHMM). Phylogenetic tree was constructed using MEGA 5.03 by a neighbor-joining (NJ) method and measured by bootstrap analysis with 1,000 replicates.
Amplification of the 5′-promoter region of GbELIP
The promoter region was isolated by genomic walk strategy using a Universal Genomewalker™ kit (Clontech, USA) according to the manufacturer’s instructions. Gene specific primer (ELIP1) and nested primer (ELIP2) were shown in Table S1. The nested PCR products were purified using the TaKaRa Agarose Gel DNA Purification Kit Ver.2.0 (Takara, Japan) and cloned into PMD18-T Vector (Takara, Japan) for sequencing (Sangon, China). The cis-element in the promoter region was predicted using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al. 2002).
Genomic DNA isolation and DNA gel blot analysis
Genomic DNA was extracted from ginkgo leaves according to the CTAB protocol (Yu et al. 2006). 30 μg DNA was digested overnight at 37 °C with BamHI, EcoRI and HindIII, separated by electrophoresis on a 0.8 % agarose gel, denatured (with 0.1 M HCl), neutralized and blotted onto the positively changed Hybond N+ membrane (Amersham, UK). Two primers, ELIPTU and ELIPTD (Table S1) were designed to amplify the cDNA sequence. Programs of PCR reactions are listed in Fig. S2. 50 ng purified PCR products were used as a template in a total volume of 20 μl for probe labeling. Probe labeling (DIG), hybridization, and signal detection were performed following the manufacturer’s instruction for the DIG High Primer DNA Labeling and Detection Starter Kit II (Roche, Germany).
RNA isolation and GbELIP expression profiling using real-time RT-PCR
Total RNA was extracted from leaves collected at different time points after stress using the CTAB method (Cai et al. 2007). Prior to reverse transcription, RNA samples were treated with DNase I (TaKaRa, Japan) according to the manual. RNA was quantified and evaluated for purity by UV spectroscopy and agarose gel electrophoresis. Reverse transcription-polymerase chain reaction was performed using Prime Script™ 1st Strand cDNA Synthesis Kit (Takara, Japan). Real-time PCR was carried out using an ABI PRISM 7500 Sequence Detection System (Applied Biosystems, USA) with SYBR Green PCR Master Mix (Roche, Germany) according to the manufacturer’s protocol. The reaction mix (20 μl) contained 10–50 ng template cDNA, 2 × SYBR Green Real-time PCR Master Mix (Roche, Germany) and 2.5 mM of each primer. The ginkgo glyceraldehydes-3-phosphate dehydrogenase gene (GbGAPDH, L26924) and histone H2A gene (GbHis2, a sequence of high-throughput RNA sequence reads, the sequence is shown in supplemental file) were used as reference genes. Primer sequences (ELIPF, ELIPR, GAPDHF, GAPDHR, HIS5F and HIS5R) designed for real-time PCR are listed in Table S1. Programs of real-time PCR reactions are listed in Table S2. Melt curves of the three primers are shown in Fig. S1. cDNA samples were diluted to 0.5, 0.25, 0.125, 0.0625-fold. The concentrations of the above five templates were between 50 and 0.05 ng/μl. These solutions (1 μl) were added to the standard curve reaction. Standard curves were shown in Fig. S2. Amplification efficiency of each primer pair was measured by comparing slopes of standard curves according to the manual of 7500 real-time PCR (Fig. S3). Real-time PCR data were calibrated relative to the corresponding gene expression level at time zero for each treatment, following the 2−ΔΔCt method for relative quantification (Livak and Schmittgen 2001). The data are presented as mean ± SE of three replicates and each replicate have three reactions performed in different 96-well plates.
Chlorophyll fluorescence
Leaves of 2–3 cm width and 15-day age on short branches of 16 acclimated grafted ramets were collected and disks of 1 cm diameter were cut from each leaf. All disks were divided into four groups and each group contains 32 disks. The four groups were, respectively, put on a wet gauze at following four different conditions (1,500 photons μmol m−2 s−1, 4 °C; 1,500 photons μmol m−2 s−1, 45 °C, 400 photons μmol m−2 s−1, 4 °C and 400 photons μmol m−2 s−1, 45 °C). Eight disks were taken out after 2, 6, 12 and 24 h, respectively, then dark-adapted in leaf-clips for 20 min before measurements. Chlorophyll fluorescence before treatment was measured as control. Chlorophyll fluorescence was measured with a FMS-2 Pulse Modulated Fluorometer (Hansatech, UK). The minimum fluorescence yield (F 0) and maximum fluorescence yield (F m) were measured under dark condition. The degree of photoinhibition was determined by the reduction in F v/F m estimates (Maxwell and Johnson 2000).
Chlorophyll content detection
Leaves were divided into three groups based on leaf width and age, respectively, small (1–1.5 cm width and 7-day age, S), middle (2–3 cm width and 15-day age, M) and large (more than 4 cm width and 30-day age, L). Leaf disks of 1 cm diameter were cut from each leaf then frozen in liquid nitrogen before pigment concentration were determined. Extraction of chlorophylls and carotenoids was carried out at low light by grinding the frozen disks in 1 ml of cold acetone (−20 °C, 80 %). Chlorophyll and carotenoid concentration were determined spectrophotometrically (Hodges et al. 1999). Contents of chlorophyll and carotenoids were measured in these three samples with five replicates.
Assay for malondialdehyde (MDA)
MDA is the ultimate product of membrane lipid peroxidation, which is often related to the extent of oxidative damage caused by ROS (active oxygen species). Leaf MDA content was assayed according to the thiobarbituric acid (TBA) method (Hodges et al. 1999; Radić et al. 2006; Tao et al. 2011). Fresh tissues (200 mg) were homogenized in 1 ml 0.25 % TBA with 10 % trichloracetic acid. The mixture was heated at 95 °C for 30 min and then quickly cooled in an ice bath. After centrifugation at 10,000g for 10 min, the absorbance of the supernatant was read at 532 nm with a spectrophotometer (Hewlett Packard, USA). Correction for nonspecific turbidity was done by subtracting absorbance at 600 nm. The MDA content was calculated using its extinction coefficient of 155 mm−1 cm−1. MDA concentration was expressed as nmol MDA per gram of fresh weight. Each sample was carried out with three biological and technical replicates.
Data analysis
Data were subjected to analysis of variance (ANOVA). Mean values were compared by the Duncan’s new multiple range test (DMRT) at 0.05 probability. Statistical analyses were performed using SPSS 17.0 and Eview 6.0 software.
Results
Characterization of GbELIP gene in ginkgo
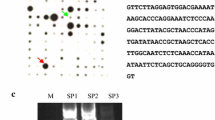
A full-length cDNA was obtained from ginkgo leaf tissue, which was 1,016 bp with a polyA tail (KC818609). The cDNA sequence contains a 570 bp open reading frame (ORF). A 177 bp 5′ untranslated region (5′ UTR) was upstream of the start codon and the coding region was followed by a 266 bp 3′ untranslated region (3′ UTR) downstream from the stop codon (Fig. 1). The genomic GbELIP gene was isolated by a pair of specific primers derived from the cDNA sequence containing the start and stop codon region. The full-length genomic DNA of GbELIP gene was 821 bp and identical with the coding region of the full-length cDNA sequence. Exon 1 (136 bp), exon 2 (90 bp) and exon 3 (347 bp) were separated by intron 1 (143 bp) and intron 2 (105 bp). The putative splicing site also obeyed the GU/AG rule (Fig. 1). Genomic DNA blot analysis of ginkgo using a DIG-labelled probe sequence showed only one band from the restriction digests of BamH I, EcoR I and Hind III (Fig. S4).
The full-length GbELIP sequence, promoter region sequence, intron sequence and deduced amino acid sequence of GbELIP gene. The exons are indicated in capital letters and underlined. The introns are indicated in italic letters. The start and stop codons are in red. The putative protein sequence is in blue. The putative chloroplast transit peptide is in shaded. Cis-elements of the GbELIP promoter region are shown in black box (colour figure online)
By genome walking, we isolated a 444 bp fragment up stream of the ATG codon which corresponded to the partial putative promoter region (Fig. 1). Computational analysis revealed five potential TATA box sequences within the upstream region of GbELIP, at positions -20, -185, -392, -414, and -429. Several conserved eukaryotic promoter elements, including a CAAT box, were also observed at positions -52, -234, and -376. Several light responsive elements were identified: the Box II located at position -432; the G-box located at positions -137, -292 and -343 and the sp1 located at position -93. The defense and stress responsiveness element was found at positions -211.
Theoretical molecular weight of putative GbELIP (190 amino acids) was 19.9 kDa and pI value was 9.47. A 17 amino acid N-terminal domain was predicted as a putative chloroplast transit peptide (Fig. 1 and Fig. S5). Hydrophobicity plot analysis (Fig. S6) and transmembrane topology prediction (Fig. S7) revealed three hydrophobic domains in the putative GbELIP protein, which may be the transmembrane regions. A multiple alignment of GbELIP with other ELIPs is shown in Fig. S7. N-terminal domains of ELIPs exhibit low similarity while the C-terminal domains have high conservation. Sequence analysis showed that GbELIP shared moderate homology with three unknown proteins (ABR16737, 81 %; ACN40010, 76 % and ACN39996, 73 %) in Picea sitchensis, two ELIP s in A.thaliana (AAD28779, 70 % and AAB88391, 69 %) and ELIP in Glycine max (AAC16403, 68 %).
A phylogenetic tree based on the putative amino acid sequences of GbELIP and other ELIPs from different species was constructed to investigate the evolutionary relationships among various ELIPs. An unrooted neighbor-joining cladogram is shown in Fig. 2. Relationships shown in this tree generally reflect standard relationships of phycophytas, pteridophytes, ginkgo, conifer, monocotyledon and eudicotyledons. GbELIP belongs to the ginkgo group and has a relatively close relationship with ELIP from the conifer Picea sitchensis.
Phylogenetic relationships between the putative amino acid sequence of GbELIP and identified ELIPs. The tree was constructed using the software Molecular Evolutionary Genetics Analysis (MEGA) Version 5.03 by the neighbor-joining method with pairwise deletion and the poisson correction model. Bootstrap value of the phylogenetic tree was set as 1,000. Bootstrap support values (percentage of bootstrap trees supporting the node) for each node are shown. The boot letters A to F indicate different ELIPs groups: A eudicotyledon, B ginkgo, C conifers, D monocotyledon, E phycophyta, F pteridophyte. Accession numbers for all ELIP sequences from different species are listed here. Trifolium pratense ELIP (AAQ21120), Medicago sativa ELIP (AAK63815), Pisum sativum ELIP (CAA29399), Glycine max ELIP (AAC16403), Ricinus communis ELIP (XP_002517068), Populus trichocarpa ELIP (XP_002312552), Arabidopsis thaliana ELIP1 and ELIP2 (AAD28779 and AAB88391), Camellia sinensis ELIP (ACB20694), Solanum lycopersicum ELIP (AAS92268), Vitis vinifera ELIP (XP_002283398), Rhododendron catawbiense ELIP (ABU98944), Ginkgo biloba ELIP (KC818609), Picea sitchensis ELIP (ACN39996), Hordeum vulgare ELIP1, ELIP2 and ELIP3 (CAA33726, CAA33727 and CAA33728), Oryza sativa ELIP (NP_001059030), Triticum aestivum ELIP (BAA76309), Dunaliella bardawil ELIP (SP_P27516), Syntrichia ruralis ELIP1 and ELIP2 (AAK59376 and AAK59377), Onoclea sensibilis ELIP (AAB25012)
Expression of GbELIP during one photoperiod and in different growth stages of leaves
GbELIP expression in one light period (12 h light/12 h dark) was detected in immature (1.5 width and 10-day age) ginkgo leaves. The expression patterns of GbELIP transcript in the immature leaves at different time points are shown in Fig. 3. The expression of GbELIP at the end of darkness is included as a control. After light was turned on, the transcripts of GbELIP were increased rapidly, reaching a maximum (almost 29-fold) at 4 h, and dropping two-threefold before light was turned down. The expression decreased steadily to the basal level under dark period. Overall, significant (P ≤ 0.05) differences in the level of GbELIP expression exist among different time points under illumination. On the contrary, no obviously difference (P ≤ 0.05) in expression was found under dark condition.
GbELIP expression in young leaves at different time points during one day at the end of April (1,000 photons μmol m−2s−1; 25 °C). Plant was maintained in 12 h light/12 h dark cycles and leaves were sampled every 2 h. The X axis shows sampling time, with hours indicated after the beginning of the light (L) and dark (D) periods. D12 was the reference sample. Data are means of three replicates. Errors bars show standard errors (SE). Lowercase letters show significant differences at P ≤ 0.05 in ELIP expression at different time points during the photoperiod (12 h light/12 h dark). Different letters represent significant or greatly significant differences between the different time points
Based on the above results, the relative expression of GbELIP was tested after 4 h illumination in three kinds of leaves on May 15th. The expressions of GbELIP in S (1–1.5 cm width and 7-day age) and M (2–3 cm width and 15-day age) were significantly (P ≤ 0.01) higher than in L (more than 4 cm width and 30-day age). GbELIP mRNA in S exhibited expression levels of 5.3-fold higher, while expression in M was 1.9-fold greater than L (Fig. 4).
GbELIP expression in different size leaves after 4 h illumination at the end of April. Data are means of three replicates. Errors bars show standard errors (SE). S (width: 1–1.5 cm), M (width: 2–3 cm), L (width: 4 cm, as control). Lowercase letters show significant difference of GbELIP at P ≤ 0.05. Different letters represent significant or greatly significant differences between different size of plant leaves
Contents of chlorophyll and carotenoids in different size of leaves
Chlorophylls content in S is lower than M and L, but Carotenoids contents in S is higher than M and L. Contents of all three pigments in S were significantly (P ≤ 0.01) different with M and L. However, there was no significant difference (P ≤ 0.01) between M and L (Table 1).
Expression of GbELIP under different temperature and light intensity
Heating and chilling influence on GbELIP expression are shown in Fig. 5a, b, respectively. Under low temperature, both high and low light increased the GbELIP expression during testing period, and the value reached a maximum at the same time (12 h), but the peak values differed significantly with light intensity (6.85-fold under low light and 9.98-fold under high light intensity; P ≤ 0.01). After 12 h treatment, GbELIP expression under low light decreased to 5.98-fold at 24 h. However, no significant difference existed between 12 and 24 h under high light (P ≤ 0.05). Under high temperature, expressions peaked suddenly at 6.71-fold (low light) and 8.79-fold (high light) at 0.5 h, then gradually decreased to 2.56-fold (low light) and 4.55-fold (high light) between 0.5 and 2 h, and maintained twofold (low light) and threefold expression (high light) at 4 h. After 4 h, the decline of expression of GbELIP under high light was significant between 12 and 24 h, but not significant under low light.
Expression of GbELIP over time under different light intensity (400 photons μmol m−2 s−1 and 1,500 photons μmol m−2 s−1) and temperature (4 °C in a and 45 °C in b) at the end of April. Solid and hollow squares showed GbELIP expression values at 4 °C in high light (1,500 photons μmol m−2 s−1) and low light (400 photons μmol m−2 s−1). Solid and hollow triangles showed GbELIP expression values at 45 °C in high light and low light. Data were means of three replicates. Error bars showed standard errors (SE). Multiple comparisons were preformed by two-way ANOVA. Lowercase letters showed significant differences of GbELIP expression between time points and light intensities at P ≤ 0.05
Photoinhibition under light intensity and temperature stress
To test whether the accumulation of GbELIP in ginkgo correlates with photoinactivation and photodamage of PSII, in vitro leave disks were exposed to high or low light intensities (400 and 1,500 photons μmol m−2 s−1) under 4 or 45 °C, respectively, and PSII activity was measured by assaying the maximum quantum yield of PSII (F v/F m, dark adaption). Under chilling stress (4 °C), F v/F m declined considerably to 0.397 ± 0.015 (400 photons μmol m−2 s−1) and 0.256 ± 0.030 (1,500 photons μmol m−2 s−1) in 12 h, and then remained at a low level of 0.427 ± 0.021 and 0.236 ± 0.015 at 24 h, respectively (Fig. 6). Under heat stress (45 °C), F v/F m decreased to the bottom value faster than 4 °C, and then kept lower level of 0.207 ± 0.021 (400 photons μmol m−2 s−1) and 0.173 ± 0.020 (1,500 photons μmol m−2 s−1) at 24 h. All parameters indicate that photoinhibition and photodamage were induced by high temperature stress faster than chilling stress, and high light accelerated the decline of F v/F m at both high and low temperatures.
Effect of light intensity and temperature stress on the changes of F v/F m at different time points (at the end of April). Solid and hollow squares showed GbELIP expression values at 4 °C in high light (1,500 photons μmol m−2 s−1) and low light (400 photons μmol m−2 s−1), respectively. Solid and hollow squares showed GbELIP expression values at 4 °C in high light (1,500 photons μmol m−2 s−1) and low light (400 photons μmol m−2 s−1). Solid and hollow triangles showed GbELIP expression values at 45 °C in high light and low light. Data are means of eight replicates. Positive errors bars showed standard errors (SE). Differences in time points and treatments were analyzed by one-way ANOVA. Lowercase letters and Capital letters show, respectively, the significant (P ≤ 0.05) differences of F v/F m value in time points and treatments
Expression of GbELIP under Me-JA and wound treatment
Expression profiles of GbELIP at different stages of Me-JA and wound treatment are shown in Fig. 7a, b, respectively. Transcripts of GbELIP increased suddenly by Me-JA application and reached an expression peak (5.4-fold) at 2 h, and then decreased sharply to basal level (0.85-fold) at 12 h, after 12 h kept a low rate, and finally maintained a 0.36-fold expression level relative the control until 24 h (Fig. 7). In contrast, the GbELIP expression increased gradually after wounding in the first 4 h, maintained a one to twofold increase at 4 h, and then increased quickly to an expression peak at 12 h, and finally dropped to 2.54-fold increase relative to the control (0 h) (Fig. 7).
GbELIP expression following Me-JA application (a) and wounding treatment (b) at the end of April. Data were means of three replicates. Positive errors bars showed standard errors (SE). Multiple comparisons were preformed by one-way ANOVA. Lowercase letters indicate significant differences between time points at P ≤ 0.05
Production of malondialdehyde (MDA) under Me-JA and wound treatment
Temperature and light can result in the generation of reactive oxygen species (ROS) like hydrogen peroxide and superoxide radicals that cause oxidative stress in the cells. In this experiment, the amount of MDA was measured to reflect the lipid peroxidation caused by Me-JA and wound treatment. The results suggest that Me-JA and wound treatments can significantly increase the content of MDA by 12 h, then the value decreased to relatively low level at 24 h, which might be caused by death of mesophyll cells (Fig. 8). The influence of Me-JA treatment on membrane lipid peroxidation was stronger and faster than wounding. Expression profiles of GbELIP did not show the same tendency as the dynamical changes of MDA under Me-JA and wound treatments.
Effect of Me-JA application (A) and wounding treatment on the changes of MDA concentration at different time points (at the end of April). Solid and hollow squares showed MDA concentration values under Me-JA application and wounding treatment, respectively. Data are means of five replicates. Errors bars showed standard errors (SE). Differences in time points and treatments were analyzed by one-way ANOVA. Lowercase letters and capital letters show significant differences of Fv/Fm value at different time points and treatment at P ≤ 0.05
Discussion
The ELIP in G. biloba was found to share many characteristics with ELIPs in other plant species, such as the transit peptide sequence, three hydrophobic helices and a Chlorophyll a/b binding protein motif. But the N-terminal domains of the putative GbELIP protein are longer and show the greatest divergence relative to the other plant peptides. The number of genes encoding ELIP is different in various organisms. Like the putative single-copy gene in ginkgo, ELIP is also encoded by one gene in P. sativum (Sävenstrand et al. 2004), Nicotiana tabacum (Binyamin et al. 2001), Spirogyra varians (Han and Kim 2013), Medicago truncatula (Araújo et al. 2013), Solanum lycopersicum (Bruno and Wetzel 2004), C. annuum (Tao et al. 2011) and V. vinifera (Pinto et al. 2011). However, two ELIPs were reported in A. thaliana (Tzvetkova-Chevolleau et al. 2007), T. aestivum (Zeng et al. 2002) and Tortula rur ails (Casazza et al. 2005; Tzvetkova-Chevolleau et al. 2007; Wierstra and Kloppstech 2000), and more than two ELIPs were reported in H. vulgare (Wierstra and Kloppstech 2000) and R. catawbiense (Peng et al. 2008).
ELIPs were first reported to be transiently expressed during the greening process of etiolated plants and consequently be found in the thylakoid membrane (Grimm and Kloppstech 1987). Previous studies revealed the ELIP (RNA and protein) expression patterns might be varied with leaves types and sampling time in different species (Heddad and Adamska 2000; Tao et al. 2011). In C. annuum, ELIP expression was under the control of circadian rhythm during the 16/8 h light and dark cycle (Tao et al. 2011). In this report, the expression of GbELIP is variable at different time points (12 h light/12 h dark) and increasing significantly at the commencement of light illumination (Fig. 3). Combined with several light-response elements in promoter region, it was deduced that GbELIP expression was regulated by illumation. At the same time, this research might provide a foundation for selecting the best time to analyze ELIP expression under stress experiments.
The content of GbELIP mRNA in S (1–1.5 cm width and 7-day age) leaves was higher than M (2–3 cm width and 15-day age) and L (more than 4 cm width and 30-day age) leaves that contained a high level of chlorophylls (Fig. 4; Table 1). Similar expression patterns of ELIP were found in P. sativum (Noren et al. 2003) and V.vinifera (Pinto et al. 2011). It is consistence with the former deduction that ELIP might play a role in chlorophyll accumulation, protecting chlorophyll against degradation during the assembly of the PSII reaction centers (Tzvetkova-Chevolleau et al. 2007). In young developing leaves, expression of ELIP increased as the rate of chlorophyll synthesis exceeded its rate of degradation. It is similar with the greening process of etiolated plants. Based on this hypothesis, the rate of chlorophyll synthesis and degradation was in balance in mature leaves, so chlorophyll accumulation stopped and ELIP expression stayed low.
In former researches, ELIPs belong to a family of proteins that are structurally related to the light-harvesting complex (LHCs) and speculated to function as a transient pigment carriers or assist in energy dissipation to protect chloroplasts from light-induced damage (Tao et al. 2011; Tzvetkova-Chevolleau et al. 2007). Excessive light and extreme temperature can reduce the process of light absorption and conversion then lead to photoinhibition or photodamage. In this report, under the two different temperatures (4 and 45 °C.), both high and low light intensities (400 and 1,500 photons μmol m−2 s−1) increased expression of GbELIP. At the same temperature, GbELIP expression under high light is higher than low light. Hence, it was speculated that the expression of GbELIP may respond to photoinhibition (Fig. 8) under temperature and light stress. The expression of ELIP induced by high light was also found in various species (A. thaliana (Heddad et al. 2006), H. vulgare (Wierstra and Kloppstech 2000), T. aestivum (Shimosaka et al. 1999), S. oleracea (Lindahl et al. 1997) and S. varians (Han and Kim 2013), but some studies have reported that ELIP was just tested in the plant growing above one light intensity (Heddad et al. 2006; Shimosaka et al. 1999). Based on the results of our study, we deduced that the lowest inducing light intensity may depend on other environmental factors, such as temperature. When the light intensity is too low to cause photoinhibition under the tested temperature, the ELIP may not accumulate. On the contrary, the expression of ELIP was induced at the same light intensity under extreme temperature resulted in photoinhibition (Fowler and Thomashow 2002; Heddad et al. 2006; Hutin et al. 2003; Lindahl et al. 1997; Montané et al. 1999; Pinto et al. 2011; Wierstra and Kloppstech 2000).
The expression patterns of ELIP (RNA and protein) are not consistent in different species under chilling and heating stress. ELIP cDNA in C. annuum increased quickly and reached a maximum at the first hour of illumination then gradually decreasing, while under chilling stress it was gradually increasing over 24 h (Tao et al. 2011). It is consistent with ELIP expression patterns of the above results of G.biloba under temperature extremes. However, ELIP content of V. vinifera was higher at high temperature (42 °C, 1,000 photons μmol m−2 s−1) than low temperature (15 °C, 1,000 photons μmol m−2 s−1) after 6 h treatment. Four ELIP homologue proteins increased under high light in A. thaliana show two different expression patterns. Two of them were not detected under other stress condition. The other two was increased by cold (4 °C, 2,500 photons μmol m−2 s−1) and heat (42 °C, 2,500 photons μmol m−2 s−1), but there is no significant influence of ELIP expressions after 3 h under the above mentioned two stresses conditions (Heddad and Adamska 2000). It has been reported that ELIP expression patterns may be under the control of circadian rhythm (Tao et al. 2011), developmental stage (Tao et al. 2011), treatment time and species. Difference caused by the first two factors can be reduced by selecting suitable sampling time and leaf-age. To clarify the impaction of extreme temperature or other stresses on GbELIP expression, it is necessary to describe the dynamic expression patterns of ELIP in G. biloba under various stresses. If ELIP expression values related to the extent of photoinhibition, different species suffered different level of inhibition under same condition, and then it is common that the ELIP expression levels were inconsistent.
In addition to high light and temperature extremes, wounding, dehydration, salt and heavy metal exposure can also inhibit the downstream processes of light absorption and lead to photoinhibition or photodamage. If the expression of ELIP was related to photoinhibition, could the transcripts of ELIP be up regulated by other abiotic stresses? In this paper, we focused on the expression patterns of ELIP under wounding and Me-JA treatments. The result is consistent with the rapid ELIP accumulations in P. sativum (Sävenstrand et al. 2004) and H. vulgare (Wierstra and Kloppstech 2000) under wound or Me-JA treatment. It has also been reported that ELIP (RNA and protein) expression was increased by heavy metals, salinity and dehydration (Tao et al. 2011; Zeng et al. 2002). However, the corresponding time to peak ELIP value was earlier or later than the corresponding time to peak ELIP value, so expression of ELIP did not seem to correspond with MDA accumulation or play a role in protection for oxidative stress in this treatment (Shimosaka et al. 1999; Zeng et al. 2002).
Although the biological functions of ELIPs have not been firmly established, data are accumulating that point to a role in photo protection, especially under stressful conditions. In former researches, ELIP mRNAs or proteins expression pattern was mainly tested in herbaceous plants, but rarely tested in woody perennials species. Ginkgo, an ancient species, is evolutionarily distant to all other plants on earth. In this paper, the ELIP expression pattern in ginkgo under stress condition is similar to other plants and ELIP expression may also relate to the extent of photoinhibition. It indicated the role of ELIPs appears to have been largely conserved over hundreds of millions of years of evolution between ginkgo, and angiosperms (herbage and xylophyta). It provides a reference for the future studies on the other species.
Author contribution statement
H Wang, F Cao, G Li and W Yu participated in the design of the study. H Wang and G Li carried out most of the experiments. H Wang achieved most of the data analysis and drafted the manuscript. S N Aitken revised the whole paper. F Cao provided the financial support for this research. All authors have read and approved the final manuscript.
References
Adamska I, Kloppstech K, Ohad I (1992) UV light stress induces the synthesis of the early light-inducible protein and prevents its degradation. J Biol Chem 267:24732–24737
Araújo SS, Duque AS, Silva JM, Santos D, Silva AB, Fevereiro P (2013) Water deficit and recovery response of Medicago truncatula plants expressing the ELIP-like DSP22. Biol Plantarum 57:159–163
Bartels D, Hanke C, Schneider K, Michel D, Salamini F (1992) A desiccation related Elip-like gene from the resurrection plant Craterostigma plantagineum is regulated by light and ABA. EMBO J 11:2771–2778
Binyamin L, Falah M, Portnoy V, Soudry E, Gepstein S (2001) The early light-induced protein is also produced during leaf senescence of Nicotiana tabacum. Planta 212:591–597
Bruno AK, Wetzel CM (2004) The early light-inducible protein (ELIP) gene is expressed during the chloroplast-to-chromoplast transition in ripening tomato fruit. J Exp Bot 55:2541–2548
Cai R, Xu F, Chen LJ, Cheng SY (2007) Modification of total RNA isolation method from different Ginkgo biloba organs. Biotechnology 17:38–41
Casazza A, Rossini S, Rosso M, Soave C (2005) Mutational and expression analysis of ELIP1 and ELIP2 in Arabidopsis thaliana. Plant Mol Biol 58:41–51
Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14:1675–1690
Grimm B, Kloppstech K (1987) The early light-inducible proteins of Barley. Eur J Biochem 167:493–499
Grimm B, Kruse E, Kloppstech K (1989) Transiently expressed early light-inducible thylakoid proteins share transmembrane domains with light-harvesting chlorophyll binding proteins. Plant Mol Biol 13:583–593
Han J, Kim G (2013) An ELIP-like gene in the freshwater green alga, Spirogyra varians (Zygnematales), is regulated by cold stress and CO2 influx. J Appl Phycol 25:1297–1307
Heddad M, Adamska I (2000) Light stress-regulated two-helix proteins in Arabidopsis thaliana related to the chlorophyll a/b-binding gene family. P Natl Acad Sci USA 97:3741–3746
Heddad M, Noren H, Reiser V, Dunaeva M, Andersson B, Adamska I (2006) Differential expression and localization of early light-induced proteins in Arabidopsis. Plant Physiol 142:75–87
Hodges DM, DeLong JM, Forney CF, Prange RK (1999) Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207:604–611
Hutin C, Nussaume L, Moise N, Moya I, Kloppstech K, Havaux M (2003) Early light induced proteins protect Arabidopsis from photooxidative stress. P Natl Acad Sci USA 100:4921–4926
Kolanus W, Scharnhorst C, Kühne U, Herzfeld F (1987) The structure and light-dependent transient expression of a nuclear-encoded chloroplast protein gene from pea (Pisum sativum L.). Mol Gen Genet 209:234–239
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Lindahl M, Funk C, Webster J, Bingsmark S, Adamska I, Andersson B (1997) Expression of ELIPs and PS II-S protein in spinach during acclimative reduction of the Photosystem II antenna in response to increased light intensities. Photosynth Res 54:227–236
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using Real-time quantitative PCR and the 2−ΔΔCt Method. Methods 25:402–408
Maxwell K, Johnson GN (2000) Chlorophyll fluorescence—a practical guide. J Exp Bot 51:659–668
Meyer G, Kloppstech K (1984) A rapidly light-induced chloroplast protein with a high turnover coded for by pea nuclear DNA. Eur J Biochem 138:201–207
Montané M-H, Dreyer S, Triantaphylides C, Kloppstech K (1997) Early light-inducible proteins during long-term acclimation of barley to photooxidative stress caused by light and cold: high level of accumulation by posttranscriptional regulation. Planta 202:293–302
Montané M-H, Petzold B, Kloppstech K (1999) Formation of early-light-inducible-protein complexes and status of xanthophyll levels under high light and cold stress in barley (Hordeum vulgare L.). Planta 208:519–527
Noren H, Svensson P, Stegmark R, Funk C, Adamska I, Andersson B (2003) Expression of the early light-induced protein but not the PsbS protein is influenced by low temperature and depends on the developmental stage of the plant in field-grown pea cultivars. Plant, Cell Environ 26:245–253
Osipenkova OV, Odintsova MS, Yurina NP (2010) The influence of light, hormonal, and carbohydrate signal systems on ELIP gene expression in gun-mutants Arabidopsis thaliana. Appl Biochem Micro 46:331–338
Peng Y, Lin W, Wei H, Krebs SL, Arora R (2008) Phylogenetic analysis and seasonal cold acclimation-associated expression of early light-induced protein genes of Rhododendron catawbiense. Physiol Plantarum 132:44–52
Pinto F, Berti M, Olivares D, Sierralta WD, Hinrichsen P, Pinto M (2011) Leaf development, temperature and light stress control of the expression of early light-inducible proteins (ELIPs) in Vitis vinifera L. Environ Exp Bot 72:278–283
Radić S, Radić-Stojković M, Pevalek-Kozlina B (2006) Influence of NaCl and mannitol on peroxidase activity and lipid peroxidation in Centaurea ragusina L. roots and shoots. J Plant Physiol 163:1284–1292
Sävenstrand H, Olofsson M, Samuelsson M, Strid Å (2004) Induction of early light-inducible protein gene expression in Pisum sativum after exposure to low levels of UV-B irradiation and other environmental stresses. Plant Cell Rep 22:532–536
Shimosaka E, Sasanuma T, Handa H (1999) A Wheat Cold-Regulated cDNA Encoding an Early Light-Inducible Protein (ELIP): its Structure, Expression and Chromosomal Location. Plant Cell Physiol 40:319–325
Tao L, Zeba N, Ashrafuzzaman M, Hong CB (2011) Heavy metal stress-inducible early light-inducible gene CaELIP from hot pepper (Capsicum annuum) shows broad expression patterns under various abiotic stresses and circadian rhythmicity. Environ Exp Bot 72:297–303
Tzvetkova-Chevolleau T, Franck F, Alawady AE, Dall’Osto L, Carrière F, Bassi R, Grimm B, Nussaume L, Havaux M (2007) The light stress-induced protein ELIP2 is a regulator of chlorophyll synthesis in Arabidopsis thaliana. Plant J 50:795–809
Wang XA, Peng YH, Singer JW, Fessehaie A, Krebs SL, Arora R (2009) Seasonal changes in photosynthesis, antioxidant systems and ELIP expression in a thermonastic and non-thermonastic Rhododendron species: a comparison of photoprotective strategies in overwintering plants. Plant Sci 177:607–617
Wei H, Dhanaraj A, Rowland L, Fu Y, Krebs S, Arora R (2005) Comparative analysis of expressed sequence tags from cold-acclimated and non-acclimated leaves of Rhododendron catawbiense Michx. Planta 221:406–416
Wierstra I, Kloppstech K (2000) Differential effects of methyl jasmonate on the expression of the early light-inducible proteins and other light-regulated genes in barley. Plant Physiol 124:833–844
Yu LH, Zhang CS and Liang H (2006) Extract of DNA from ginkgo leaves and its RAPD amplification. Biotech Bull:81–84
Zarter CR, Adams WW, Ebbert V, Cuthbertson DJ, Adamska I, Demmig-Adams B (2006) Winter down-regulation of intrinsic photosynthetic capacity coupled with up-regulation of Elip-like proteins and persistent energy dissipation in a subalpine forest. New Phytol 172:272–282
Zeng Q, Chen X, Wood AJ (2002) Two early light-inducible protein (ELIP) cDNAs from the resurrection plant Tortula ruralis are differentially expressed in response to desiccation, rehydration, salinity, and high light. J Exp Bot 53:1197–1205
Acknowledgments
This research was supported by the foundation of excellent doctoral dissertation in Nanjing Forestry University, the plan of scientific research innovation of graduate students in general college (CXZZ12_0546), national nature science foundation (31170627) and nature science foundation of Jiangsu province (BK2010019). We thank Tongli Wang, a research associate in the University of British Columbia’s Department of Forest and Conservation Science, for giving us so many suggestions on structure arrangement of this paper. Thanks for Robin Mellway, a post-doctor fellow with the University of British Columbia’s Department of Forest and Conservation Sciences, on editing of wording and grammar in the paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Abe.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, H., Cao, F., Li, G. et al. The transcript profiles of a putative early light-induced protein (ELIP) encoding gene in Ginkgo biloba L. under various stress conditions. Acta Physiol Plant 37, 1720 (2015). https://doi.org/10.1007/s11738-014-1720-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-014-1720-8