Abstract
Multiple diversification rate shifts explain uneven clade richness in muroid rodents. Previous muroid studies have shown that extrinsic factors, notwithstanding ecological opportunity, are poor predictors of clade diversity. Here, we use a 297-muroid species chronogram that is sampled proportional to total clade diversity, along with various trait-dependent diversification approaches to investigate the association between diversification rates with intrinsic attributes—diet, habitat, body mass, and relative tail length. We found some association between both dietary specialization and body mass, as well as between habitat specialization with relative tail lengths using phylogenetic analyses of variance. However, there was no significant association between diversification rates with the evolution of these traits in muroid rodents. We also show that several of the state-dependent diversification approaches are highly susceptible to Type I error—a result that is in accordance with recent criticisms of these methods. Finally, we discuss several potential causes for the lack of association between the examined trait data with diversification rates, ranging from methodological biases (e.g. method conservativism) to biology (e.g. behavioral plasticity and ecological opportunism of muroid rodents).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Muroid rodents (rats, mice, gerbils, hamsters, and close relatives) constitute > 25% of mammal species, making this superfamily (Muroidea) the most diverse in its class (Musser and Carleton 2005). Muroid clades are vastly uneven in diversity (Steppan et al. 2004; Fabre et al. 2012; Schenk et al. 2013). After accounting for clade age, uneven diversity is mostly a consequence of differential speciation and/or extinction rates, which in turn could be influenced by such traits as geography, ecology, morphology, climate, habitat, demography, reproduction, and development (Maddison et al. 2007; FitzJohn et al. 2009; FitzJohn 2010; Rabosky and McCune 2010; Goldberg et al. 2011; Ng and Smith 2014; Rabosky and Goldberg 2015; Beaulieu and O’Meara 2016; Rabosky and Huang 2016).
The influence of geography on lineage diversification, particularly the role of ecological opportunity mediated by biogeographic transitions, have been investigated in various taxa [e.g. plants (Hagen and Kadereit 2003; Moore and Donoghue 2007); lizards (Harmon et al. 2008; Pinto et al. 2008); snails (Parent and Crespi 2009); fish (Langerhans 2010); rodents (Schenk et al. 2013; Parada et al. 2015; Alhajeri et al. 2016); ants (Price et al. 2014a); mammals (Rolland et al. 2014); primates (Tran 2014)]. In muroids, with the exception of the primary colonization of South America, biogeographic transitions between continents do not seem to greatly influence diversification rates (Schenk et al. 2013; Alhajeri et al. 2016).
The association between lineage diversification rates with ecological and morphological traits have also been examined in various taxa [e.g. insects (Mitter et al. 1988; Rabosky and Matute 2013); plants (Farrell et al. 1991; Moore and Donoghue 2007; Goldberg et al. 2010; Igea et al. 2016); birds (Ricklefs 2006; Rabosky and Matute 2013; Price et al. 2014b); fishes (Collar et al. 2009); bats (Rojas et al. 2012); reptiles (Pyron and Burbrink 2014)]. As a consequence of adapting to widely different habitats worldwide, from tropical rainforest canopies to arctic tundra, muroids have highly variable ecological and morphological attributes (Nowak 1999; Musser and Carleton 2005), each with the potential to influence rates of speciation and/or extinction—this possibility has yet to be fully examined.
A common explanation for elevated diversification rates is the exploitation of relatively vacant ecological niches—this could occur via adaptive specialization (e.g. Sacks et al. 2008; Price et al. 2012; Rojas et al. 2012; Vamosi et al. 2014; Walker et al. 2014; Ebel et al. 2015; Tran 2016). Adaptive specialization could spur diversification rates by allowing specialists to outcompete generalists in certain niche axes, such as in resource acquisition (Wilson and Yoshimura 1994; Hunter 1998; Schluter 2000, 2001; Büchi and Vuilleumier 2014). However, under the “survival of the unspecialized” hypothesis, generalists are expected to have greater survival (lower extinction rates) than specialist species, presumably due to specialists being generally poor at adapting to changing conditions across time (Simpson 1944; Liow 2004). Thus, while specialized species may be more efficient in exploiting certain ecological niches (leading them to be more competitive), they can be more prone to extinction than the less competitive, but more flexible generalists (Simpson 1944). This trend could translate into higher diversification rates in generalists, if speciation rates are similar in generalists versus specialists.
Dietary specialization has been shown to be associated with higher diversification rates in various taxa [mammals (Price et al. 2012); bats (Rojas et al. 2012); reef fishes (Lobato et al. 2014); birds (Burin et al. 2016)], presumably through trophic niche subdivision. Price et al. (2012) found that in mammals, diversification rates were highest in herbivores (specialist), followed by carnivores (specialist), and lowest, omnivores (generalist). However, Cantalapiedra et al. (2014), found a different pattern in ruminants—where grazing and mixed-feeding (i.e. generalization) had higher diversification rates than browsing. When comparing the patterns of expected duration of mammalian dietary categories, Smits (2015) found evidence consistent with the “survival of the unspecialized” hypothesis, where generalist omnivores had an extinction risk lower than the specialist carnivores and herbivores. Because muroids have widely varied dietary habits (Stuart and Landry 1970; Nowak 1999; Martin et al. 2016), with omnivory considered to be the ancestral dietary condition of rodents (Stuart and Landry 1970), transitions to more specialized feeding modes could in theory impact their diversification rates, and partly explain diversification patterns in this superfamily.
By also reducing competition in poorly utilized niches, habitat use specializations such as arboreality or fossoriality, may increase diversification rates relative to the more generalized terrestrial (i.e. surface-dwelling) condition. For example, fossorial mammals exhibited elevated speciation rates that involve chromosomal rearrangements, presumably instigated by both population structure and isolation by distance (Corti and Loy 1987). In the case of the blind mole rats Spalax, their diversification has been argued to be partly a consequence to adaptively specializing to their subterranean niche (Nevo 1985). An alternative hypothesis is that animals do not disperse much after they colonize a subterranean habitat, and thus speciation occurs by multiple colonizations of subterranean habitats and subsequent isolation (i.e. vicariance) (Culver and Pipan 2014). On the other hand, in response to the Cenozoic climate change, arboreality may have led to increased extinction risk (relative to terrestriality) due to the replacement of closed habitats with open ones (Janis 1993; Blois and Hadly 2009)—Smits (2015) found evidence for this in the form of lower expected duration in arboreality relative to terrestriality. Moreover, arboreal mammals may exhibit lower speciation rates than terrestrial mammals through the association of this habitat preference with K-selected traits (Lowman and Rinker 2004). Because muroids vary greatly in habitat preferences, including species that live predominantly underground, on the surface, as well as above the surface (i.e. on trees) (Carleton and Musser 1984; Nowak 1999), this variation could in theory explain some of the diversification rate patterns in this group.
Ecological specialization is often associated with morphological specialization, which in turn could show a pattern of association with diversification rates. One measure of ecomorphological specialization—as defined by Liow (2004)—is the divergence of a focal character from the mean value of a given taxonomic group (i.e. the more divergent, the more specialized). Using this definition, Liow (2004) found that low extinction rates in crinoid taxa tend to be associated with decreased specialization in a suite of examined characters. As such, directional evolution in ecomorphology (towards or away from the mean of a given taxonomic group), could be associated with speciation and/or extinction rates in a continuous fashion. For example, dietary specialization is associated with overall dental morphology in murine rodents (Martin et al. 2016), as well as with both skull size and body mass in bats (Santana and Cheung 2016). In theory, the association between body size and diet is relatively straightforward—the larger the animal, the larger the prey size it can consume. Empirically, in terrestrial mammals, omnivorous mammals (generalists) tend to have low body mass, while both carnivorous and herbivorous mammals (both specialists) are associated with higher body masses (Pineda-Munoz et al. 2016).
In addition to being associated with diet, body size is related to many other life history traits—including productivity, clutch size, maturation time, gestation time, and many others (Blueweiss et al. 1978; Peters 1983; Swihart 1984; Read and Harvey 1989; Promislow and Harvey 1990; LaBarbera 1989; Gardezi and da Silva 1999; Isaac et al. 2005; Sibly and Brown 2007). Therefore, body size could potentially influence diversification rates of certain taxa. The association between speciation and/or extinction rates with body size has been demonstrated at various taxonomic scales—ranging from primates (Paradis 2005; Freckleton et al. 2008; FitzJohn 2010; Matthews et al. 2011), snakes (Feldman et al. 2016), and ray-finned fishes (Rabosky et al. 2013), to mammals (Monroe and Bokma 2009) and metazoans in general (Etienne et al. 2012). Body size could influence diversification rates several ways; for example, increased body size is associated with both increased speciation and extinction rates in mammals (Liow et al. 2008). In general, a positive association between body size and extinction rates is often attributed to decreased reproductive rates in large species (Liow et al. 2008, 2009), whereas a negative association could be explained by the former’s correlation with geographic range (Tomiya 2013).
Even before the advent of phylogenetic methods to test the association between body mass and diversification rates, many studies found an association between body mass and overall extant clade richness in many taxonomic groups (e.g. Hutchinson and MacArthur 1959; May 1986; Dial and Marzluff 1988; Kochmer and Wagner 1988; Martin 1992; Gittleman and Purvis 1998; Cardillo et al. 2005; Paradis 2005; Clauset and Erwin 2008; Freckleton et al. 2008; Wollenberg et al. 2011). Although the direction of the association between body size and diversity varies in different groups, an inverse correlation between body size and diversity is often attributed to the ability of small species to subdivide the environment more finely, permitting them to speciate and/or avoid extinction more readily (Dial and Marzluff 1988). Because muroid rodents vary nearly 1000-fold in body mass (Nowak 1999), some of their diversity patterns could be explained by size variation.
Habitat specialization is also associated with morphological variation. For example, different habitats are associated with different relative tail lengths, with arboreal mammals having relatively elongated tails that aid in balance (Lemen 1980; Fooden and Albrecht 1999; Hayssen 2008), while burrowers tend to have relatively short tails, because they are of little utility underground (Shimer 1903; Khanna and Yadav 2005; Withers et al. 2016). Tail length variation is also associated with various other characteristics, such as with locomotory mode (e.g. cursorial species use long tails for as a counter balance while hopping; Alexander and Vernon 1975; Mares 2009), and with thermoregulation (Little and Stoner 1968; Dawson and Keber 1979; Whittow 2013). The highly variable tail lengths in muroid rodents, because it illustrates ecomorphological specialization (0–150% of body length; Nowak 1999; Steppan unpubl. data) may influence some of the diversification rates in this group.
The main objective of this study is to test the association between muroid rodent diversification rates with ecomorphological specialization, specifically: (1) diet, (2) habitat, (3) body mass, and (4) relative tail length. These associations were tested using trait-dependent-diversification methods to determine the relationship between rates of speciation and extinction (along with transitions) with trait evolution (FitzJohn 2010, 2012). Another objective was testing the association between ecological specialization and morphological variation (i.e. diet vs. body mass, habitat vs. relative tail length). We further tested the propensity of several commonly-used trait-dependent diversification methods to yield Type I errors using our dataset.
Materials and Methods
Data Collection
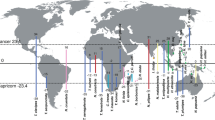
Ecological and morphological data were collected for each muroid species in the Schenk et al. (2013) species-level chronogram (Fig. 1, see the Electronic Supplementary Material Fig. S1). Schenk et al. (2013) chronogram, which was used in all the analyses below, included 297 of the over 1600 muroid species, where they sampled species proportional to total clade diversity.
Summarized version of Schenk et al. (2013) muroid rodent chronogram, indicating some of the major taxonomic groups. A full version of the chronogram, including all the tip labels appears in Fig. S1
Data on dietary habits and habitat use were assembled from Nowak (1999), field guides, as well as monographic sources from the literature (see the Electronic Supplementary Material Table S1). Based on each species predominant dietary preference, species were assigned to one of three dietary categories (carnivore, omnivore, or herbivore). These categories were similar to those that appear in Williams and Kay (2001), Samuels (2009), Pineda-Munoz and Alroy (2014), and Martin et al. (2016). Carnivores are species whose diet is composed mostly of animal material, including vertebrates, aquatic or terrestrial arthropods, worms, and other invertebrates. Omnivores are defined as species who consume plant and animal material at roughly equal amounts, whereas herbivores are defined as those whose diet is composed mostly plant or fungal material, including seeds, leaves, fruit, grass, mushrooms, and/or roots.
Species were also assigned to one of three habitat-use guilds (arboreal, terrestrial, or fossorial), using the same sources as indicated above (e.g. Nowak 1999) (see the Electronic Supplementary Material Table S1). Habitat categories refer to each species’ dominant (not exclusive) microhabitat use, refuge area, or general ‘habits’, defined as the tendency of each species to live in trees, on the ground, or underground, each of which is associated with a suite of behavioral, physiological, and ecomorphological adaptations, which allows each species to specialize to each lifestyle (see “Introduction”). Our habitat categories roughly correspond to Shimer’s (1903) “habit” categories, as well as to the “locomotor” categories in Smits (2015). We define arboreal species as those that spend much of their time above the surface, mostly in trees—these species are not necessarily exclusive to canopies, and may include those that move on the surface, but shelter in trees (i.e. includes ‘semi-arboreal’ species, see Withers et al. 2016). Terrestrial species are ‘ground-dwellers’ and are defined here as those that perform most of their activities on the ground’s surface, and those with little, to no climbing or burrowing ability (Withers et al. 2016). Fossorial species are those that are specialized for living underground, but are not necessarily fully subterranean, nor must they dig their own burrows, and may include those that perform some of their activities, such as foraging, on the surface (i.e. includes ‘semi-fossorial’ species, see Withers et al. 2016). Habitat use is a complex character, and the use of only three categories could obscure subtle variation or lump together distinctly different ecological conditions—reducing the power to find a correlation (see “Discussion”). However, subdividing habitat into additional categories necessarily reduces power by having too few species per category for accurate rate estimates.
The proportion of species sampled from each diet and habitat category appear in Table 1, and a detailed account of the diet, as well as the habitat of each species appears in the Electronic Supplementary Material Table S1.
In addition to diet and habitat, size (log-transformed adult body mass in grams) and relative tail length (ratio of head-body length to tail length, both in millimeters) were extracted from Alhajeri et al. (2016; see the Electronic Supplementary Material Table S2). For details on the collection of these ecomorphological data, see Alhajeri et al. (2016).
Association Between Ecological and Ecomorphological Specialization
The significance of size variation among dietary categories, and relative tail length variation among habitat categories, were both assessed using a standard analysis of variance (ANOVA) and phylogenetic ANOVA and visualized using boxplots. For the standard ANOVA, the significance of the pairwise differences in the means were determined via a post-hoc Tukey honest significant difference (HSD) test. In the phylogenetic ANOVA, the significance of the differences between the groups, as well as the post hoc pairwise comparisons of group means (with a Holm [1979] adjustment for multiple comparisons), were determined using 100,000 simulations (Garland et al. 1993). The standard ANOVA (including Tukey’s HSD test) was conducted with the base package in R (R Development Core Team 2016). The phylogenetic ANOVA and the associated post hoc tests and simulations were all conducted in the PHYTOOLS library (Revell 2012) in R.
Diet- and Habitat-Dependent Diversification: Multistate Approach
We first tested the association between diet and habitat data (with three states each) with diversification rates using the multistate speciation and extinction model (MuSSE; FitzJohn 2012), a generalization of the binary state speciation and extinction model (BiSSE; Maddison et al. 2007). Because trait-dependent diversification models tend to be poorly behaved when character states are missing (FitzJohn et al. 2010), we pruned species with missing data from the tree prior to analyses (Table 1; see the Electronic Supplementary Material Table S1). We then corrected for unsampled species in the phylogeny (‘skeleton tree’ correction, FitzJohn 2012) by using a sampling proportion equal to the total number of species in the phylogeny divided by the total number of muroid species (e.g. 266 sampled species out of ~ 1600 total muroid species for diet) following FitzJohn et al. (2009).
We compared the fit of eight nested models that test the relationship between traits and diversification rates—in all these models, transitions were only allowed between the generalist and the specialist conditions (i.e. carnivory ↔ omnivory ↔ herbivory; fossoriality ↔ terrestriality ↔ arboreality; see “Introduction”). (1) A fully constrained, character-independent (null) model, where speciation (λ), extinction (µ), and transition (q) rates for and between all character states are constrained to be the same (three parameters: one each for λ, µ, and q). (2) A five-parameter model, where only λ can vary (three parameters for λ, and one each for µ and q). (3) A five-parameter model, where only µ can vary (three parameters for µ, and one each for λ and q). (4) A six-parameter model, where only q can vary (four parameters for q, and one each for λ and µ). (5) A seven-parameter model, where both λ and µ can vary (three parameters each for λ and µ, and one for q). (6) An eight-parameter model, where both λ and q can vary (three parameters for λ, four for q, and one for µ). (7) An eight-parameter model, where both µ and q can vary (three parameters for µ, four for q, and one for λ). (8) A ten-parameter model, where all parameters can vary (three parameters each for λ and µ, and four for q). The MuSSE analyses were conducted in the DIVERSITREE library (FitzJohn 2012) in R.
For each of these models, we used the character-independent birth–death model to make a heuristic estimate of starting points for each parameter, a method that is reasonably successful in estimating the maximum likelihood (ML) point value (FitzJohn 2012). Model fit was assessed by comparing their corrected Akaike information criterion (AICc) scores using Akaike weights (wi), with the model receiving the highest wi considered to be the best fit (Akaike 1973, 1974; Bozdogan 1987; Burnham and Anderson 2002; Wagenmakers and Farrell 2004). AICc, ∆AICc, and wi were all computed in the QPCR package (Pabinger et al. 2014) in R.
State-dependent diversification models, including BiSSE, MuSSE, and the quantitative state speciation and extinction model (QuaSSE; FitzJohn 2010), have recently been criticized for having high Type I error rates (FitzJohn 2012; Machac 2014; Rabosky and Goldberg 2015; Rabosky and Huang 2016; but see Gamisch 2016). Therefore, the robustness of the MuSSE results were tested via ten randomized runs, where the same analyses were performed after the taxon labels (tips) were shuffled on the phylogeny, as implemented in the PICANTE library (Kembel et al. 2010) in R. The randomized analyses indicated that MuSSE might not be well suited for our dataset (see “Results”), therefore, the diet and habitat datasets were re-analyzed using alternative, less problematic approaches (see below).
Diet- and Habitat-Dependent Diversification: Binary Approaches
One such alternative to MuSSE to test the association of diet and habitat to diversification rates is the newly developed hidden state speciation and extinction model (HiSSE; Beaulieu and O’Meara 2016). HiSSE is a modified version of MuSSE that allows for the possibility that diversification rates may be influenced by an unobserved (i.e. hidden) trait, which addresses the major criticism of MuSSE (and BiSSE) by relaxing its assumption that all diversification rate variation is associated with the observed trait (e.g. diet or habitat) (Rabosky and Goldberg 2015; Beaulieu and O’Meara 2016; Gamisch 2016; Rabosky and Huang 2016). However, unlike MuSSE, the current implementation of HiSSE can only be conducted on binary traits. As such, we conducted pairwise comparisons between the generalist and the specialist conditions for each of diet (carnivory ↔ omnivory; omnivory ↔ herbivory) and habitat (fossoriality ↔ terrestriality; terrestriality ↔ arboreality). This means that for each analysis, species belonging to the unexamined specialist condition must be pruned from the tree prior to the analysis. As in the MuSSE analysis above, species with missing data were also pruned from the tree prior to the analyses–unsampled species were accounted for using the same method describe above.
The fit of the same eight nested models of trait-dependent diversification (described above for MuSSE) were compared. However, instead of λ and µ rates, HiSSE optimizes orthogonal transformations of these variables—net turnover (τ) and extinction fraction (ε) respectively. τ is defined as the sum of birth and death rates, whereas ε is defined as the death rate divided by the birth rate; both these changes reduce the problem of overfitting, which is encountered when birth and death rates are highly correlated (Beaulieu and O’Meara 2016). Moreover, in all the compared models, the default 12 parameter transition matrix have been modified to disallow (remove) dual transitions between both the observed trait and the hidden trait (q0A ↔ q1B), leading to an eight-parameter model, as recommended by the authors (Beaulieu and O’Meara 2016).
In addition to the eight variations of the HiSSE model described above, we also tested the fit of three associated models nested within HiSSE (each of which were tested for the same eight variations of HiSSE described above). (1) The BiSSE model, which is the same as HiSSE when the hidden state is absent (i.e. HiSSE is an expansion of BiSSE that adds a hidden state). In addition, variations of two character-independent diversification (CID) models were tested that assign diversification rates in such a way as to maximize the likelihood of the model, regardless of character states (i.e. unlinked to observed traits). These were (2) CID-2, a two-state trait independent HiSSE (“null-two”) model that has the same number of free parameters as BiSSE, and (3) CID-4, a four-state trait-independent HiSSE (‘null-four’) model with the same number of parameters as HiSSE. CID-4 assumes that τ and ε are linked between the two observed states. CID models provide more reasonable nulls for which to test the fit of the BiSSE and HiSSE models, over more traditional equal-rates models (which are also included), that force diversification rates to be constant across the entire tree. In this way, including CID models in the list of alternative models for comparison makes it much less likely to accept either trait-dependent model (i.e. BiSSE or HiSSE), and thus reduce the Type I error rate (Beaulieu and O’Meara 2016).
The same eight variations of the BiSSE, CID-2, and CID-4 models were used as the ones in HiSSE, for a total of 32 nested models compared (with the eight variations of the HiSSE model)—AICc scores were used to select the best of the 32 models. The HiSSE, BiSSE, CID-2, and the CID-4 model comparisons were conducted in the HISSE library (Beaulieu and O’Meara 2016) in R—for both diet and habitat.
An adaptive search procedure was used to sample the likelihood surface to generate a measure of the confidence for the parameters estimated in the best-fit model. We obtained confidence intervals by adaptively sampling 1000 points that are within two ln L units away from the ML estimate for each parameter.
Diet- and Habitat-Dependent Diversification: Structured Rate Permutations Approach
Finally, the association between diet and habitat with diversification rates was tested using Structured Rate Permutations on Phylogenies (STRAPP, Rabosky and Huang 2016). This semiparametric method can be applied to both binary and multistate discrete data, as well as to continuous traits. STRAPP differs from traditional state-dependent diversification methods in that diversification rates are first estimated across a phylogenetic tree independent of character states. The association between diversification rates and trait data is then tested by comparing the value of the empirically calculated test statistic of the association to a null distribution generated by permuting the evolutionary rates at the tips of the tree in structured fashion that maintains the location of the rate shift events in the phylogeny. We tested the association between both diet and habitat evolution with each of λ, µ, and r (net diversification = λ − µ) rates, both by treating them as multistate and as binary traits (see above).
Diversification rates across the tree were first estimated using Bayesian analysis of macroevolutionary mixtures (BAMM; Rabosky 2014), that uses reversible jump Markov chain Monte Carlo (rjMCMC) to find the locations for shifts in evolutionary dynamics that are maximally supported by the data, following the settings used in Alhajeri et al. (2016). In order to account for incomplete taxon sampling within muroid clades, we specified clade-specific sampling probabilities, as implemented in BAMM (Rabosky 2014) [see Alhajeri et al. (2016) for more information]. The starting parameters for the priors were estimated using the BAMMTOOLS library (Rabosky et al. 2014) in R. We ran the rjMCMC chain for 107 generations while sampling every 2000 steps, which yielded 5000 posterior samples, and the convergence of the chain was assessed using the CODA (Plummer et al. 2010) in R.
The STRAPP analysis was then applied to the BAMM diversification output after the first 10% of the rjMCMC chain posterior samples were discarded as burn-in, and the tips with missing data were removed. The Mann–Whitney U-test statistic was used to compare the diversification rates (each of λ, µ, and r) among the pairwise comparisons of diet and habitat (i.e. the binary analysis). The differences in the diversification rates in each pairwise comparison was tested by comparing the empirical value of the U-test statistic to a null distribution generated by a structured permutation of 100,000 randomly drawn posterior samples (with replacement). The significance of the differences in the diversification rates of the two different states of the binary trait is measured as the proportion of posterior samples with a U-test statistic value more extreme than those of its permuted sample (= the P value of the two-tailed test). The same process was repeated using a Kruskal–Wallis rank sum statistic, when diet and habitat were treated as multistate categorical traits.
Body Mass- and Relative Tail Length-Dependent Diversification
The association between body mass and relative tail length with diversification rates was first tested using the QuaSSE model (FitzJohn 2010). A flat null (character-independent) speciation rates likelihood function was contrasted against linear, step, sigmoidal, and modal functions, for each of log body mass and relative tail length. These list of alternative models, while not comprehensive, provided a reasonable set of hypotheses to test against the null. The linear, step, and sigmoidal functions could be an indication of higher speciation rates for certain specialists (i.e. most divergent from the mean log body mass or relative tail length), whereas the modal function could indicate that the highest speciation rates occurs for the generalists (i.e. closer to the phylogenetic mean). For each model, fit was tested both with the drift parameter (µd; an estimate of the directional tendency of Brownian motion evolution) allowed to vary, and with a constrained µd. In all models, the extinction rate was set as constant (constrained), and only the speciation rate can vary. QuaSSE requires the user to specify a universal or a tip-specific standard error around the mean of the traits (an estimate of measurement error)—because our data were average values obtained from the literature, we set a universal standard error to the lowest possible value (= 0.0001). As in MuSSE (see above): (1) a character-independent birth–death model was used to make a heuristic estimate of starting points for each parameter for each model, (2) the models were compared using the AICc scores and wi, and (3) a ‘skeleton tree’ correction was applied to account for unsampled species in the phylogeny, using the sane sampling proportion discussed above. QuaSSE analyses were conducted in the DIVERSITREE library (FitzJohn 2012) in R.
Like MuSSE, QuaSSE has been shown to be subject to high Type I error rates (FitzJohn 2012; Machac 2014; Rabosky and Goldberg 2015; Rabosky and Huang 2016; but see Gamisch 2016), and thus the robustness of the QuaSSE results were tested via ten randomized runs, using the same scheme as discussed above for MuSSE. The randomized analyses indicated that QuaSSE too might not be well suited for our data (see “Results”), for this reason, the body mass and the relative tail length data were re-analyzed using an alternative approach (see below).
The association between body mass and relative tail length with diversification rates was also tested using STRAPP (Rabosky and Huang 2016), using the same BAMM diversification output discussed above for diet and habitat. We used the Spearman correlation coefficient as a test statistic for the relationship between diversification rates (each of λ, µ, and r) and each of body mass and relative tail length values. The null distribution of the test statistic was constructed using structured block permutations of evolutionary rates across 100,000 samples from the BAMM posterior on a phylogenetic tree. The significance of the relationship was assessed by tabulating the percentage of the 100,000 permutations where the null correlation was more extreme than the observed correlation (i.e. a two-tailed test).
Results
Association Between Ecology and Morphological Specialization
The standard ANOVA indicated a significant difference in the body size between dietary categories (F = 6.39, P = 0.0019), however the significance was marginal after phylogenetic correction (F = 6.38, P = 0.0578). Tukey’s HSD test indicated that herbivores were significantly larger than omnivores (P = 0.0013), whereas there was no significant difference between carnivores and herbivores (P = 0.2203) or between carnivores and omnivores (P = 0.8473). Phylogenetically corrected post-hoc analyses further indicated that all pairwise comparisons were not significant (P = 0.0926–0.6648).
The standard ANOVA indicated a significant difference in relative tail length between habitats (F = 37.84, P < 0.0001), a result that was retained after phylogenetic correction (F = 37.83, P < 0.0001; data not shown). Tukey’s HSD test indicates that arboreal species had relative tail lengths that were significantly greater than both terrestrial (P < 0.0001) and fossorial (P < 0.0001) species, and that terrestrial species had relative tail lengths that were significantly greater than fossorial species (P < 0.0001). Phylogenetically corrected post-hoc analyses indicated that all pairwise comparisons were significant (all P < 0.0019), and in the same direction discussed above for Tukey’s HSD.
Diet- and Habitat-Dependent Diversification
Among the eight nested MuSSE models of the association between the three dietary specializations with diversification rates, two models received 90.9% of the total weight (see Electronic Supplementary Material Table S3a). The eight-parameter model (λ and q can vary) seem to fit the data best (ΔAICc = 0.00, wi = 0.670) followed by the eight-parameter model (µ and q can vary) (ΔAICc = 2.06, wi = 0.239, see the Electronic Supplementary Material Table S3a). When tip data were randomized, the same eight-parameter model, where both λ (speciation rates) and q (transition rates) can vary, fit the data best (wi = 0.609–0.881, see the Electronic Supplementary Material Table S3b–k), followed by either the eight-parameter model, where both µ (extinction rates) and q can vary (ΔAICc = 1.56, wi = 0.279, see the Electronic Supplementary Material Table S3b), or the ten parameter model (all parameters can vary) (ΔAICc = 4.00, wi = 0.116–0.119, see the Electronic Supplementary Material Table S3c–k).
Among the eight nested MuSSE models of the association between the three habitat categories with diversification rates, two models received 99.8% of the total weight (see the Electronic Supplementary Material Table S4a). The eight-parameter model (λ and q can vary) fit the data best (wi = 0.721) followed by the ten-parameter model (all parameters can vary) (ΔAICc = 1.92, wi = 0.277, see the Electronic Supplementary Material Table S4a). When the tip data were randomized, for nine out of the ten randomizations, the same eight-parameter model fit the data best (wi = 0.570–0.990), followed by the same ten-parameter model (ΔAICc = 0.57–9.33, wi = 0.009–0.429, see the Electronic Supplementary Material Table S4b–c, e–k). In the tenth tip-randomized data, the eight-parameter model (µ and q can vary) fit the data best (wi = 0.725) followed by the eight-parameter model (λ and q can vary) (ΔAICc = 3.18, wi = 0.148, see the Electronic Supplementary Material Table S4d).
The fact that both the observed data and the tip-randomized data for both diet and habitat favored the same or equally complex models indicated that MuSSE may not be well suited for our data. As such, although λ, µ, and q rates varied across both diet and habitat groups in their respective best-fit model (of the empirical data), we do not report nor describe the parameter estimates of the best models (but see Alhajeri 2014 for those results).
Of the 32 nested HiSSE models (including BiSSE, CID-2, and CID-4), for the binary association between diversification rates and omnivorous versus carnivorous diet, the four-parameter CID-2 model, where τ (net turnover) can vary, fit the data best (wi = 0.331, Table 2a). This model received much more support that all character-dependent BiSSE and HiSSE models, none of which had a wi greater than 0.04 (they all sum up to 0.053). Of the remaining character-independent models, the second-best fit was the nine-parameter CID-4 model (τ and q can vary) (ΔAICc = 0.96, wi = 0.193, Table 2a). This result suggests that trait-independent diversification may explain diversification patterns better than the pairwise difference in omnivorous versus carnivorous diet rates.
For the binary association between diversification rates and omnivorous versus herbivorous diet, the 13-parameter HiSSE model, where ε (extinction fraction) and q can vary, fit the data best (wi = 0.671, Table 2b). This model was a marked improvement over all the other variations of the HiSSE model, as well as all the included BiSSE and CID-4 models, none of which had a wi greater than 0.001. The second-best model was the 11-parameter CID-2 model (τ and q can vary; ε constrained) (ΔAICc = 1.94, wi = 0.254, Table 2b). The best-fit HiSSE model suggested that a hidden state nested within some of the omnivorous lineages was associated with reduced net diversification rates (romnivores+ = 0.136, support region: [0.075,0.187]). The model indicates that omnivorous lineages not associated with the reduced-diversification hidden state had a net diversification rate similar to that of herbivorous lineages (romnivores− = rherbivores− = 0.771, support region: [0.694, 0.905]). On the other hand, herbivorous lineages associated with the reduced-diversification hidden state also had reduced diversification rates (rherbivores+ = 0.024, support region: [− 0.024, 0.057]), when compared to the herbivorous lineages showing the alternative hidden state, suggesting strong rate heterogeneity. Overall, based on the comparisons of the net-diversification rates of dietary categories, we can conclude that dietary transitions alone are not strongly correlated with diversification patterns in muroid rodents.
Of the 32 compared nested variations of the HiSSE model (including BiSSE, CID-2, and CID-4), for the binary association between diversification rates and terrestriality versus arboreality, the 11-parameter CID-2 model (ε and q can vary) fit the data best (wi = 0.800, Table 3a). This model indicated that trait-independent diversification explained diversification patterns better than the pairwise difference in terrestriality versus arboreality.
For the binary association between diversification rates and terrestriality versus fossoriality, the 13-parameter HiSSE model (τ and q can vary; ε constrained) was by far the best fitting model (wi = 0.934, Table 3b). The parameters estimated under this model indicated a generally low net diversification rates in both terrestrial (rterrestrial− < 0.001, support region: [< 0.001, 0.002]) and fossorial lineages (rfossorial− = 0.163, support region: [0.139, 0.190]). However, a hidden state nested within some of the lineages led to a marked increase in the diversification rates of both habitat categories, but more so in fossorial lineages (rfossorial+ = 0.920, support region: [0.788, 1.122]) when compared with the terrestrial lineages (rterrestrial+ = 0.479, support region: [0.416, 0.557]), also suggesting strong rate heterogeneity. Based on the comparisons of the net-diversification rates of habitat categories, we can conclude that habitat transitions, without the consideration of associated hidden states, do not clearly explain variation in diversification rate patterns in muroid rodents.
The STRAPP permutation test indicated that when treating both diet and habitat as binary traits, in both pairwise comparisons, no significant differences between rates with respect to each character state was found for any of λ rates (all P > 0.233), µ rates (all P > 0.402), and r rates (all P > 0.724, Table 4a). Similarly, when both diet and habitat categories were treated as multistate categorical traits, the STRAPP permutation test recovered no significant differences between rates with respect to each character state for each of λ rates (both P > 0.365), µ rates (both P > 0.542), and r rates (both P > 0.355, Table 4b).
Body Mass- and Relative Tail Length-Dependent Diversification
Of the ten nested QuaSSE models of the association between body mass and λ rates, the seven-parameter modal model that includes a drift parameter fit the data best (wi = 0.881) followed by the six-parameter modal model that does not include drift (ΔAICc = 4.04, wi = 0.117, see the Electronic Supplementary Material Table S5a). When the tip data were randomized, the same two models received the highest wi (see the Electronic Supplementary Material Table S5b–k). Similarly, of the ten nested QuaSSE models of the association between relative tail length with λ rates, the seven-parameter modal model that included drift fit the data best (wi = 0.901) followed by the six-parameter modal model that did not (ΔAICc = 4.70, wi = 0.086, see the Electronic Supplementary Material Table S6a). When the tip data were randomized, the same two models received the highest wis (see the Electronic Supplementary Material Table S6b–k).
Thus, the QuaSSE outcome was similar to that of MuSSE—nearly identical results for empirical and tip-randomized data, both of which prefer complex trait-dependent λ models over the null, trait-independent λ model. Consequently, the QuaSSE results also indicated that this method may not be well suited for our data. Therefore, we do not report nor describe the parameter estimates of the best models (but see Alhajeri 2014).
In contrast, the STRAPP permutation test recovered no significant correlations between the estimated log body mass and relative tail length evolution with each of λ rates (both P > 0.676), µ rates (both P > 0.568), and r rates (both P > 0.679, Table 4c).
Discussion
Specialization is not Associated with Diversification Rates in Muroid Rodents
After correcting for phylogenetic relationships, we found no significant differences in log body mass between dietary categories, a result that was in contrast to both bats (examined at the order level, Santana and Cheung 2016) and mammals (examined at the class level, Pineda-Munoz et al. 2016). This discrepancy may be explained by the application of the phylogenetic correction, because without this correction, we did find significant differences among the dietary categories. However, for habitat preference categories, significant differences in relative tail length were detected both without and with phylogenetic correction. This result is concordant with the general observation that arboreal mammals have elongated tails that aid in balance in the canopy (Lemen 1980; Fooden and Albrecht 1999; Hayssen 2008) while fossorial mammals have reduced tails, that are of little utility underground (Shimer 1903; Khanna and Yadav 2005; Withers et al. 2016).
Both when ecological specialization was examined directly (diet and habitat) or indirectly through morphological specialization (body mass and relative tail length), we find that specialization is not associated with diversification rates in muroid rodents. This contrasts with previous results in other taxa, where most of these traits have been associated with diversification rates and general extant diversity.
The general pattern that emerged was that when ecological trait categories (diet and habitat) were coded as multi-state characters and analyzed using MuSSE, an association with diversification rates was found—an association that was maintained even when tip data were randomized. Likewise, when continuous ecomorphological data (log body mass and relative tail length) were analyzed using QuaSSE, an association was found with speciation rates, both in empirical and in tip-randomized data. These results corroborate the criticisms raised about this class of state-dependent speciation and extinction models (i.e. BiSSE, MuSSE, QuaSSE), regarding their inflated Type I error rates (FitzJohn 2012; Machac 2014; Rabosky and Goldberg 2015; Rabosky and Huang 2016; but see Gamisch 2016). More specifically, Rabosky and Goldberg (2015) found that for neutral traits simulated on real phylogenetic trees, diversification rate shifts that are independent of the focal trait frequently led to elevated false positive associations between the focal trait and diversification rates. Beaulieu and O’Meara (2016) argued that this shortcoming is mostly attributable to the commonly used unrealistically trivial null models that assume that all character states have equal diversification rates—not taking into account the possibility of complex CID. In response, Beaulieu and O’Meara (2016) recently developed more complex character-independent null models (CID-2 and CID-4), adding the possibility that a “hidden” unmeasured factor may impact diversification dynamics of the observed character states (the HiSSE model), which relaxes BiSSE’s assumption that all diversification rate heterogeneity is associated with the focal trait.
Accordingly, when the same two traits (diet and habitat) were reanalyzed more conservatively using pairwise comparisons of diversification rates using more conservative models confirmed that the MuSSE results were false-positives. None of the pairwise analyses preferred a strictly character-dependent model of diversification (i.e. BiSSE)—CID-2 and HiSSE models fit the data best for the two pairwise dietary comparisons, as well as the two pairwise habitat comparisons, suggesting either purely trait-independent diversification, or diversification that is influenced by an unmeasured hidden state.
The more conservative STRAPP permutation test, that unlike HiSSE, also addresses the issue of phylogenetic pseudoreplication (Rabosky and Huang 2016) and can be used to analyze multistate and continuous data, likewise found no significant associations with diversification rates in ecology or morphology. Therefore, taken together, the tests indicate no significant association between diversification rates and ecological/ecomorphological specialization in muroid rodents.
Methodology Could Explain the Lack of Associations with Diversification Rates
One possible explanation for the lack of association between the examined traits with diversification rates is methodological limits. For example, the failure to detect significant trait-dependent diversification using STRAPP could reflect low statistical power, because the power to detect diversification rate shifts using BAMM depends on the size of the phylogeny (Rabosky and Huang 2016). Thus, while MuSSE may be prone to type I error inflation (see above), the more conservative method, STRAPP, could be susceptible to type II error, which is something that we do not directly test in this study. However, STRAPP was not the only method used in this study, and its results agreed with those from other methods.
While not associated in muroid rodents, diet and diversification rates seem to be associated in almost all the recently published studies—including in birds (Burin et al. 2016), reef fishes (Lobato et al. 2014), bats (Rojas et al. 2012), ruminants (Cantalapiedra et al. 2014), and in the class Mammalia (Price et al. 2012). One possible explanation for the lack of association in muroid rodents, is the different methods used—for example, most of the aforementioned studies that report an association were conducted using MuSSE (i.e. Price et al. 2012; Rojas et al. 2012; Cantalapiedra et al. 2014; Burin et al. 2016), and most without testing for Type I errors as we did (but see Burin et al. 2016 for an exception). For example, Rojas et al. (2012) used a 60-tip genus level phylogeny of phyllostomid bats, that they grouped into four dietary categories, and found that the group with the lowest diversity (the most specialized form, the strictly frugivorous genera; representing 15% of the total diversity) had the highest diversification rates. We found a similar result to Rojas et al. (2012) when we used MuSSE, with the least common diet (carnivory, 10% of species) and habitat (arboreal, 18%) categories having the highest diversification rates (3–4 times greater than the most common state; Fig. 2). Our apparently incongruous result was partially explained by very high transition rates out of carnivory and habitat, wherein the model essentially concluded that carnivores, for example, diversified most rapidly, but even more rapidly evolved to other diets, leaving isolated terminal clades of low diversity. When we used more conservative methods (HiSSE and STRAPP), the diet-dependent diversification signal that was detected by MuSSE disappeared. While we could not find studies that compare diversification rates among species that occupy different habitats (arboreal, terrestrial, or fossorial) using MuSSE, we expect that our analysis of this trait would follow the same pattern as that for diet. We suggest that researchers consider the transition rates in more detail rather than focusing exclusively on the diversification rates, and while some Type I errors may result from overestimated transition rates, it is also possible that those results reflect biologically important asymmetries in those rates. We also suggest reexamining some published studies based on state-dependent speciation models.
Posterior probability distributions of speciation rates estimated by MuSSE for a diet and b habitat. Distributions based on a 10,000 step MCMC simulation. Bars below the distributions indicate the 95% credibility intervals. Similar results are obtained with randomized-tips data and are likely spurious
Far fewer studies have used QuaSSE on morphological data. The two studies that that we could find that test for the association between body size and diversification rates using QuaSSE both find a significant association (primates, FitzJohn 2010; snakes, Feldman et al. 2016). Both studies found that a modal function fit the data best for the association between speciation and log body mass—the same as our results in muroid rodents. QuaSSE has been criticized for the same high propensity for Type I error rates (e.g. FitzJohn 2012; Machac 2014; Rabosky and Goldberg 2015; Rabosky and Huang 2016) as MuSSE. We confirmed this with the tip-randomizations, that yielded similar significant results as the empirical data. However, the association between body size and diversification has also been shown to be significant using various other methods [e.g. primates (Paradis 2005; Freckleton et al. 2008; Matthews et al. 2011); ray-finned fishes (Rabosky et al. 2013); mammals (Monroe and Bokma 2009; metazoans; Etienne et al. 2012)]. We could not find studies that analyzed relative tail lengths using QuaSSE for comparisons.
The lack of trait-dependent diversification signal detected in this study could also be a consequence of trait-category classifications (i.e. carnivore, omnivore, or herbivore; and arboreal, terrestrial, or fossorial). In other words, when examining the same trait (e.g. diet) different studies use different categories in which to divide taxa. For example, Rojas et al. (2012) used four categories to divide bats (animalivore, sanguinivore, nectarivore, and frugivore), while Cantalapiedra et al. (2014) used three categories for ungulates (browser, mixed feeder, and grazer). Alternatively, diet can be divided into finer categories than used in this study—for example, within herbivores, granivores can be separated into those that consume soft and/or abrasive vegetable matter; and within carnivores, insectivores can be separated from those that specialize on worms. Similarly, omnivory can be subdivided by the proportion of animal versus plant material (e.g. Martin et al. 2016). Some categorizations may not isolate the critical factor. The same reasoning can be applied to how habitat is coded—e.g. fully subterranean can be separated from fossorial.
Finally, an alternative mode of relationship between ecomorphology and diversification rate may be beyond the state-dependent effect experienced by each species, but instead be that the evolvability of traits may affect diversification rates. For example, clades that contain species who can adapt more quickly might have be more prone to speciate or less susceptible to extinction. Trait evolvability, however, is beyond the scope of this study.
Biological Explanations for Lack of Associations with Diversification Rates
Assuming that methodology alone does not explain the lack of association between the examined traits with diversification rates, we can explain some of the results in biological terms—real differences in the ecology of muroid rodents (and rodents in general) when compared with clades examined in previous studies. While we divided muroids into general diet and habitat categories, the lack of signal for diet-dependent and habitat-dependent diversification in muroids could be explained by the overall opportunism of this group. Thus, while it was necessary to divide species into discrete categories (in order to test their association with diversification rates), based on their primary diet and habitat use, the general opportunistic behavior of this group means that many of the examined species can freely switch categories, depending on the situation.
For example, despite most rodents being herbivores and omnivores, nearly all will consume animal matter opportunistically (Stuart and Landry 1970), and many rodents show seasonal variation in diet, with herbivores consuming insects at certain times of the year, and insectivores supplementing their diet with vegetation and other material (Nowak 1999; Samuels 2009). Similarly, many rodents show generalized habitat use (Nowak 1999)—thus, even species that we assigned to the terrestrial and arboreal habitat categories (they primarily use these habitats), do show some burrowing behavior (Nowak 1999; Deacon 2006), though not as extensive as those that we categorize as fossorial (nor those that others classify as subterranean). The generality of flexibility of rodents’ diet and habitat use (behavioral plasticity) is exemplified by their generalized and conserved body plan and feeding apparatus (Stuart and Landry 1970; Samuels 2009; Verde Arregoitia et al. 2017). The opportunism and flexibility of muroid rodents could dilute the signal for diet- and habitat-dependent diversification, as there is no discrete transition to a novel ecological category, but rather, small shifts in preference or behavior.
The independence between body mass and relative tail length with diversification rates is supported by fewer analyses than diet- and habitat-dependent diversification, and thus no strong inference can be made concerning the lack of association in muroid rodents. This is especially the case for body mass, considering that the association between size and diversification rates is an ubiquitous phenomenon, and supported by a wide range of analyses that differ from the present study. Moreover, there is a robust theoretical foundation to explain an association between diversification rates and body size—since this trait is associated with generation time and population size (Peters 1983; Gardezi and Silva 1999; Isaac et al. 2005) as well as the ability to subdivide the environment (Dial and Marzluff 1988). Therefore, it is particularly surprising that we do not find an effect of size given the nearly 1000-fold range in mass, but perhaps the relationship is less pronounced at the small extreme of mammalian variation, which is occupied by muroids. Still, it remains true that despite the lack of strong evidence for the influence of size, diet, habitat (this study) or even geographic opportunity (Schenk et al. 2013) to explain diversification rate shifts in muroid rodents, muroids remain exceptionally diverse among mammals and contain significant diversification rates within them (Schenk et al. 2013).
Conclusion
Overall, muroid rodent lineage diversification rates were not associated with the evolution of diet, habitat preference, log body mass, nor relative tail length—traits that theoretically could enhance diversification rates by reducing crowding in niche space in specialized lineages. Therefore, it seems that the diversification dynamics that have led to the uneven clade diversity in muroid rodents may have been directed by other mechanisms. Moreover, the diversification of muroid rodents, as in other clades, is a complex process, that is likely influenced by various processes working simultaneously, that could not be clearly detected when examining each trait in isolation (as hinted at by the HiSSE results).
Despite the negative result recovered by this study, it still serves a valuable function as to eliminate some common traits from consideration in the long quest to explain the unique diversification dynamics of the most specious superfamily of mammals. Moreover, this study could serve as a case study in the importance of testing trait-dependent diversification using more than one method, as method conservativism clearly has the potential to drastically influence the resulting outcome—where less conservative methods could have arguable led to spurious associations of traits with diversification rates in previous studies. This is especially exemplified in our analysis of diet-dependent diversification, where to our knowledge, no previously published study found a lack of association between diet and diversification rates; this outcome was largely dependent on the use of newer, more conservative trait-dependent diversification approaches, as the use of a less conservative method led to a (arguably spurious) association, matching previous studies.
Data Accessibility
Trait data was uploaded to the electronic supplementary material.
References
Akaike, H. (1973). Information theory and an extension of the maximum likelihood principle. In B. N. Petrov & F. Csaki (Eds.), 2nd International symposium on information theory (pp. 267–281). Budapest: Akademiai Kiado.
Akaike, H. (1974). A new look at statistical model identification. IEEE Transactions on Automatic Control, 19, 716–723.
Alexander, R. M., & Vernon, A. (1975). The mechanics of hopping by kangaroos (Macropodidae). Journal of Zoology, 177(2), 265–303. https://doi.org/10.1111/j.1469-7998.1975.tb05983.x.
Alhajeri, B. H. (2014). Adaptation, diversification, and desert ecology of the most diverse order of mammals (Mammalia, Rodentia). Tallahassee, FL: Department of Biological Science, Florida State University.
Alhajeri, B. H., Schenk, J. J., & Steppan, S. J. (2016). Ecomorphological diversification following continental colonization in muroid rodents (Rodentia: Muroidea). Biological Journal of the Linnean Society, 117(3), 463–481. https://doi.org/10.1111/bij.12695.
Beaulieu, J. M., & O’Meara, B. C. (2016). Detecting hidden diversification shifts in models of trait-dependent speciation and extinction. Systematic Biology, 65(4), 583. https://doi.org/10.1093/sysbio/syw022.
Blois, J. L., & Hadly, E. A. (2009). Mammalian response to Cenozoic climatic change. Annual Review of Earth and Planetary Sciences, 37, 181–208. https://doi.org/10.1146/annurev.earth.031208.100055.
Blueweiss, L., Fox, H., Kudzma, V., Nakashima, D., Peters, R., & Sams, S. (1978). Relationships between body size and some life history parameters. Oecologia, 37(2), 257–272. https://doi.org/10.1007/BF00344996.
Bozdogan, H. (1987). Model selection and Akaike’s information criterion (AIC): The general theory and its analytical extensions. Psychometrika, 52(3), 345–370. https://doi.org/10.1007/BF02294361.
Büchi, L., & Vuilleumier, S. (2014). Coexistence of specialist and generalist species is shaped by dispersal and environmental factors. The American Naturalist, 183(5), 612–624. https://doi.org/10.1086/675756.
Burin, G., Kissling, W. D., Guimarães, P. R. Jr, Şekercioğlu, Ç. H., & Quental, T. B. (2016). Omnivory in birds is a macroevolutionary sink. Nature Communications, 7, 11250. https://doi.org/10.1038/ncomms11250.
Burnham, K. P., & Anderson, D. R. (2002). Model selection and multimodel inference: A practical information-theoretic approach. New York: Springer.
Cantalapiedra, J. L., Fitzjohn, R. G., Kuhn, T. S., Fernández, M. H., DeMiguel, D., Azanza, B., et al. (2014). Dietary innovations spurred the diversification of ruminants during the Caenozoic. Proceedings of the Biological Sciences/The Royal Society, 281(1776), 20132746. https://doi.org/10.1098/rspb.2013.2746.
Cardillo, M., Mace, G. M., Jones, K. E., Bielby, J., Bininda-Emonds, O. R. P., Sechrest, W., et al. (2005). Multiple causes of high extinction risk in large mammal species. Science, 309(5738), 1239–1241. https://doi.org/10.1126/science.1116030.
Carleton, M. D., & Musser, G. G. (1984). Muroid rodents. In S. Anderson & J. K. Jones Jr. (Eds.), Orders and families of recent mammals of the world (pp. 289–379). New York: Wiley.
Clauset, A., & Erwin, D. H. (2008). The evolution and distribution of species body size. Science, 321(5887), 399–401. https://doi.org/10.1126/science.1157534.
Collar, D. C., O’Meara, B. C., Wainwright, P. C., & Near, T. J. (2009). Piscivory limits diversification of feeding morphology in centrarchid fishes. Evolution, 63(6), 1557–1573. https://doi.org/10.1111/j.1558-5646.2009.00626.x.
Corti, M., & Loy, A. (1987). Morphometric divergence in southern European moles (Insectívora, Talpidae). Bolletino di Zoologia, 54(2), 187–191. https://doi.org/10.1080/11250008709355580.
Culver, D. C., & Pipan, T. (2014). Shallow Subterranean habitats: Ecology, evolution, and conservation. Oxford: Oxford University Press.
Dawson, N. J., & Keber, A. W. (1979). Physiology of heat loss from an extremity: The tail of the rat. Clinical and Experimental Pharmacology and Physiology, 6(1), 69–80. https://doi.org/10.1111/j.1440-1681.1979.tb00009.x.
Deacon, R. M. J. (2006). Burrowing in rodents: A sensitive method for detecting behavioral dysfunction. Nature Protocols, 1(1), 118–121. https://doi.org/10.1038/nprot.2006.19.
Dial, K. P., & Marzluff, J. M. (1988). Are the smallest organisms the most diverse? Ecology, 69(5), 1620–1624. https://doi.org/10.2307/1941660.
Ebel, E. R., DaCosta, J. M., Sorenson, M. D., Hill, R. I., Briscoe, A. D., Willmott, K. R., et al. (2015). Rapid diversification associated with ecological specialization in Neotropical Adelpha butterflies. Molecular Ecology, 24(10), 2392–2405. https://doi.org/10.1111/mec.13168.
Etienne, R. S., de Visser, S. N., Janzen, T., Olsen, J. L., Olff, H., & Rosindell, J. (2012). Can clade age alone explain the relationship between body size and diversity? Interface Focus, 2(2), 170–179.
Fabre, P. H., Hautier, L., Dimitrov, D., Douzery, P., & Emmanuel, J. (2012). A glimpse on the pattern of rodent diversification: a phylogenetic approach. BMC Evolutionary Biology, 12. https://doi.org/10.1186/1471-2148-12-88
Farrell, B. D., Dussourd, D. E., & Mitter, C. (1991). Escalation of plant defense: Do latex and resin canals spur plant diversification?. The American Naturalist, 138(4), 881–900.
Feldman, A., Sabath, N., Pyron, R. A., Mayrose, I., & Meiri, S. (2016). Body sizes and diversification rates of lizards, snakes, amphisbaenians and the tuatara. Global Ecology and Biogeography, 25(2), 187–197. https://doi.org/10.1111/geb.12398.
FitzJohn, R. G. (2010). Quantitative traits and diversification. Systematic Biology, 59(6), 619–633. https://doi.org/10.1093/sysbio/syq053.
FitzJohn, R. G. (2012). Diversitree: Comparative phylogenetic analyses of diversification in R. Methods in Ecology and Evolution, 3(6), 1084–1092. https://doi.org/10.1111/j.2041-210X.2012.00234.x.
FitzJohn, R. G., Maddison, W. P., & Otto, S. P. (2009). Estimating trait-dependent speciation and extinction rates from incompletely resolved phylogenies. Systematic Biology, 58(6), 595–611. https://doi.org/10.1093/sysbio/syp067.
Fooden, J., & Albrecht, G. H. (1999). Tail-length evolution in fascicularis -group Macaques (Cercopithecidae: Macaca). International Journal of Primatology, 20, 431–440.
Freckleton, R. P., Phillimore, A. B., & Pagel, M. (2008). Relating traits to diversification: A simple test. The American Naturalist, 172(1), 102–115. https://doi.org/10.1086/588076.
Gamisch, A. (2016). Notes on the statistical power of the binary state speciation and extinction (BiSSE) model. Evolutionary Bioinformatics, 12, 165–174. https://doi.org/10.4137/EBO.S39732.
Gardezi, T., & da Silva, J. (1999). Diversity in relation to body size in mammals: A comparative study. The American Naturalist, 153(1), 110–123. https://doi.org/10.1086/303150.
Garland, T., Dickerman, A. W., Janis, C. M., & Jones, J. A. (1993). Phylogenetic analysis of covariance by computer simulation. Systematic Biology, 42(3), 265–292. https://doi.org/10.1093/sysbio/42.3.265.
Gittleman, J. L., & Purvis, A. (1998). Body size and species-richness in carnivores and primates. Proceedings of the Biological sciences/The Royal Society, 265(1391), 113–119. https://doi.org/10.1098/rspb.1998.0271.
Goldberg, E. E., Kohn, J. R., Lande, R., Robertson, K. A., Smith, S. A., & Igić, B. (2010). Species selection maintains self-incompatibility. Science, 330(6003), 493–495. https://doi.org/10.1126/science.1194513.
Goldberg, E. E., Lancaster, L. T., & Ree, R. H. (2011). Phylogenetic inference of reciprocal effects between geographic range evolution and diversification. Systematic Biology, 60(4), 451–465. https://doi.org/10.1093/sysbio/syr046.
Harmon, L. J., Melville, J., Larson, A., & Losos, J. B. (2008). the role of geography and ecological opportunity in the diversification of day geckos (Phelsuma). Systematic Biology, 57(4), 562–573. https://doi.org/10.1080/10635150802304779.
Hayssen, V. (2008). Patterns of body and tail length and body mass in Sciuridae. Journal of Mammalogy, 89(4), 852–873. https://doi.org/10.1644/07-MAMM-A-217.1.
Holm, S. (1979). A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics, 6(2), 65–70. https://doi.org/10.2307/4615733.
Hunter, J. P. (1998). Key innovations and the ecology of macroevolution. Trends in Ecology & Evolution, 13, 31-36.
Hutchinson, G. E., & MacArthur, R. A. (1959). A theoretical ecological model of size distributions among species of animals. American Naturalist, 93, 117–125.
Igea, J., Miller, E. F., Papadopulos, A. S. T., & Tanentzap, A. J. (2016). Seed size drives species diversification across angiosperms. bioRxiv. http://biorxiv.org/content/early/2016/05/12/053116.abstract.
Isaac, N. J., Jones, K. E., Gittleman, J. L., & Purvis, A. (2005). Correlates of species richness in mammals: Body size, life history, and ecology. The American Naturalist, 165(5), 600–607.
Janis, C. M. (1993). Tertiary mammal evolution in the context of changing climates, vegetation, and tectonic events. Annual Review of Ecology and Systematics, 24, 467–500. https://doi.org/10.1146/annurev.es.24.110193.002343.
Kembel, S. W., Cowan, P. D., Helmus, M. R., Cornwell, W. K., Morlon, H., Ackerly, D. D., et al. (2010). Picante: R tools for integrating phylogenies and ecology. Bioinformatics, 26(11), 1463–1464. https://doi.org/10.1093/bioinformatics/btq166.
Khanna, D. R., & Yadav, P. R. (2005). Biology of mammals. New Delhi: Discovery Publishing House.
Kochmer, J. P., & Wagner, R. H. (1988). Why are there so many kinds of passerine birds? Because they are small. a reply to raikow. Systematic Biology, 37(1), 68–69. https://doi.org/10.2307/2413193.
LaBarbera, M. (1989). Analyzing body size as a factor in ecology and evolution. Annual Review of Ecology and Systematics, 20(1), 97–117. https://doi.org/10.1146/annurev.es.20.110189.000525.
Langerhans, R. B. (2010). Predicting evolution with generalized models of divergent selection: A case study with Poeciliid Fish. Integrative and Comparative Biology, 50(6), 1167–1184. https://doi.org/10.1093/icb/icq117.
Lemen, C. (1980). Relationship between relative brain size and climbing ability in peromyscus. Journal of Mammalogy, 61(2), 360–364.
Liow, L. H. (2004). A test of Simpson’s “rule of the survival of the relatively unspecialized” using fossil crinoids. The American Naturalist, 164(4), 431–443. https://doi.org/10.1086/423673.
Liow, L. H., Fortelius, M., Bingham, E., Lintulaakso, K., Mannila, H., Flynn, L., & Stenseth, N. C. (2008). Higher origination and extinction rates in larger mammals. Proceedings of the National Academy of Sciences of the United States of America, 105(16), 6097–6102. https://doi.org/10.1073/pnas.0709763105.
Liow, L. H., Fortelius, M., Lintulaakso, K., Mannila, H., & Stenseth, N. C. (2009). Lower extinction risk in sleep-or-hide mammals. The American Naturalist, 173(2), 264–272. https://doi.org/10.1086/595756.
Little, R. A., & Stoner, H. B. (1968). The measurement of heat loss from the rat’s tail. Quarterly Journal of Experimental Physiology and Cognate Medical Sciences, 53(1), 76–83. https://doi.org/10.1113/expphysiol.1968.sp001947.
Lobato, F. L., Barneche, D. R., Siqueira, A. C., Liedke, A. M. R., Lindner, A., Pie, M. R., et al. (2014). Diet and diversification in the evolution of coral reef fishes. PLoS ONE, 9(7), e102094.
Lowman, M., & Rinker, H. B. (2004). Forest canopies. Cambridge: Academic Press.
Machac, A. (2014). Detecting trait-dependent diversification under diversification slowdowns. Evolutionary Biology, 41(2), 201–211. https://doi.org/10.1007/s11692-013-9258-z.
Maddison, W. P., Midford, P. E., & Otto, S. P. (2007). Estimating a Binary character’s effect on speciation and extinction. Systematic Biology, 56(5), 701–710. https://doi.org/10.1080/10635150701607033.
Mares, M. A. (2009). A desert calling: Life in a forbidding landscape. Cambridge: Harvard University Press.
Martin, R. A. (1992). Generic species richness and body mass in North American mammals: Support for the inverse relationship of body size and speciation rate. Historical Biology, 6(2), 73–90. https://doi.org/10.1080/10292389209380420.
Martin, S. A., Alhajeri, B. H., & Steppan, S. J. (2016). Dietary adaptations in the teeth of murine rodents (Muridae): A test of biomechanical predictions. Biological Journal of the Linnean Society, 119(4), 766–784. https://doi.org/10.1111/bij.12822.
Matthews, L. J., Arnold, C., Machanda, Z., & Nunn, C. L. (2011). Primate extinction risk and historical patterns of speciation and extinction in relation to body mass. Proceedings of the Royal Society B: Biological Sciences, 278(1709), 1256–1263. https://doi.org/10.1098/rspb.2010.1489.
May, R. M. (1986). The search for patterns in the balance of nature advances and retreats. Ecology, 67, 1115–1126.
Mitter, C. B., Farrell, B., & Wiegmann, B. (1988). The phylogenetic study of adaptive zones: Has phytophagy promoted insect diversification? American Naturalist, 132(1), 107–128.
Monroe, M. J., & Bokma, F. (2009). Do speciation rates drive rates of body size evolution in mammals? The American naturalist, 174(6), 912–918. https://doi.org/10.1086/646606.
Moore, B. R., & Donoghue, M. J. (2007). Correlates of diversification in the plant clade dipsacales: Geographic movement and evolutionary innovations. The American Naturalist, 170, S28–S55.
Musser, G. G., & Carleton, M. D. (2005). Superfamily Muroidea. In D. E. Wilson & D. M. Reeder (Eds.), Mammal species of the world (3rd ed., pp. 894–1531). Baltimore: The Johns Hopkins University Press.
Nevo, E. (1985). Speciation in action and adaptation in subterranean mole rats: Patterns and theory. Bolletino di Zoologia, 52(1–2), 65–95. https://doi.org/10.1080/11250008509440344.
Ng, J., & Smith, S. D. (2014). How traits shape trees: New approaches for detecting character state-dependent lineage diversification. Journal of Evolutionary Biology, 27(10), 2035–2045. https://doi.org/10.1111/jeb.12460.
Nowak, R. M. (1999). Walker’s mammals of the world. Volume 1 and 2 (6th ed.). Baltimore: John Hopkins University Press.
Pabinger, S., Rödiger, S., Kriegner, A., Vierlinger, K., & Weinhäusel, A. (2014). A survey of tools for the analysis of quantitative PCR (qPCR) data. Biomolecular Detection and Quantification, 1(1), 23–33. https://doi.org/10.1016/j.bdq.2014.08.002.
Parada, A., D’Elía, G., & Palma, R. E. (2015). The influence of ecological and geographical context in the radiation of Neotropical sigmodontine rodents. BMC Evolutionary Biology, 15(1), 172. https://doi.org/10.1186/s12862-015-0440-z.
Paradis, E. (2005). Statistical analysis of diversification with species traits. Evolution, 59(1), 1–12. https://doi.org/10.1111/j.0014-3820.2005.tb00889.x.
Parent, C. E., & Crespi, B. J. (2009). Ecological opportunity in adaptive radiation of Galápagos endemic land snails. The American Naturalist, 174, 898–905.
Peters, R. H. (1983). The ecological implications of body size. Cambridge: Cambridge University Press.
Pineda-Munoz, S., & Alroy, J. (2014). Dietary characterization of terrestrial mammals. Proceedings of the Royal Society B: Biological Sciences, 281(1789), 20141173
Pineda-Munoz, S., Evans, A. R., & Alroy, J. (2016). The relationship between diet and body mass in terrestrial mammals. Paleobiology, 42(4), 659–669.
Pinto, G., Mahler, D. L., Harmon, L. J., & Losos, J. B. (2008). Testing the island effect in adaptive radiation: Rates and patterns of morphological diversification in Caribbean and mainland Anolis lizards. Proceedings of the Royal Society B: Biological Sciences, 275(1652), 2749–2757. https://doi.org/10.1098/rspb.2008.0686.
Plummer, M., Best, N., Cowles, K., & Vines, K. (2010). Coda: Output analysis and diagnostics for MCMC. R package version 0.14-2
Price, S. A., Hopkins, S. S. B., Smith, K. K., & Roth, V. L. (2012). Tempo of trophic evolution and its impact on mammalian diversification. Proceedings of the National Academy of Sciences, 109(18), 7008–7012. https://doi.org/10.1073/pnas.1117133109.
Price, S. L., Powell, S., Kronauer, D. J. C., Tran, L. A. P., Pierce, N. E., & Wayne, R. K. (2014a). Renewed diversification is associated with new ecological opportunity in the Neotropical turtle ants. Journal of Evolutionary Biology, 27(2), 242–258. https://doi.org/10.1111/jeb.12300.
Price, T. D., Hooper, D. M., Buchanan, C. D., Johansson, U. S., Tietze, D. T., Alstrom, P., et al. (2014b). Niche filling slows the diversification of Himalayan songbirds. Nature, 509(7499), 222–225. https://doi.org/10.1038/nature13272.
Promislow, D. E. L., & Harvey, P. H. (1990). Living fast and dying young: A comparative analysis of life-history variation among mammals. Journal of Zoology, 220(3), 417–437. https://doi.org/10.1111/j.1469-7998.1990.tb04316.x.
Pyron, R. A., & Burbrink, F. T. (2014). Early origin of viviparity and multiple reversions to oviparity in squamate reptiles. Ecology Letters, 17(1), 13–21. https://doi.org/10.1111/ele.12168.
Rabosky, D. L. (2014). Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS ONE, 9(2), e89543. https://doi.org/10.1371/journal.pone.0089543.
Rabosky, D. L., & Goldberg, E. E. (2015). Model inadequacy and mistaken inferences of trait-dependent speciation. Systematic Biology, 64(2), 340–355. https://doi.org/10.1093/sysbio/syu131.
Rabosky, D. L., Grundler, M., Anderson, C., Title, P., Shi, J. J., Brown, J. W., et al. (2014). BAMMtools: An R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods in Ecology and Evolution. https://doi.org/10.1111/2041-210X.12199.
Rabosky, D. L., & Huang, H. (2016). A robust semi-parametric test for detecting trait-dependent diversification. Systematic Biology, 65(2), 181. https://doi.org/10.1093/sysbio/syv066.
Rabosky, D. L., & Matute, D. R. (2013). Macroevolutionary speciation rates are decoupled from the evolution of intrinsic reproductive isolation in Drosophila and birds. Proceedings of the National Academy of Sciences. https://doi.org/10.1073/pnas.1305529110.
Rabosky, D. L., & McCune, A. R. (2010). Reinventing species selection with molecular phylogenies. Trends in Ecology & Evolution, 25(2), 68–74. https://doi.org/10.1016/j.tree.2009.07.002.
Rabosky, D. L., Santini, F., Eastman, J., Smith, S. A., Sidlauskas, B., Chang, J., & Alfaro, M. E. (2013). Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation. Nature Communications, 4, 1958. https://doi.org/10.1038/ncomms2958.
Read, A. F., & Harvey, P. H. (1989). Life history differences among the eutherian radiations. Journal of Zoology, 219(2), 329–353. https://doi.org/10.1111/j.1469-7998.1989.tb02584.x.
Revell, L. J. (2012). phytools: An R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution, 3(2), 217–223. https://doi.org/10.1111/j.2041-210X.2011.00169.x.
Ricklefs, R. E. (2006). Global variation in the diversification rate of passerine birds. Ecology, 87(10), 2468–2478.
Rojas, D., Vale, Á, Ferrero, V., & Navarro, L. (2012). The role of frugivory in the diversification of bats in the Neotropics. Journal of Biogeography, 39(11), 1948–1960. https://doi.org/10.1111/j.1365-2699.2012.02709.x.
Rolland, J., Condamine, F. L., Jiguet, F., & Morlon, H. (2014). Faster speciation and reduced extinction in the tropics contribute to the Mammalian latitudinal diversity gradient. PLoS Biology, 12(1), e1001775. https://doi.org/10.1371/journal.pbio.1001775.
Sacks, B. N., Bannasch, D. L., Chomel, B. B., & Ernest, H. B. (2008). coyotes demonstrate how habitat specialization by individuals of a generalist species can diversify populations in a heterogeneous ecoregion. Molecular Biology and Evolution, 25(7), 1384–1394. https://doi.org/10.1093/molbev/msn082.
Samuels, J. X. (2009). Cranial morphology and dietary habits of rodents. Zoological Journal of the Linnean Society, 156(4), 864–888. https://doi.org/10.1111/j.1096-3642.2009.00502.x.
Santana, S. E., & Cheung, E. (2016). Go big or go fish: Morphological specializations in carnivorous bats. Proceedings of the Royal Society B: Biological Sciences, 283(1830), 20160615.
Schenk, J. J., Rowe, K. C., & Steppan, S. J. (2013). Ecological opportunity and incumbency in the diversification of repeated continental colonizations by muroid rodents. Systematic Biology, 62(6), 837–864. https://doi.org/10.1093/sysbio/syt050.
Schluter, D. (2000). The ecology of adaptive radiation. Oxford: Oxford University Press.
Schluter, D. (2001). Ecology and the origin of species. Trends in Ecology & Evolution, 16(7), 372–380.
Shimer, H. W. (1903). Adaptations to aquatic, arboreal, fossorial and cursorial habits in mammals. III. Fossorial adaptations. The American Naturalist, 37(444), 819–825.
Sibly, R. M., & Brown, J. H. (2007). Effects of body size and lifestyle on evolution of mammal life histories. Proceedings of the National Academy of Sciences of the United States of America, 104(45), 17707–17712. https://doi.org/10.1073/pnas.0707725104.
Simpson, G. G. (1944). The tempo and mode in evolution. New York: Columbia University Press.
Smits, P. D. (2015). Expected time-invariant effects of biological traits on mammal species duration. Proceedings of the National Academy of Sciences, 112(42), 13015–13020. https://doi.org/10.1073/pnas.1510482112.
Steppan, S., Adkins, R., & Anderson, J. (2004). Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Systematic Biology, 53(4), 533–553. https://doi.org/10.1080/10635150490468701.
Stuart, O., & Landry, J. (1970). The Rodentia as omnivores. The Quarterly Review of Biology, 45(4), 351–372. https://doi.org/10.1086/406647.
Swihart, R. K. (1984). Body size, breeding season length, and life history tactics of Lagomorphs. Oikos, 43(3), 282–290. https://doi.org/10.2307/3544145.
Team, R. D. C. (2016). R: A language and environment for statistical computing. Vienna: R Core Team
Tomiya, S. (2013). Body size and extinction risk in terrestrial mammals above the species level. The American Naturalist, 182(6), E196–E214. https://doi.org/10.1086/673489.
Tran, L. A. P. (2014). The role of ecological opportunity in shaping disparate diversification trajectories in a bicontinental primate radiation. Proceedings of the Royal Society of London B: Biological Sciences, 281(1781), 20131979.
Tran, L. A. P. (2016). Interaction between digestive strategy and niche specialization predicts speciation rates across herbivorous mammals. The American Naturalist, 187(4), 468–480. https://doi.org/10.1086/685094.
Vamosi, J. C., Armbruster, W. S., & Renner, S. S. (2014). Evolutionary ecology of specialization: Insights from phylogenetic analysis. Proceedings of the Royal Society of London B: Biological Sciences, 281(1795), 20142004. https://doi.org/10.1098/rspb.2014.2004.
Verde Arregoitia, L. D., Fisher, D. O., & Schweizer, M. (2017). Morphology captures diet and locomotor types in rodents. Royal Society Open Science, 4(1), 160957. https://doi.org/10.1098/rsos.160957.
von Hagen, K. B., & Kadereit, J. W. (2003). The diversification of Halenia (Gentianaceae): Ecological opportunity versus key innovation. Evolution, 57(11), 2507–2518. https://doi.org/10.1111/j.0014-3820.2003.tb01495.x.
Wagenmakers, E.-J., & Farrell, S. (2004). AIC model selection using Akaike weights. Psychonomic Bulletin & Review, 11(1), 192–196. https://doi.org/10.3758/BF03206482.
Walker, D. M., Castlebury, L. A., Rossman, A. Y., & Struwe, L. (2014). Host conservatism or host specialization? Patterns of fungal diversification are influenced by host plant specificity in Ophiognomonia (Gnomoniaceae: Diaporthales). Biological Journal of the Linnean Society, 111(1), 1–16. https://doi.org/10.1111/bij.12189.
Whittow, G. C. (2013). Comparative physiology of thermoregulation: Mammals. Waltham: Academic Press.
Williams, S., & Kay, R. (2001). A comparative test of adaptive explanations for hypsodonty in ungulates and rodents. Journal of Mammalian Evolution, 8(3), 207–229. https://doi.org/10.1023/A:1012231829141.
Wilson, D. S., & Yoshimura, J. (1994). On the coexistence of specialists and generalists. The American Naturalist, 144(4), 692–707. https://doi.org/10.1086/285702.
Withers, P. C., Cooper, C. E., Cruz-Neto, A. P., & Bozinovic, F. (2016). Ecological and environmental physiology of mammals. Oxford: Oxford University Press.
Wollenberg, K. C., Vieites, D. R., Glaw, F., & Vences, M. (2011). Speciation in little: The role of range and body size in the diversification of Malagasy mantellid frogs. BMC Evolutionary Biology, 11(1), 217. https://doi.org/10.1186/1471-2148-11-217.
Acknowledgements
Earlier versions of the manuscript benefited from comments by Gregory Erickson, Joseph Travis, Thomas Miller, William Parker, and especially John Schenk. We also appreciate correspondence with Daniel Rabosky concerning the general issues with state-dependent diversification methods. An anonymous reviewer contributed useful comments that improved the final version of the manuscript. Financial support for this work was provided by a fellowship from Kuwait University to BHA.
Funding
This project was partially funded by NSF Grant DEB-0841447 to SJS.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest regarding the publication of this article.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11692_2018_9449_MOESM1_ESM.eps
Full version of Schenk et al. (2013) muroid rodent chronogram (showing the tip labels), with some of the major taxonomic groups indicated. (EPS 4078 KB)
11692_2018_9449_MOESM2_ESM.xlsx
Diet and habitat data for muroid species used in the analyses. For diet, C = carnivore, O = omnivore, H = herbivore. For habitat, A = arboreal, F = fossorial, T = terrestrial. Diet description and references are also included. Missing data are designated with (−). See Materials and Methods for more information. (XLSX 55 KB)
11692_2018_9449_MOESM3_ESM.xlsx
Log-transformed adult body mass (log mass) and relative tail lengths (rtl) of muroid rodents used in the analyses. Mass is in grams and lengths are in millimeters. Data extracted from Alhajeri et al. (<link rid="bib5">2016</link>). See Material and methods for more information. (XLSX 23 KB)
11692_2018_9449_MOESM4_ESM.xlsx
Summary of alternative multistate-dependent speciation and extinction (MuSSE) model fits for the association between diet and diversification rates in muroid rodents. Both the results for the (a) empirical (observed) analysis, and the (b–k) ten tip-shuffled (randomized) analyses are shown. The best-fit model based on ∆AICc and Akaike weights (wi) are denoted in bold. n = number of estimated parameters; ln L = log-likelihood score; ∆AICc = fit relative to the model with the lowest AICc score (italicized). (XLSX 17 KB)
11692_2018_9449_MOESM5_ESM.xlsx
Summary of alternative multistate-dependent speciation and extinction (MuSSE) model fits for the association between habitat and diversification rates in muroid rodents. Both the results for the (a) empirical (observed) analysis, and the (b–k) ten tip-shuffled (randomized) analyses are shown. The best-fit model based on ∆AICc and Akaike weights (wi) are denoted in bold. n = number of estimated parameters; ln L = log-likelihood score; ∆AICc = fit relative to the model with the lowest AICc score (italicized). (XLSX 17 KB)
11692_2018_9449_MOESM6_ESM.xlsx
Summary of alternative quantitative state-dependent speciation and extinction (QuaSSE) model fits for the association between log body mass and diversification rates in muroid rodents. The extinction rate (µ) is set as constant (constrained) in all the models, and only the speciation (λ) rate is allowed to vary. Both the results for the (a) empirical (observed) analysis, and the (b–k) ten tip-shuffled (randomized) analyses, are shown. The best-fit model based on ∆AICc and Akaike weights (wi) are denoted in bold. n = number of estimated parameters; ln L = log-likelihood score; ∆AICc = fit relative to the model with the lowest AICc score (italicized). (XLSX 19 KB)
11692_2018_9449_MOESM7_ESM.xlsx
Summary of alternative quantitative state-dependent speciation and extinction (QuaSSE) model fits for the association between relative tail length and diversification rates in muroid rodents. The extinction rate (µ) is set as constant (constrained) in all the models, and only the speciation (λ) rate is allowed to vary. Both the results for the (a) empirical (observed) analysis, and the (b–k) ten tip-shuffled (randomized) analyses, are shown. The best-fit model based on ∆AICc and Akaike weights (wi) are denoted in bold. n = number of estimated parameters; ln L = log-likelihood score; ∆AICc = fit relative to the model with the lowest AICc score (italicized). (XLSX 19 KB)
Rights and permissions
About this article
Cite this article
Alhajeri, B.H., Steppan, S.J. Ecological and Ecomorphological Specialization Are Not Associated with Diversification Rates in Muroid Rodents (Rodentia: Muroidea). Evol Biol 45, 268–286 (2018). https://doi.org/10.1007/s11692-018-9449-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11692-018-9449-8