Abstract
Background
Eimeria spp. are coccidian protozoan parasites of domestic and wild animals. Pelecaniform birds are hosts of some Eimeria spp., however, from the family Threskiornithidae only one eimerian species is recorded, Eimeria bazi Chauhan et Bhatia, 1970 which was described from red-naped ibises Pseudibis papillosa (Temminck, 1824) in India. In this study, in turn, this species is morphologically and molecularly identified from buff-necked ibises Theristicus caudatus (Boddaert, 1783) in Brazil.
Purpose
This study aimed to report E. bazi from buff-necked ibises T. caudatus in southeastern Brazil, revealing the worldwide distribution of this coccidian species, in addition to providing preliminary genotypic identification via sequencing of the mitochondrial cytochrome c oxidase subunit 1 (COI) gene.
Methods
A total of 73 fecal samples were collected from a flock of buff-necked ibises, which remained on the campus of the Federal Rural University of Rio de Janeiro (Universidade Federal Rural do Rio de Janeiro—UFRRJ) from March 2019 to August 2020. Fecal samples were processed by the Sheather’s method to recover oocysts. The morphological and morphometrical studies of the oocysts were performed using an optical microscope and graphic editing software. Molecular analysis was performed by sequencing of the COI gene, and the phylogenetic analysis was based in the neighbor-joining and maximum likelihood estimates.
Results
Forty-five fecal samples were positive for oocysts identified as E. bazi. This oocysts are ovoidal, 26.2 × 18.9 μm, with smooth to slightly rough wall, c.1.7 μm thick. Micropyle robust and protruding, sometimes with a polar body attached. Oocyst residuum absent, but one or two small polar granules are present. Sporocysts ovoidal to lemon-shaped, 14.2 × 8.7 μm. The Stieda body is knob-like to rounded and sub-Stieda body is absent or vestigial. Sporocyst residuum is composed of granules often membrane-bound. Sporozoites are vermiform, with refractile bodies. This morphology was consistent with the original description of E. bazi from P. papillosa in India. Molecular analysis at the COI gene exhibited low similarity with coccidians sequenced for the same genic region deposited in GenBank, sitting E. bazi separately on the cladogram.
Conclusions
The morphological and molecular studies support the identification of E. bazi from T. caudatus in South America, thus revealing the wide distribution of this eimerian species in the world provided by migratory birds and/or with intercontinental distribution.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Coccidia are a highly diverse subclass of protozoan obligate intracellular parasites of domestic and wild animals [1, 2]. Eimeria Schneider, 1875 is the genus most predominant and diverse, being identified from mammals, birds, reptiles, amphibians and fish, in addition to invertebrates [3]. From birds, the most prevalent coccidian genera are Isospora Schneider, 1881 and Eimeria; however, Isospora spp. occur more frequently in passeriform birds, while Eimeria spp. are more frequent in non-passeriform birds [3].
The buff-necked ibis Theristicus caudatus (Boddaert, 1783) is a non-passeriform bird of the order Pelecaniformes, family Threskiornithidae. This species is among the ibises with the widest geographic distribution in the Neotropical Region [4]. It is a bird typical of grasslands and occurs in a wide variety of open and semi-open habitats, such as grasslands and savannas, in addition to lawns on the shores of lakes, ponds and other watercourses [5]. It forages in flocks, but also occasionally solitary, in soft soils and grasslands. It feeds on small reptiles, amphibians and arthropods, mainly insects including their larvae, which are captured directly from the substrate and through the deep penetration of its long beak into the soil [6]. It is a mainly sedentary species with predominance of local movements; however, it has expanded its geographic distribution, especially in southeastern Brazil [7, 8].
The habits of the buff-necked ibises living in flocks, foraging in contact with the ground and having a predominance of local movements, favor the biology of coccidian parasites [9]. In general, infected birds shed coccidian oocysts into the environment when defecating, which will become sporulated/viable depending on the conditions of humidity, temperature and atmospheric oxygen [2]. Then, the habits of the buff-necked ibises would ensure the continuity of the life cycle of its coccidians, since the presence of other susceptible ibises in the same environment would be constant [9].
Despite this favorable context for coccidiosis in buff-necked ibises, there are no descriptions of coccidian species parasitizing T. caudatus; however, there is one eimerian species recorded from Threskiornithidae, Eimeria bazi Chauhan et Bhatia, 1970 described from red-naped ibises Pseudibis papillosa (Temminck, 1824) in India [10]. Consequently, in this study, fecal samples were collected from a flock of buff-necked ibises T. caudatus that remained on the campus of the Federal Rural University of Rio de Janeiro (Universidade Federal Rural do Rio de Janeiro—UFRRJ) from March 2019 to August 2020, aiming to recover, isolate and identify coccidian oocysts.
Materials and Methods
Sample Collection
Eight fieldworks were carried out to collect fecal samples from the flock of buff-necked ibises T. caudatus that remained on the Seropédica campus of UFRRJ (22° 45′ 46″ S, 43° 41′ 18″ W), from March 2019 to August 2020. The monospecific flock of ibises were observed from a distance until defecations were seen, which were then sought and found. Depending on the type of surface that the feces were found, they were discarded, giving preference to those droplets of feces shed on leaves, rocks, or other surfaces less susceptible to contamination. Each fresh droplet of feces from each individual ibis was placed individually in a centrifuge tube with a potassium dichromate 2.5% (K2Cr2O7) solution at 1:6 (v/v).
Morphological Analyses
Fecal samples were transported to the Laboratório de Biologia de Coccídios, Universidade Federal Rural do Rio de Janeiro (UFRRJ). In laboratory, the samples were kept at room temperature (~ 25 °C) for 7 days in the same centrifuge tubes which were collected, being oxygenated daily by shaking until the oocysts were sporulated. Oocysts were recovered and isolated using the Sheather’s method and were morphologically studied following the guidelines of Duszynski and Wilber [11] and Berto et al. [12]. Optical microscopy, photomicrography and morphometry were performed using an Olympus BX binocular microscope (Olympus Optical, Tokyo, Japan) equipped with a digital camera Eurekam 5.0 (BEL Photonics, Monza, Italy). Line drawings were performed using the applications Corel DRAW and Corel PHOTO-PAINT from CorelDRAW® Graphics Suite (Version 2020, Corel Corporation, Canada). Measurements (in micrometers) are given as the range followed by the mean in parentheses.
Molecular Analyses
A single oocyst from each sample was individualized from serial dilutions of the oocysts in drops on a microscope slide using an automatic micropipette. The individualized oocyst was transferred to a microtube containing PBS and washed by centrifuging until the solution is clear [13]. Four freeze–thaw cycles were performed to achieve complete lysis of the oocyst. DNA extraction was performed using the Qiagen DNeasy Blood and Tissue Kit (Qiagen, São Paulo, Brazil). Amplification by PCR of a partial fragment of the mitochondrial cytochrome c oxidase subunit 1 (COI) gene (c.250 bp) was performed using nested PCR, according Dolnik et al. [13] and Yang et al. [14]. The nested PCR amplicons were purified using the Qiagen MinElute PCR Purification (Qiagen, São Paulo, Brazil).
DNA Sequence Analyses
Nested PCR amplicons were sequenced using an ABI-Prism 3500 Genetic Analyzer (Applied Biosystems, Foster City, California). The newly generated sequence was compared to those for coccidian species available in the GenBank database using the Basic Local Alignment Search Tool (BLAST). The phylogenetic tree was constructed using the newly generated COI sequence aligned with coccidian species available on GenBank. Distance analyses and phylogenies were conducted using MEGA X [15]. Briefly, the nucleotide sequences were imported into MEGA X and aligned with reference sequences from GenBank using Clustal W (http://www.clustalw.genome.jp). Maximum likelihood (ML) and neighbor-joining (NJ) trees were constructed, and the distances computed using the Tamura-Nei method based on model selection using ModelTest in MEGA X. Bootstrap analyses were conducted using 1000 replicates to assess the reliability of inferred tree topologies.
Results
A total of 73 fecal samples were collected from buff-necked ibises and 45 were positive for coccidia. All observed oocysts were morphologically identified as E. bazi. This material is described below.
Family Eimeriidae Minchin, 1903
Genus Eimeria Schneider, 1875
Eimeria bazi Chauhan et Bhatia, 1970 (Figs. 1, 2)
Photomicrographs of sporulated oocysts of Eimeria bazi from the buff-necked ibis Theristicus caudatus. inner (il) and outer (ol) layers of the oocyst wall; micropyle (m) and polar body attached to the micropyle (pb/m); polar granule (pg); Stieda (sb) and vestigial sub-Stieda (ssb) bodies; sporocyst residuum (sr); anterior (arb) e posterior (prb) refractile bodies. All to same scale. Scale-bar: 10 µm
Oocysts (n = 88) ovoidal, 21–30 × 16–21 (26.2 × 18.9); length/width (L/W) ratio 1.2–1.6 (1.39). Wall bi-layered, 1.5–1.9 (1.7) thick, outer layer smooth to slightly rough, c.2/3 of total thickness. Micropyle present, robust and protruding, 4.1–5.4 (4.8) wide; occasionally with a rounded polar body attached to the micropyle, 1.6–2.3 × 1.5–2.1 (2.0 × 1.7). Oocyst residuum absent, but one or two small polar granules are present. Sporocysts (n = 30) ovoidal to lemon-shaped, 13–15 × 7–10 (14.2 × 8.7); L/W ratio 1.4–1.8 (1.65). Stieda body present, knob-like to rounded, 0.7–1.4 × 1.3–1.9 (1.1 × 1.5); sub-Stieda body absent or vestigial; para-Stieda body absent; sporocyst residuum present, composed of granules often membrane-bound in the center of the sporocyst, 3.4–5.5 × 3.5–4.3 (4.7 × 4.0), but also scattered among the sporozoites. Sporozoites vermiform, with anterior and posterior refractile bodies, but nucleus is indiscernible.
Host: Theristicus caudatus (Boddaert, 1783) (Aves: Pelecaniformes: Threskiornithidae), buff-necked ibis.
Locality: Campus of the Federal Rural University of Rio de Janeiro (22° 45′ 46″ S, 43° 41′ 18″ W), southeastern Brazil.
Representative specimens: Photomicrographs, line drawing and oocysts in 2.5% K2Cr2O7 solution (Williams et al. 2010) are deposited and available (http://r1.ufrrj.br/labicoc/colecao.html) in the Parasitology Collection of the Laboratório de Biologia de Coccídios, at UFRRJ, under the repository number 128/2022. Photographs of the host specimen are deposited in the same collection.
Site in host: Unknown.
Representative DNA sequence: One representative COI sequence was deposited in the GenBank database under the accession number OM933654.
Remarks: The morphology of Eimeria spp. recorded from the order Pelecaniformes, in addition to the orders Suliformes and Phaethontiformes, which were related to Pelecaniformes, and of E. bazi identified from T. caudatus in this study are shown in Table 1. Nine Eimeria spp. are recorded in these orders: Eimeria ardeae Dubinin, 1939, E. bazi, Eimeria pelecani Courtney et Ernst, 1975, Eimeria ardae Shamsuddin et Jasim, 1980 and Eimeria garzettae Golemansky et Kuldjieva, 1980 from Pelecaniformes and Eimeria roscoviensis (Labbé, 1893), Eimeria urnula Hoare, 1933, Eimeria auritusi Yabsley, Gottdenker et Fisher, 2002 and Eimeria phalacrocoraxae Yabsley et Gibbs, 2006 from Suliformes. From Phaethontiformes there are no described species. As shown in Table 1, the oocysts recovered from T. caudatus in this study were morphologically similar with the original description of E. bazi from P. papillosa in India. In addition, only E. roscoviensis, E. urnula, E. garzettae and E. phalacrocoraxae have a micropyle and oocyst shape that are reasonably similar to the oocysts of this study; however, these species can be distinguished by their smaller sizes, pear-shaped oocysts, non-protruding micropyle and absence of polar granules in the sporulated oocyst (Table 1).
Phylogenetic analysis: DNA amplification of the oocyst of E. bazi showed a clear band of c.250 bp. Phylogenetic analysis included 20 sequences for coccidians available on GenBank (Fig. 3). Hepatozoon canis (James, 1905) was used as the outgroup. Eimeria bazi sat separately on the cladogram for having low similarity with coccidians sequenced for the same genic region deposited in GenBank. The highest similarity was only 88.4% with Isospora oliveirai Ortúzar-Ferreira et Berto, 2020, which is a coccidian parasite of Neotropical passerines [16]. The closest eimerian sequence was of Eimeria bubonis Cawthorn et Stockdale, 1981, an owl parasite, with 86.3% similarity [17]; followed by Eimeria cahirinensis Couch, Blaustein, Duszynski, Shenbrot et Nevo, 1997, Eimeria columbinae Ortúzar-Ferreira et Berto, 2020, Eimeria purpureicephali Yang, Brice et Ryan, 2016 and Eimeria dispersa Tyzzer, 1929, which are parasites of rodents, columbiformes, psittaciformes and galliformes, respectively, with 85–86% similarity [18,19,20,21].
Discussion
The study of parasitology presupposes, among other things, the biological understanding of both the parasite and its host, because the dynamics of this ecological interaction between parasite–host can occur in a variety of ways [22, 23]. There are parasites that are specific to a group of susceptible hosts, while others parasitize a wide range of hosts, thus being generalists [22]. The overwhelming part of the scientific literature on eimeriid coccidia follows the guidelines of Duszynski and Wilber [11], that these parasites are minimally specific at the familial level of their hosts; therefore, to describe a new species, it is necessary to have a detailed morphological comparison with other congeneric species already described for the same host family [11].
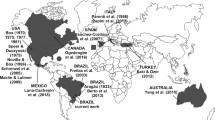
So far only E. bazi has been recorded in the Threskiornithidae family. This coccidian species was originally described in Asia, from red-naped ibises P. papillosa, which have a geographic distribution restricted to India, Nepal and Pakistan [24]. In this context, it is unlikely that E. bazi was directly transmitted from P. papillosa to T. caudatus, as these birds are allopatric inhabiting distinct and distant continents. On the other hand, many ibises are migrants and/or have very wide geographic distributions, being observed on all continents, with the exception of Antarctica [25]. The glossy ibis Plegadis falcinellus (Linnaeus, 1766), which is observed in Oceania, Asia, Africa, Europe and in South, Central and North America, can be highlighted by its migratory potential and wide intercontinental distribution [24]. The Eurasian spoonbill Platalea leucorodia Linnaeus, 1758, in turn, is distributed as resident in Europe, Asia and Africa, but is reported as vagrant in Brazil [24]. Thus, these and other migrant and/or intercontinental ibises could be transmissors/dispersers of coccidians for susceptible threskiornithid birds on all continents, including P. papillosa and T. caudatus, which have more restricted distributions [26]. Studies such as those by Silva et al. [27] and Silva-Carvalho et al. [28] exemplify the dispersal potential of coccidian parasites. In addition, the report of eimerian oocysts from northern bald ibises Geronticus eremita (Linnaeus, 1758) and African sacred ibises Threskiornis aethiopicus (Latham, 1790), which records at least one Eimeria sp. on the African continent from Threskiornithidae reinforces the assumption of the intercontinental dispersion of E. bazi; however, it would be unwise to identify these eimerian oocysts of these african ibises as E. bazi outside India, as in these studies none of the oocysts were specifically identified, described, or named [29,30,31].
Another possibility that should be considered is the anthropogenic dispersion of coccidia through the trade/trafficking of wild animals [26]. An example is the zoos and breeding sites that are qualified to receive wild animals from the most diverse parts of the world. Isospora araponga Doležalová, Torres, Fernández et Modrý, 2004 was described from bare-throated bellbirds Procnias nudicollis (Vieillot, 1817) imported to Barcelona Zoo from Brazil [32]. Berto et al. [33] reported Tyzzeria parvula (Kotlán, 1933) in Brazil from greylag geese Anser anser (Linnaeus, 1758), an Asian species that is commonly traded as a domestic animal in several regions of the world [24]. In this way, the trade/trafficking of wild animals can allow the dispersion/transmission of coccidia among naturally allopatric birds.
Still on the geografic distribution of ibises, it is worth mentioning that T. caudatus, despite being typical of open environments, such as cerrado and fields, has expanded its geographic distribution in recent years to other biomes, which were originally forested, such as the Atlantic Forest [5, 7, 34]. This biogeographic expansion has been potentiated by changes in these habitats resulting from deforestation, in addition to the large increase in the creation of pasture fields, providing the emergence of environments similar to the original habitat of this bird [35]. Thus, these new open areas of anthropogenic origin provide an ideal habitat for the permanence of T. caudatus. One of the consequences of this expansion of the geographic distribution of T. caudatus is the dispersion of its coccidia in new areas, including for new host species that previously would not have sympatry with this bird in its native habitat. Along with that, the very high densities of oocysts (above 100,000), observed in some droplets of feces collected from T. caudatus in the present study may cause epizootic diseases in host populations, especially those without previous contact with the parasite [26]. This may occur considering that the symptoms of coccidiosis must be closely related to the amount of oocysts ingested, in addition to other factors [2, 36].
In the absence or when there is a low number of descriptions of coccidian species in a host family is suitable the comparison with coccidian species recorded in higher taxa of the host [16]. Thus, although the oocysts recovered from T. caudatus in the current study are morphologically identifiable as E. bazi, which is the only recorded species of the host family Threskiornithidae, these were also compared with the coccidians described in the next higher taxon, in this case, the order Pelecaniformes. Recent studies have shown that the traditional Pelecaniformes are actually a polyphyletic group; therefore, several taxonomic rearrangements were proposed in a new classification that included two new orders, Suliformes and Phaethontiformes, from the traditional Pelecaniformes. Furthermore, it is noteworthy that the family Threskiornithidae belonged to the order Ciconiiformes; however, currently the order Ciconiiformes has only the family Ciconiidae, and Threskiornithidae along with other families were reclassified to the order Pelecaniformes [4, 6, 37,38,39]. Thus, aiming to expand the comparative morphology of E. bazi, Eimeria spp. recorded from Pelecaniformes, Suliformes and Phaethontiformes were included in this study, obtaining nine Eimeria spp. for morphological comparison (Table 1).
The first described species of these traditional Pelecaniformes was Eimeria roscoviensis (Labbé, 1893) from European shags Gulosus aristotelis (Linnaeus, 1761) in Roscoff, France. This species was originally described as Coccidium roscoviense Labbé, 1893 before being correctly classified in the genus Eimeria [40]. Two later described species of birds of the same host family (Phalacrocoracidae), Eimeria urnula Hoare, 1933 and Eimeria phalacrocoraxae Yabsley et Gibbs, 2006, are very similar to each other, both morphometrically and morphologically, mainly due to the typical micropyle with a polar body (Table 1) [41, 42]. This observation shows the possibility of these coccidians being a single species, or being closely related species that have co-evolved from an ancestral eimeriid that parasitized an ancestral bird of the Phalacrocoracidae, in the process named co-speciation [26, 43]. In the oocysts identifiable as E. bazi in this study, as well as in its original description [10] and in E. garzettae [44], this same type of oocyst shape and size, including the micropyle with polar body, are observed (Table 1), showing that perhaps the common ancestor of these Eimeria spp. may have started parasitism even earlier in the evolution of Pelecaniformes and Suliformes [26, 43]. This assumption is reinforced by the findings of Labbé from the description of E. roscoviensis, still in the late nineteenth century, who observed oocysts of the same morphotype as E. roscoviensis from Charadriiformes of the genera Charadrius Linnaeus, 1758, Arenaria Brisson, 1760, Numenius Brisson, 1760, Tringa Linnaeus, 1758 and Calidris Merrem, 1804, showing that the diversity of hosts susceptible to these Eimeria spp. with polar bodies attached to the micropyle may be even larger [40].
The other species Eimeria ardeae Dubinin, 1939, Eimeria pelecani Courtney et Ernst, 1975, Eimeria ardae Shamsuddin et Jasim, 1980 and Eimeria auritusi Yabsley, Gottdenker et Fisher, 2002 do not have the typical morphology observed in the previous species, as they do not have micropyle and are subspherical as in the case of E. auritusi and E. ardae; therefore, these must have another evolutionary origin [45,46,47,48]. The answers to these phylogenetic questions of these Eimeria spp. of Pelecaniformes, Suliformes, and also Charadriiformes, could be conclusive by means of phylogenetic molecular analyses. However, of all Eimeria spp. related in this study, only E. auritusi and E. phalacrocoraxae were sequenced for a region of the 18S small subunit ribosomal RNA (18S) gene, unlike in this study, where E. bazi was sequenced for a genic region of the COI, and therefore, these species did not appear in the cladogram of Fig. 3. The sequencing of the 18S gene was intended in this study, but its amplification was not successful, perhaps due to the use of DNA extraction methodology from an individual oocyst that provides few copies of nuclear DNA, unlike the greater amount of mitochondrial DNA available in each oocyst. In any case, the use of the 18S gene for species differentiation and detection of recent evolutionary events has been shown to be unsuitable, whereas the COI gene has been considered the most suitable in this sense [49, 50]. Thus, when a greater number of Eimeria spp. of these birds have been sequenced, the phylogenetic analysis should be more conclusive, as, so far, the phylogenetic analysis for the COI gene has only shown that E. bazi is distant and paraphyletic from some coccidia groups of passeriform and non-passeriform birds (Fig. 3).
Finally, after the detailed morphological study of the oocysts recovered from T. caudatus in the current study and considering the migratory potential and worldwide distribution of ibises, in addition to other possibilities, E. bazi is reported for the first time in South America from a new host, T. caudatus. As an additional element, this study provided the first genetic sequencing by the COI gene for this coccidian species.
References
Ruggiero MA, Gordon DP, Orrell TM, Bailly N, Bourgoin T, Brusca RC, Cavalier-Smith T, Guiry MD, Kirk PM (2015) Correction: a higher level classification of all living organisms. PLoS ONE 10:e0130114. https://doi.org/10.1371/journal.pone.0119248
Fayer R (1980) Epidemiology of protozoan infection the coccidia. Vet Parasitol 6:75–103. https://doi.org/10.1016/0304-4017(80)90039-4
Duszynski DW, Couch L, Upton SJ (2000) The Coccidia of the World. https://www.k-state.edu/parasitology/worldcoccidia. Accessed 1 Mar 2022
Clements JF, Schulenberg TS, Iliff MJ, Billerman SM, Fredericks TA, Gerbracht JA, Lepage D, Sullivan BL, Wood CL (2021) The eBird/clements checklist of birds of the World: version 2021. https://www.birds.cornell.edu/clementschecklist/download/. Accessed 01 March 2022.
Matheu E, delHoyo J, Garcia EFJ, Boesman PFD (2020) Buff-necked Ibis (Theristicus caudatus), version 1.0. In: delHoyo J, Elliott A, Sargatal J, Christie DA, de Juana E (eds) Birds of the World. Cornell Lab of Ornithology, Ithaca. https://doi.org/10.2173/bow.bunibi1.01. Accessed 1 Mar 2022.
Sick H (1997) Ornitologia Brasileira. Nova Fronteira, Rio de Janeiro
Serpa GA, Pacheco JF, Lima LM, Parrini R, Pimentel LS, Pinto MFR, Antonini RD, Rajão H, Oliveira AH, Tavares DC, Siciliano S, Mallet-Rodrigues F, Luz HR, Soares BR, Crud N (2010) A curicaca, Theristicus caudatus (Ciconiiformes: Threskiornithidae) no estado do Rio de Janeiro: revisão dos registros e novas observações. Atual Ornitol 153:62–68
Cabral RBG, Ferreira I (2012) Primeiro registro de Theristicus caudatus (Pelecaniformes, Threskiornithidae) em Seropédica. RJ Atual Ornitol 165:10–11
Dolnik OV, Dolnik VR, Bairlein F (2010) The effect of host foraging ecology on the prevalence and intensity of coccidian infection in wild passerine birds. Ardea 98:97–103. https://doi.org/10.5253/078.098.0112
Chauhan PPS, Bhatia BB (1970) Eimerian oocysts from Pseudibis papillosa. Indian J Microbiol 10:53–54
Duszynski D, Wilber P (1997) A guideline for the preparation of species descriptions in the Eimeriidae. J Parasitol 83:333–336. https://doi.org/10.2307/3284470
Berto BP, McIntosh D, Lopes CWG (2014) Studies on coccidian oocysts (Apicomplexa: Eucoccidiorida). Rev Bras Parasitol Vet 23:1–15. https://doi.org/10.1590/S1984-29612014001
Dolnik OV, Palinauskas V, Bensch S (2009) Individual oocysts of Isospora (Apicomplexa: Coccidia) parasites from avian faeces: from photo to sequence. J Parasitol 95:169–174. https://doi.org/10.1645/GE-1873.1
Yang R, Brice B, Elliot A, Ryan U (2015) Isospora serinuse n. sp. (Apicomplexa: Eimeriidae) from a domestic canary (Serinus canaria forma domestica) (Passeriformes: Fringillidae) in Western Australia. Exp Parasitol 159:59–66. https://doi.org/10.1016/j.exppara.2015.08.020
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Ortúzar-Ferreira CN, Genovez-Oliveira JL, Oliveira MS, Mello ER, Thode-Filho S, Oliveira ÁA, Lima VM, Ferreira I, Berto BP (2020) Isospora oliveirai n. sp. (Chromista: Miozoa: Eimeriidae) from the greenish schiffornis Schiffornis virescens (Lafresnaye, 1838) (Passeriformes: Tyranni: Tityridae) in South America. Acta Parasitol 65:843–851. https://doi.org/10.1007/s11686-020-00237-8
McAllister CT, Hnida JA, Woodyard ET, Rosser TG (2019) Eimeria spp. (Apicomplexa: Eimeriidae) from great horned owls, Bubo virginianus (Gmelin) (Aves: Strigiformes) from Arkansas and Oklahoma, USA, with novel molecular information on Eimeria bubonis Cawthorn & Stockdale, 1981. Syst Parasitol 96:695–702. https://doi.org/10.1007/s11230-019-09881-9
Kvičerová J, Hypša V (2013) Host-parasite incongruences in rodent Eimeria suggest significant role of adaptation rather than cophylogeny in maintenance of host specificity. PLoS ONE 8:e63601. https://doi.org/10.1371/journal.pone.0063601
Vrba V, Pakandl M (2014) Coccidia of turkey: from isolation, characterisation and comparison to molecular phylogeny and molecular diagnostics. Int J Parasitol 44:985–1000. https://doi.org/10.1016/j.ijpara.2014.06.004
Yang R, Brice B, Ryan U (2016) Morphological and molecular characterization of Eimeria purpureicephali n. sp. (Apicomplexa: Eimeriidae) in a red-capped parrot (Purpureicephalus spurius, Kuhl, 1820) in Western Australia. Int J Parasitol Parasites Wildl 5:34–39. https://doi.org/10.1016/j.ijppaw.2016.01.003
Ortúzar-Ferreira CN, Oliveira MS, Genovez-Oliveira JL, Franco HA, Thode-Filho S, Cardozo SV, Oliveira ÁA, Lima VM, Ferreira I, Berto BP (2020) Coccidia of Columbiformes: a taxonomic review of its Eimeriidae species and Eimeria columbinae n. sp. from Columbina talpacoti (Temminck, 1809) from Brazil. Parasitol Res 119:267–281. https://doi.org/10.1007/s00436-019-06514-4
Ferreira LF (1973) O fenômeno parasitismo. Rev Soc Bras Med Trop 3:261–277. https://doi.org/10.1590/S0037-86821973000400006
Atkinson CT, Thomas NJ, Hunter DB (2008) Parasitic diseases of wild birds. Wiley-Blackwell, Singapore
BirdLife International (2016) The IUCN red list of threatened species. http://www.iucnredlist.org. Accessed 23 May 2022
Billerman M, Keeney BK, Rodewald PG, Schulenberg TS (2022) Birds of the World. Cornell Lab of Ornithology, Ithaca. https://birdsoftheworld.org/bow/home. Accessed 23 May 2022
Berto BP, Lopes CWG (2020) Coccidia of wild birds as ecological biomarkers: some approaches on parasite-host-environment interaction. J Parasitol 106:707–713. https://doi.org/10.1645/19-148
Silva LM, Rodrigues MB, Pinho IF, Lopes BB, Luz HR, Ferreira I, Lopes CWG, Berto BP (2017) Some remarks on the distribution and dispersion of coccidia from icterid birds in South America: Isospora guaxi n. sp. and Isospora bellicosa Upton, Stamper & Whitaker, 1995 (Apicomplexa: Eimeriidae) from the red-rumped cacique Cacicus haemorrhous (L.)(Passeriformes: Icteridae) in Southeastern Brazil. Syst Parasitol 94:151–157. https://doi.org/10.1007/s11230-016-9688-y
Silva-Carvalho LM, Pastura DG, Rodrigues MB, Gomes JV, Oliveira MS, Siqueira PB, Genovez-Oliveira JL, Soares SS, Oliveira ÁA, Lima VM, Ferreira I, Berto BP (2018) Isospora sagittulae McQuistion & Capparella, 1992 (Apicomplexa: Eimeriidae) from antbirds (Passeriformes: Thamnophilidae) in the Amazon and Atlantic Forest of Brazil: with notes on its distribution and dispersion in the Neotropical region. Parasitol Res 117:2635–2641. https://doi.org/10.1007/s00436-018-5955-y
Frigerio D, Cibulski L, Ludwig SC, Campderrich I, Kotrschal K, Wascher CAF (2016) Excretion patterns of coccidian oocysts and nematode eggs during the reproductive season in Northern Bald Ibis (Geronticus eremita). J Ornithol 157:839–851. https://doi.org/10.1007/s10336-015-1317-z
Parsani HR, Momin RR, Sahu RK, Patel BG (2003) Prevalence of gastro-intestinal parasites in captive birds at Kalma Nehru Zoological Garden, Kankaria Zoo, Ahmedabad, Gujarat. Zoo’s Print J 18:987–992
Bastian S, Yesou P, Clergeau P, Laroucau K, Pellerin JL, Hars J, Bazus J, Passet A, Lagrange P, L’Hostis M (2010) Eléments pour l’évaluation des risques sanitaires liés aux Ibis sacrés (Threskiornis aethiopicus) en France. Rapport d’étude pour la Direction Régionale de l’Environnement Bretagne et la Direction Régionale de l’Environnement, de l’Aménagement et du Logement des Pays de la Loire. https://controverses.minesparis.psl.eu/public/promo15/promo15_G6/www.oncfs.gouv.fr/IMG/pdf/rapport_ibis_pathogenes.pdf. Accessed 1 Mar 2022
Doležalová M, Torres J, Fernández H, Modrý D (2004) Isospora araponga sp. n. (Apicomplexa: Eimeriidae), a new species of Isospora Schneider from a bare-throated bellbird, Procnias nudicollis (Vieillot, 1817) (Passeriformes: Cotingidae) from Brazil. Mem Inst Oswaldo Cruz 99:829–830. https://doi.org/10.1590/S0074-02762004000800008
Berto BP, Teixeira M, Lopes CWG (2007) Tyzzeria parvula (Kotlán, 1933) Klimes, 1963 (Apicomplexa: Eimeriidae) in the greylag goose (Anser anser Linnaeus, 1758) in southeastern Brazil. Rev Bras Parasitol Vet 16:156–158. https://doi.org/10.1590/S1984-29612007000300008
WikiAves (2022) Wiki Aves—A Enciclopédia das Aves do Brasil. https://www.wikiaves.com.br/wiki/curicaca. Accessed 01 Mar 2022
Sigrist T (2014) Guia de Campo: Avifauna Brasileira. Avis Brasilis, São Paulo
Eckert J, Braun R, Shirley MW, Coudert P (1995) Biotechnology—guidelines on techniques in Coccidiosis Research. European Commission, Luxembourg
Ericson PGP, Anderson CL, Britton T, Elzanowski A, Johansson US, Källersjö M, Ohlson JL, Parsons TJ, Zuccon D, Mayr G (2006) Diversification of Neoaves: integration of molecular sequence data and fossils. Biol Lett 2:543–547. https://doi.org/10.1098/rsbl.2006.0523
Hackett SJ, Kimball RT, Reddy S, Bowie RCK, Braun EL, Braun MJ, Chojnowski JL, Cox WA, Han KL, Harshman J, Huddleston CJ, Marks BD, Miglia KJ, Moore WS, Sheldon FH, Steadman DW, Witt CC, Yuri T (2008) A phylogenomic study of birds reveals their evolutionary history. Science 320:1763–1768. https://doi.org/10.1126/science.1157704
Mayr G (2011) Metaves, Mirandornithes, Strisores and other novelties—a critical review of the higher-level phylogeny of neornithine birds. J Zool Syst Evol Res 49:58–76. https://doi.org/10.1111/j.1439-0469.2010.00586.x
Labbé A (1893) Sur les coccidies des oiseaux. C R Seances Acad Sci 117:407–409
Hoare CA (1933) Studies on some new ophidian and avian coccidian from Uganda, with a revision of the classification of the Eimeriidae. Parasitol 24:359–388. https://doi.org/10.1017/S0031182000019569
Yabsley MJ, Gibbs SE (2006) Description and phylogeny of a new species of Eimeria from double-crested cormorants (Phalacrocorax auritus) near Fort Gaines, Georgia. J Parasitol 92:385–388. https://doi.org/10.1645/GE-592R.1
Gardner SL, Duszynski DW (1990) Polymorphism of eimerian oocysts can be a problem in naturally infected hosts: an example from subterranean rodents in Bolivia. J Parasitol 76:805–811 (3282798)
Golemansky V, Kuldjieva D (1980) Eimeria garzettae sp. n. (Coccidia: Eimeriidae) in the little egret (Egretta garzetta) from Bulgaria. Acta Protozoologica 19:177–179
Dubinin VB (1939) The coccidiosis of the herons in the state Astrakhan reservate. Seriya Biologischeskikh Nauk 11:44–57
Courtney CH, Ernst JV (1975) Eimeria pelecani sp. n. from the brown pelican, Pelecanus occidentalis carolinensis, from Florida. J Parasitol 61:1081–1082. https://doi.org/10.2307/3279380
Shamsuddin M, Jasim MK (1980) Coccidia of some birds and mammals from Iraq. Bull Nat Hist Res Centre 7:81–109
Yabsley MJ, Gottdenker NL, Fischer JR (2002) Description of a new Eimeria sp. and associated lesions in the kidneys of double-crested cormorants (Phalocrocorax auritus). J Parasitol 88:1230–1233. https://doi.org/10.1645/0022-3395(2002)088[1230:DOANES]2.0.CO;2
Ogedengbe JD, Hanner RH, Barta JR (2011) DNA barcoding identifies Eimeria species and contributes to the phylogenetics of coccidian parasites (Eimeriorina, Apicomplexa, Alveolata). Int J Parasitol 41:843–850. https://doi.org/10.1016/j.ijpara.2011.03.007
Genovez-Oliveira JL, Oliveira MS, Thode-Filho S, Cardozo SV, Oliveira ÁA, Lima VM, Ferreira I, Berto BP (2020) Morphological and molecular identification of Isospora massardi Lopes, Berto, Luz, Galvão, Ferreira & Lopes, 2014 (Chromista: Miozoa: Eimeriidae) from thrushes Turdus spp. (Passeriformes: Turdidae) in South America. Parasitol Int 75:102040. https://doi.org/10.1016/j.parint.2019.102040
Acknowledgements
We thank Dr. Donald W. Duszynski (Emeritus Professor of Biology, University of New Mexico), the Biblioteca de Ciências Biomédicas of the Fundação Oswaldo Cruz, the Central Library of the Universidade Federal Rural do Rio de Janeiro, and the Library of the Museu Nacional of the Universidade Federal do Rio de Janeiro, for providing us with old and rare scientific articles that were essential for the conclusion of this study.
Funding
This study was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ). RBGC has a scholarship from CNPq (Grant/Award number: 170041/2018-2). CNO-F has a scholarship from CNPq/UFRRJ (Grant/Award Number: PVBS2485-2021). MSO has a post-doctoral scholarship from FAPERJ (Grant/Award number: E-26/204.228/2021). BPB has fellowships from CNPq (Grant/Award number: 303899/2019-0) and from FAPERJ (Grant/Award number: E-26/202.797/2019).
Author information
Authors and Affiliations
Contributions
The study was designed by VML, IF and BPB. Field work was performed by IF and RBGC. Laboratory procedures for maintenance, recovery, measurements, photomicrographs and isolation of oocysts were performed by RBGC, CNO-F and MSO. DNA extraction, amplification and sequencing were performed by ERM, AAO and VML. BPB analyzed the data and drew the coccidian oocyst. The manuscript was written by RBGC, CNO-F and BPB and subsequently revised by all other authors. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no confict of interest.
Ethical approval
Field-collecting permits were issued by SISBIO/ICMBio (license 42798) and CEUA/IV/UFRRJ (protocol 6606250616). All applicable institutional, national and international guidelines for the care and use of animals were followed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Cabral, R.B.G., Ortúzar-Ferreira, C.N., Oliveira, M.d.S. et al. Worldwide Dispersion of Coccidia from Migratory Birds: First Report of Eimeria bazi Chauhan et Bhatia, 1970 (Eimeriidae) Outside Asia from Buff-Necked Ibises Theristicus caudatus (Boddaert, 1783) (Threskiornithidae) in South America. Acta Parasit. 67, 1343–1353 (2022). https://doi.org/10.1007/s11686-022-00585-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11686-022-00585-7