Abstract
GRM5 (coding for metabotropic glutamate receptor 5, mGluR5) is a promising target for the treatment of cognitive deficits in schizophrenia, but there has been little investigation of its association with cognitive and brain phenotypes within this disorder. We examined the effects of common genetic variation in GRM5 with cognitive function, hippocampal volume, and hippocampal mGluR5 protein levels in schizophrenia patients relative to healthy controls. Two independent GRM5 variants rs60954128 [C>T] and rs3824927 [G>T] were genotyped in a schizophrenia case/control cohort (n=249/261). High-resolution anatomical brain scans were available for a subset of the cohort (n=103 schizophrenia /78 control). All participants completed a standard set of neuropsychological tests. In a separate postmortem cohort (n=19 schizophrenia/20 controls), hippocampal mGluR5 protein levels were examined among individuals of different GRM5 genotypes. Schizophrenia minor allele carriers of rs60954128 had reduced right hippocampal volume relative to healthy controls of the same genotype (−12.3%); this effect was exaggerated in males with schizophrenia (−15.6%). For rs3824927, compared to major allele homozygotes, minor allele carriers with schizophrenia had lower Intelligence Quotients (IQ). Examination in hippocampal postmortem tissue showed no difference in mGluR5 protein expression according to genotype for either rs60954128 or rs3824927. While these genetic variants in GRM5 were associated with cognitive impairments and right hippocampal volume reduction in schizophrenia, they did not affect protein expression. Further study of these mechanisms may help to delineate new targets for the treatment of cognitive deficits in schizophrenia, and may be relevant to other disorders.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Schizophrenia is a complex psychiatric disorder, with genetic understanding inherently challenged by its heterogeneous clinical presentation and polygenic origin, as highlighted by recent genome-wide association studies (GWAS; Schizophrenia Working Group of the Psychiatric Genomics Consortium 2014). Despite the success of these GWAS involving large global consortia, there remains a need for candidate gene approaches to determine the functional significance of implicated genes, particularly when there is strong a priori evidence for potential mechanisms of interest based on other biological data (Tabor et al. 2002; Wilkening et al. 2009). Aberrant glutamate transmission has been implicated in the pathophysiology of schizophrenia for several decades (Beck et al. 2016; Tsai et al. 1998) and specifically cognitive function in the disorder (Yang et al. 2016). More recently the role of glutamate signaling has been supported by the association of several common genetic variants (including GRM3, GRIN2A and GRIA1) with schizophrenia within large-scale GWAS (Schizophrenia Working Group of the Psychiatric Genomics Consortium 2014). This also includes the detection of rare copy number variations affecting post-synaptic N-methyl-D-aspartate receptor (NMDAR) function (Kirov et al. 2012). Among relevant genetic candidates affecting glutamate transmission, GRM5 encodes for the postsynaptic metabotropic glutamate receptor 5 (mGluR5). mGluR5 is a compelling drug target for the treatment of cognitive deficits in schizophrenia because of its ability to mediate post-synaptic NMDA receptor (NMDAR) currents (Attucci et al. 2001; Jia et al. 1998; Mannaioni et al. 2001), with proper function of NMDARs being critical for cognition (Malhotra et al. 1996). However, there has been little investigation of novel GRM5 variants in relation to cognitive and brain features of schizophrenia that would assist to determine its utility as a therapeutic target. The goal of the present study was thus to investigate the association of GRM5 polymorphisms with specific cognitive, molecular and morphological brain indices, as well as cognitive subtypes of schizophrenia.
Relatively few studies have examined genetic variation within GRM5 in schizophrenia, despite the first report being over a decade ago. This study showed genetic linkage of a long-range restriction map covering the GRM5 locus was associated with schizophrenia within a large Scottish pedigree (Millar et al. 1998). A novel intragenic microsatellite in this gene was subsequently associated with schizophrenia in a Caucasian case-control study (Devon et al. 2001). Alongside these human studies, GRM5 knockout or pharmacological blockade of mGluR5 in rodent models were shown to induce a wide range of schizophrenia-like behaviors. In particular deficits in hippocampal-dependent functions were reported, including, spatial learning and memory (Ayala et al. 2009; Manahan-Vaughan and Braunewell 2005). mGluR5 activity in the hippocampus is critical for modulation of the molecular mechanisms underlying hippocampal synaptic plasticity (Manahan-Vaughan and Braunewell 2005), and recent reports indicate that the expression of mGluR5 and mGluR5 regulatory proteins may be altered in hippocampal tissues from schizophrenia patients (Matosin et al. 2015a). Although no single GRM5 variant has reached the high threshold of significance in recent genome-wide association studies (Ripke et al. 2011; Ripke et al. 2013; Schizophrenia Working Group of the Psychiatric Genomics Consortium 2014), a convergent functional genomics study ranked GRM5 as a top candidate gene for schizophrenia (Ayalew et al. 2012). The effects of GRM5 on cognitive and brain phenotypes of schizophrenia require further exploration.
In this study, we examined association of GRM5 variants with schizophrenia, as well as a range of cognitive functions typically impaired in schizophrenia. Particular focus was given to the hippocampus following evidence from postmortem brain studies indicating alterations to mGluR5 expression and regulation in this region (Matosin et al. 2015a). While other GRM5 variants have been previously examined in schizophrenia (Ayalew et al. 2012; Devon et al. 2001; Timms et al. 2013), we were specifically interested in variants located in the 3′ untranslated region (3′ UTR) of GRM5, as this region is involved in post-transcriptional regulation of protein expression, and is highly relevant considering mGluR5 protein levels are affected in the postmortem schizophrenia brain (Matosin, Fernandez-Enright, Lum, and Newell 2015). Two genetically independent GRM5 variants (rs60954128 and rs3824927) were selected based on their minor allele frequency (MAF) in Caucasian populations, which was greater than 10%. The association of these variants with hippocampal volume and related cognitive functions were investigated in an established case-control cohort, and the association of these variants with mGluR5 protein abundance in the CA1 hippocampal region was examined in an independent case-control postmortem brain cohort.
Methods
Case-control cohort
Participants
DNA for 249 schizophrenia cases and 261 controls was acquired from the Australian Schizophrenia Research Bank (ASRB; Loughland et al. 2010). All subjects were Caucasian to prevent potential confounding effects of population stratification; schizophrenia cases were diagnosed according to DSM-IV criteria, and matched with controls (no prior personal or family history of mental disorders) according to sex and age (Table 1). The majority of schizophrenia subjects were medicated (58 typical antipsychotics, 211 atypical antipsychotics, 54 mood stabilizer, 92 antidepressants). This work was approved by the University of Wollongong (HE10/161) and the University of New South Wales (HC12658).
Genotyping
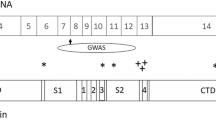
Two SNPs within the 3′ UTR of the GRM5 gene (rs60954128 [C > T], 11:88,506,288; rs3824927 [G > T], 11:88,508,028; see Fig. 1; Sherry et al. 2001) were selected on the basis of their position in the 3’UTR of GRM5 and having MAF > 10% in a Caucasian population in the National Centre for Biotechnology Information (NCBI) genetic database. High-throughput genotyping was performed using MassARRAY® (Sequenom, Inc., San Diego, CA, USA) and analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). Polymerase-chain reaction (PCR) and extension primer design, selection and multiplexing were performed using MassARRAY® Designer Software (Sequenom, Inc.).
Structural magnetic resonance imaging processing
Imaging data from the ASRB was available for a subset of 78 controls (55 males) and 103 cases (72 males). High-resolution T1-weighted structural MRI scans (MPRAGE; 176 contiguous 1 mm sagittal slices; field-of-view 250 × 250 mm2, time-to-repetition 1980 ms, time-to-echo 4.3 ms, data acquisition matrix 256 × 256, voxel size 0.98 × 0.98 × 1.0 mm3, flip angle 15°) were collected on Siemens Avanto 1.5 T scanners across five Australian research sites (Loughland et al. 2010). Scan quality control was performed by a trained researcher, whilst blind to all genetic, clinical and diagnostic data. Stringent exclusion criteria were applied for excess motion or other artifacts (Supplementary Methods M1).
Neuropsychological measures
Standardized estimates of premorbid and current intelligence quotient (IQ) were obtained using the Wechsler Tests for Adult Reading (WTAR; Wechsler 2001) and Wechsler Abbreviated Scale of Intelligence (WASI; Wechsler 1997). Executive functions were assessed using the Controlled Oral Word Association Test (COWAT; Spreen 1998) and the Letter Number Sequencing Test (LNS; Wechsler 1997). The Repeatable Battery for the Assessment of Neuropsychological Status (RBANS) was used to derive indices of attention, delayed memory, immediate memory, visuospatial construction, and language (Randolph 1998).
Postmortem cohort
Postmortem human samples from the cornu ammonis (CA)1 region of 39 subjects (19 schizophrenia, 20 control; Table 2) were obtained from the New South Wales Brain Bank Network (Sydney, Australia). mGluR5 protein levels were previously determined by quantitative immunoblot (Matosin et al. 2015a). Genomic DNA was extracted from postmortem tissues using the QIAamp DNA Mini Kit (Qiagen, Australia). This work was approved by the University of Wollongong (HE 10/161 and HE99/222).
Statistical methods
All analyses were performed using SPSS (v22, IBM). Genotypic distributions were assessed for deviation from Hardy Weinberg Equilibrium (HWE; P > 0.05). Analyses were tested against a significance level of α = 0.05 and Bonferroni-Holm adjusted P values were computed to control for multiple comparisons (Aickin and Gensler 1996). All analyses were additionally performed separately for males and females (Goldstein et al. 2013). Minor allele carriers were compared to major allele homozygotes in cases of small sample sizes (in all analyses except i), as described below. Potential effects of medication dosage could not be considered in the ASRB sample, due to limited details of medication history and dosage collected at the time of testing.
-
(i)
Allelic/genotypic frequency among cases/controls: Differences in allelic and genotypic frequencies between the schizophrenia group and controls were examined using χ2 analyses performed for each SNP. Linkage disequilibrium (LD) analyses based on absolute D’ values (pairwise comparison) and haplotype association tests (per haplotype) were also generated (v8.2 SVS Institute, Cary, NC, USA; SNP & Variation Suite, Golden Helix).
-
(ii)
SNP effects on hippocampal volume: The effects of genotype, diagnosis, and their potential interactive effects on the adjusted hippocampal volumes were examined using mixed-design multiple analyses of covariance (MANCOVAs), with the left and right adjusted hippocampal volumes as dependent variables, genotype (major allele homozygotes/minor allele carriers) and diagnosis (cases/controls) as independent factors, and age, sex and scan location as covariates. Significant effects were followed up with relevant ANCOVA models.
-
(iii)
SNP effects on cognition: For each of nine cognitive performance measures, main effects of genotype, diagnosis and their interaction were examined using a series of mixed-design multiple analyses of variance (MANOVAs), with two levels of the between-subjects factors of genotype (major allele homozygotes/minor allele carriers) and diagnosis (cases/controls). Significant interactions were followed up with analyses of variance (ANOVAs) using Tukey’s post-hoc tests.
-
(iv)
SNP effects on protein levels in postmortem brain tissue: mGluR5 protein levels were drawn from our previous immunoblot study in CA1 hippocampal tissues (Matosin et al. 2015a). Spearman correlations were initially used to identify potential correlations of mGluR5 with confounding factors, including brain pH, age at death, postmortem interval, RNA integrity, freezer storage time (months), brain volume, illness duration and lifetime antipsychotic drug exposure (chlorpromazine equivalent); ANCOVAs were then implemented to test the effect of genotype on mGluR5 protein levels in all subjects (irrespective of diagnosis due to limited sample size).
Results
Genotypic/allelic frequencies and differences among SNPs
rs60954128 and rs3824927 did not deviate from HWE. The control group showed a similar MAF as those reported in Caucasian populations (http://www.ncbi.nlm.nih.gov/projects/SNP; MAF-rs60954128 = 14.2%; MAF-rs3824927 = 51.5%; Table 3). No significant results were found from the haplotype association analysis after Bonferroni-Holm correction for multiple comparisons. Pairwise LD statistics of the sample revealed low to moderate LD between rs60954128 and rs3824927 (D’ = 0.37, R2 = 0.23; Supplementary Fig. S1) and haplotype association analysis did not reveal any significant associations, after Bonferroni-Holm correction for multiple comparisons. These results support the hypothesis of a limited effect of substructure population in our control group although further analysis will be necessary to confirm this hypothesis.
A significant difference in rs60954128 allelic frequency was observed in cases relative to controls (χ2 = 5.133, Bonferroni-Holm corrected P < 0.001; Table 3). Sex-specific analyses showed that rs60954128 minor alleles and homozygous genotypes were over-represented in male schizophrenia cases relative to male controls following Bonferroni-Holm correction (allelic frequency: χ2 = 6.106, Bonferroni-Holm corrected P = 0.004; genotypic frequency: χ2 = 9.085, P = 0.011, Bonferroni-Holm corrected P = 0.002). For rs3824927, there was no difference in genotype or allele frequency in the schizophrenia group relative to controls (Table 3).
Effects of genotypes on hippocampal volume
A significant group-by-genotype interaction for rs60954128 was observed in the right hippocampus (F1,131 = 6.512, P = 0.012, Bonferroni-Holm corrected P = 0.001). Post-hoc analyses for the right hippocampus showed schizophrenia minor allele carriers had reduced right hippocampal volume compared to control subjects of the same genotype (P < 0.001, Bonferroni-Holm corrected P < 0.001; −12.3%; Fig. 2b), but not compared to schizophrenia major allele homozygotes at the reduced significance following Bonferroni-Holm correction (P = 0.030; Bonferroni-Holm corrected P = −0.022). Sex-specific analyses for schizophrenia showed this interaction was driven by men (F1,95 = 6.507, P = 0.012; Bonferroni-Holm corrected P < 0.001), with male schizophrenia rs60954128 minor allele carriers having reduced right hippocampal volume relative to control minor allele carriers (P < 0.001; −15.8%) and compared to male schizophrenia major allele carriers (P = 0.003; −15.6%). No effects of rs3824927 were observed on hippocampal volumes.
a Increased minor allele frequency of rs60954128 with schizophrenia and specifically in males. b Reduced right hippocampal volume in schizophrenia minor allele carriers relative to control minor allele carriers. Abbreviations: Adj hippocampal vol, adjusted hippocampal volume; HC, healthy control; SZ, schizophrenia. *significance according to Bonferroni-Holm correction for multiple comparisons
Effects of genotypes on cognitive function
Schizophrenia subjects performed significantly worse than controls on all cognitive assessments (F3,451 ≥ 7.36, P ≤ 0.007; Supplementary Table S1). There were no main effects of either SNP after Bonferroni-Holm correction. For rs3824927, group-by-genotype interaction effects for rs3824927 were observed for WASI (IQ estimates; F3,457 = 10.866, P = 0.001, Bonferroni-Holm corrected P = 0.001), with scores in this test significantly lower in schizophrenia minor allele carriers compared to schizophrenia major allele homozygotes (P = 0.015; Fig. 3). Sex-specific analysis showed the same effect was present in only males (F3,300 = 11.073, P < 0.001, Bonferroni-Holm corrected P < 0.001). No other effects of rs60954128 or rs3824927 were observed following Bonferroni-Holm correction (Supplementary Table S1).
Effects of rs3814927 on WASI IQ scores, which was reduced in schizophrenia minor allele carriers relative to schizophrenia major allele carriers, and both control genotypes. Abbreviations: HC, healthy control; SZ, schizophrenia; WASI, Wechsler Abbreviated Scale of Intelligence. *significance according to Bonferroni-Holm correction for multiple comparisons
Effects of genotype on postmortem mGluR5 protein levels in CA1
One schizophrenia subject (male) was removed from the final analysis due to technical issues, and thus these results are presented for 20 control and 19 schizophrenia subjects. As distribution of mGluR5 protein was skewed to the right (Matosin et al. 2015a), normalized distribution for these proteins for parametric tests was achieved by transforming to the natural logarithm of the relative protein values. Spearman’s Rho were initially used to identify potential correlations of untransformed mGluR5 protein values with confounding factors, including brain pH, age at death, postmortem interval, RNA integrity, freezer storage time (months), brain volume, illness duration and lifetime antipsychotic drug exposure (chlorpromazine equivalent); mGluR5 protein level was significantly associated with brain pH (R = −0.355, P = 0.026) and freezer storage time (R = 0.356, P = 0.026). Subsequent ANCOVA, using brain pH and freezer storage time as covariates, showed that neither rs60954128 (n = 27 CC and n = 12 CT, no TT carriers; F1,39 = 0.038; P = 0.847) nor rs3824927 (n = 10 GG, n = 22 GT and n = 7 TT; F1,39 = 0.274; P = 0.762) significantly affected mGluR5 protein level. Genotype-by-diagnosis interactions could not be performed owing to small sample sizes.
Discussion
This study examined the association of two genetic polymorphisms (rs60954128 and rs3824927) situated in the 3’UTR of GRM5, with cognitive functions, hippocampal volume and levels of mGluR5 protein in the hippocampus, in people with schizophrenia. There have been no such investigations of these genetic variants to date, despite accumulating evidence for the role of GRM5 in schizophrenia etiology. In this study, rs60954128 minor alleles were more highly represented in patients with schizophrenia compared to controls, with these minor allele carriers also having reduced volumes of the right hippocampus compared to healthy participants of the same genotype. These effects were strongest in males with schizophrenia. Allelic frequency of rs3824927 was not different in schizophrenia relative to controls, but minor allele carriers with schizophrenia had lower IQ scores compared to major allele homozygotes with schizophrenia, and all control subjects regardless of genotype. In the postmortem samples, GRM5 genotypes were not associated with the difference of mGluR5 protein levels in the CA1 region in the tested cohort. The two SNPs examined in this study were in low to moderate LD, suggesting that they operate independently. The differential associations with brain structure and cognition observed in this study may be taken as some support for this.
That rs60954128 minor allele carriers had reduced volume of the right hippocampus compared to healthy participants of the same genotypes (in combination with lower cognitive performance of schizophrenia cases) is consistent with previous associations between hippocampal integrity and cognitive ability (Colom et al. 2013; Minh 2014), including the effects of mGluR5 on hippocampal-dependent plasticity and spatial memory (Manahan-Vaughan and Braunewell 2005; Mukherjee and Manahan-Vaughan 2013). Although we found no effects of rs60954128 and hippocampal-dependent cognitive functions (e.g. delayed memory), it remains possible that other hippocampal-dependent cognitive functions that were not tested in this sample (e.g., long-term memory function) may be associated with common variation in GRM5 genes. In contrast, a strong effect of rs3824927 was observed on general intellectual ability in schizophrenia, with minor allele carriers showing reduced IQ relative to major allele homozygotes.
The contribution of GRM5 to intellectual ability has been established in the context of other disorders for which the clinical phenotype and genetic architecture overlaps with schizophrenia (Doherty and Owen 2014). For example, in Fragile X syndrome (FXS), which is characterized by severe mental retardation, reduced GRM5 activity improves intellectual ability (Dölen and Bear 2008). This finding suggests that high levels of GRM5 (which have been reported in the hippocampus in schizophrenia) may lead to impaired IQ (Matosin et al. 2015a). FXS commonly occurs in children with attention deficit hyperactivity disorder (ADHD) (Sullivan et al. 2006), in which self-regulation deficits appear to affect IQ (Kuntsi et al. 2004). A deletion in GRM5 has been identified in a familial study of ADHD (Elia et al. 2010), providing further support for this association. The rs3824927 variant might be involved in mediating impaired intelligence in schizophrenia and other psychiatric disorders, potentially via genetic effects on epigenetic modifications that affect chromosome conformation and result in altered gene regulation (Krijger and de Laat 2016). Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR/Cas9) gene editing in rodent models would be valuable to assess the effects of these polymorphisms on behavior, particularly domains of cognition, as well as treatment response to both novel and current antipsychotic medications.
Given that the variants examined here are located within the 3′ UTR of GRM5, they might have potential regulatory functions, such as control of RNA splicing/stability, translation, microRNA interactions, and transcription. However, mGluR5 protein levels were not affected by the GRM5 genetic variants examined in this study. This finding should be considered with regards that many polymorphisms influence gene regulation, not only at the level of the full length transcript and protein, but also by altering the expression of alternative transcripts. This is important as GRM5 has (at least) two alternative transcripts encoding two separate proteins, mGluR5a and mGluR5b as indexed in RefSeq. As our protein measures did not discriminate between the mGluR5 isoforms, the molecular consequences of the genetic variants in this study require further exploration.
This study has some important limitations. Firstly, the potential effects of medication dosage could not be considered in the ASRB sample, due to limited details of medication history and dosage collected at the time of testing; we note that no influence of typical or atypical antipsychotic drug treatment on the mGluR5 protein system was reported in the prefrontal cortex and hippocampus of rodents (Matosin et al. 2015a). The limitations of single SNP studies should also be considered, and determination of how these genetic variants operate in different biological and environmental contexts (for example, in relation to other genotypes or epigenetic regulation) will be important in the future. The relatively small size of our postmortem cohort precludes observations of any possible association between GRM5 variants and mGluR5 protein expression, and lastly, no individuals were homozygous for the minor allele, which limited our ability to test group by genotype interactions. Further investigation and replication in larger cohorts will be important.
In summary, this work suggests contribution of common variants of GRM5 to impaired attention and reduced right hippocampal volume in schizophrenia, particularly in men, suggesting these variants may operate in a sex-specific manner. The effect of rs3814927 on IQ may have further implications of these findings in other disorders in which GRM5 is also associated such as FXS and ADHD. Additionally, our findings suggest that changes to hippocampal volume associated with rs60954128 variation could contribute to the cognitive decline in schizophrenia, and may therefore lead to novel therapeutic targets. Further investigation of the functional and phenotypic associations of GRM5 would be useful in the context of the potential interactive epistatic effects of these genetic variants with other GRM5 variants with this region, or other genes, which will allow determination of key pathways affected in schizophrenia.
References
Aickin, M., & Gensler, H. (1996). Adjusting for multiple testing when reporting research results: The Bonferroni vs Holm methods. American Journal of Public Health, 86(5), 726–728.
Attucci, S., Carla, V., Mannaioni, G., & Moroni, F. (2001). Activation of type 5 metabotropic glutamate receptors enhances NMDA responses in mice cortical wedges. British Journal of Pharmacology, 132(4), 799–806.
Ayala, J. E., Chen, Y., Banko, J. L., Sheffler, D. J., Williams, R., Telk, A. N., et al. (2009). mGluR5 positive allosteric modulators facilitate both hippocampal LTP and LTD and enhance spatial learning. Neuropsychopharmacology, 34(9), 2057–2071. doi:10.1038/npp.2009.30.
Ayalew, M., Le-Niculescu, H., Levey, D. F., Jain, N., Changala, B., Patel, S. D., et al. (2012). Convergent functional genomics of schizophrenia: From comprehensive understanding to genetic risk prediction. Molecular Psychiatry, 17(9), 887–905.
Beck, K., Javitt, D. C., & Howes, O. D. (2016). Targeting glutamate to treat schizophrenia: Lessons from recent clinical studies. Psychopharmacology, 233(13), 2425–2428.
Colom, R., Stein, J. L., Rajagopalan, P., Martínez, K., Hermel, D., Wang, Y., et al. (2013). Hippocampal structure and human cognition: Key role of spatial processing and evidence supporting the efficiency hypothesis in females. Intelligence, 41(2), 129–140.
Devon, Anderson S., Teague, P., Muir, W., Murray, V., Pelosi, A., et al. (2001). The genomic organisation of the metabotropic glutamate receptor subtype 5 gene, and its association with schizophrenia. Molecular Psychiatry, 6(3), 311.
Doherty, J. L., & Owen, M. J. (2014). Genomic insights into the overlap between psychiatric disorders: Implications for research and clinical practice. Genome Medicine, 6(4), 29. doi:10.1186/gm546.
Dölen, G., & Bear, M. F. (2008). Role for metabotropic glutamate receptor 5 (mGluR5) in the pathogenesis of fragile X syndrome. The Journal of Physiology, 586(6), 1503–1508.
Elia, J., Gai, X., Xie, H. M., Perin, J. C., Geiger, E., Glessner, J. T., et al. (2010). Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Molecular Psychiatry, 15(6), 637–646.
Goldstein, J. M., Cherkerzian, S., Tsuang, M. T., & Petryshen, T. L. (2013). Sex differences in the genetic risk for schizophrenia: History of the evidence for sex-specific and sex-dependent effects. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 162(7), 698–710.
Jia, Z., Lu, Y., Henderson, J., Taverna, F., Romano, C., Abramow-Newerly, W., et al. (1998). Selective abolition of the NMDA component of long-term potentiation in mice lacking mGluR5. Learning & Memory, 5(4), 331–343. doi:10.1101/lm.5.4.331.
Kirov, G., Pocklington, A. J., Holmans, P., Ivanov, D., Ikeda, M., Ruderfer, D., et al. (2012). De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Molecular Psychiatry, 17(2), 142–153. doi:10.1038/mp.2011.154.
Krijger, P.H.L., & de Laat, W. (2016). Regulation of disease-associated gene expression in the 3D genome. Nature Reviews Molecular Cell Biology. http://www.nature.com/nrm/journal/vaop/ncurrent/full/nrm.2016.138.html. Accessed 24 January 2017.
Kuntsi, J., Eley, T. C., Taylor, A., Hughes, C., Asherson, P., Caspi, A., & Moffitt, T. E. (2004). Co-occurrence of ADHD and low IQ has genetic origins. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 124(1), 41–47.
Loughland, C., Draganic, D., McCabe, K., Richards, J., Nasir, A., Allen, J., et al. (2010). Australian schizophrenia research Bank: A database of comprehensive clinical, endophenotypic and genetic data for aetiological studies of schizophrenia. Australian and New Zealand Journal of Psychiatry, 44(11), 1029–1035.
Malhotra, A. K., Pinals, D. A., Weingartner, H., Sirocco, K., Missar, C. D., Pickar, D., & Breier, A. (1996). NMDA receptor function and human cognition: The effects of ketamine in healthy volunteers. Neuropsychopharmacology, 14(5), 301–307. doi:10.1016/0893-133x(95)00137-3.
Manahan-Vaughan, D., & Braunewell, K. H. (2005). The metabotropic glutamate receptor, mGluR5, is a key determinant of good and bad spatial learning performance and hippocampal synaptic plasticity. Cerebral Cortex, 15(11), 1703–1713.
Mannaioni, G., Marino, M. J., Valenti, O., Traynelis, S. F., & Conn, P. J. (2001). Metabotropic glutamate receptors 1 and 5 differentially regulate CA1 pyramidal cell function. The Journal of Neuroscience, 21(16), 5925–5934.
Matosin, N., Fernandez-Enright, F., Lum, J. S., Andrews, J. L., Engel, M., Huang, X.-F., & Newell, K. A. (2015a). Metabotropic glutamate receptor 5, and its trafficking molecules Norbin and Tamalin, are increased in the CA1 hippocampal region of subjects with schizophrenia. Schizophrenia Research, 166(1–3), 212–218. doi:10.1016/j.schres.2015.05.001.
Matosin, N., Fernandez-Enright, F., Lum, J. S., & Newell, K. A. (2015b). Shifting towards a model of mGluR5 dysregulation in schizophrenia: Consequences for future schizophrenia treatment. Neuropharmacology. doi:10.1016/j.neuropharm.2015.08.003.
Millar, J. K., Brown, J., Maule, J. C., Shibasaki, Y., Christie, S., Lawson, D., et al. (1998). A long-range restriction map across 3 Mb of the chromosome 11 breakpoint region of a translotion linked to schizophrenia: Localization of the breakpoint and the search for neighbouring genes. Psychiatric Genetics, 8(3), 175–182.
Minh, T. N. N. (2014). Relationship between subcortical volumes and cognitive abilities in young healthy adults. Vietnam: International University HCMC Retrieved from http://csc.hcmiu.edu.vn:8080/dspace/handle/123456789/1166.
Mukherjee, S., & Manahan-Vaughan, D. (2013). Role of metabotropic glutamate receptors in persistent forms of hippocampal plasticity and learning. Neuropharmacology, 66, 65–81.
Randolph, C. (1998). RBANS manual: Repeatable battery for the assessment of neuropsychological status. San Antonio, TX: The Psychological Corporation.
Ripke, S., O’Dushlaine, C., Chambert, K., Moran, J. L., Kähler, A. K., Akterin, S., et al. (2013). Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nature Genetics, 45(10), 1150–1159. doi:10.1038/ng.2742.
Ripke, S., Sanders, A. R., Kendler, K. S., Levinson, D. F., Sklar, P., Holmans, P. A., et al. (2011). Genome-wide association study identifies five new schizophrenia loci. Nature Genetics, 43(10), 969–976. doi:10.1038/ng.940.
Schizophrenia Working Group of the Psychiatric Genomics Consortium. (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature, 511(7510), 421–427.
Sherry, S. T., Ward, M.-H., Kholodov, M., Baker, J., Phan, L., Smigielski, E. M., & Sirotkin, K. (2001). dbSNP: The NCBI database of genetic variation. Nucleic Acids Research, 29(1), 308–311.
Spreen, O. (1998). A compendium of neuropsychological tests: Administration, norms, and commentary. New York: Oxford University Press.
Sullivan, K., Hatton, D., Hammer, J., Sideris, J., Hooper, S., Ornstein, P., & Bailey, D. (2006). ADHD symptoms in children with FXS. American Journal of Medical Genetics Part A, 140(21), 2275–2288.
Tabor, H. K., Risch, N. J., & Myers, R. M. (2002). Candidate-gene approaches for studying complex genetic traits: Practical considerations. Nature Reviews. Genetics, 3(5), 391–397. doi:10.1038/nrg796.
Timms, A. E., Dorschner, M. O., Wechsler, J., Choi, K. Y., Kirkwood, R., Girirajan, S., et al. (2013). Support for the N-methyl-D-aspartate receptor hypofunction hypothesis of schizophrenia from exome sequencing in multiplex families. JAMA psychiatry (Chicago, Ill.), 70(6), 582–590. doi:10.1001/jamapsychiatry.2013.1195.
Tsai, G., Van Kammen, D. P., Chen, S., Kelley, M. E., Grier, A., & Coyle, J. T. (1998). Glutamatergic neurotransmission involves structural and clinical deficits of schizophrenia. Biological Psychiatry, 44(8), 667–674.
Wechsler, D. (1997). WAIS-III: Administration and scoring manual: Wechsler adult intelligence scale. Psychological Corporation.
Wechsler, D. (2001). Wechsler test of adult reading: WTAR. Psychological Corporation.
Wilkening, S., Chen, B., Bermejo, J. L., & Canzian, F. (2009). Is there still a need for candidate gene approaches in the era of genome-wide association studies? Genomics, 93(5), 415–419.
Yang, Y. S., Marder, S. R., & Green, M. F. (2016). Repurposing drugs for cognition in schizophrenia. Clinical Pharmacology & Therapeutics, n/a-n/a. doi:10.1002/cpt.529
Acknowledgements
This study was supported by a Grant-in-Aid held by Authors NM, KN and FF and the Daniel Beck Award held by Author NM, from the Schizophrenia Research Institute. The study used samples and data from the Australian Schizophrenia Research Bank (ASRB), funded by the Australian National Health and Medical Research Council (NHMRC) Enabling Grant (No. 386500) held by V. Carr, U. Schall, R. Scott, A. Jablensky, B. Mowry, P. Michie, S. Catts, F. Henskens and C. Pantelis (Chief Investigators), and the Pratt Foundation, Ramsay Health Care, the Viertel Charitable Foundation, as well the Schizophrenia Research Institute, using an infrastructure grant from the NSW Ministry of Health. Authors NM and JA were supported by Ian Scott Scholarships from Australian Rotary Health. Author NT was supported by an Australian Postgraduate Award. Author MG was supported by an Australian National Health and Medical Research Council (NHMRC) Biomedical Career Development Fellowship (1061875). Postmortem brain tissues were received from the NSW Tissue Resource Centre, which is supported by the National Health and Medical Research Council of Australia, Schizophrenia Research Institute and the National Institute of Alcohol Abuse and Alcoholism [NIH (NIAA) R24AA012725]. The authors wish to thank P. Bitter and B. Yamasani from the Australian Cancer Research Foundation at The Garvan Institute of Medical Research, Sydney for their assistance with the MassARRAY genotyping assays, as well as A.M. Shepherd and I.C. Gould for assistance with structural imaging and Freesurfer processing. All authors report no financial relationships with commercial interests.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
All authors declare that they have no conflicts of interest.
Informed consent
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, and the applicable revisions at the time of the investigation. Informed consent was obtained from all patients for being included in the study
Additional information
Melissa Jayne Green and Francesca Fernandez contributed equally to this work.
Electronic Supplementary Material
ESM 1
(PDF 167 KB)
Rights and permissions
About this article
Cite this article
Matosin, N., Newell, K.A., Quidé, Y. et al. Effects of common GRM5 genetic variants on cognition, hippocampal volume and mGluR5 protein levels in schizophrenia. Brain Imaging and Behavior 12, 509–517 (2018). https://doi.org/10.1007/s11682-017-9712-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11682-017-9712-0