Abstract
Coumarate 3-hydroxylase (C3h) genes participate in the synthesis of lignin and may affect the properties of wood that are important for its commercial value. A better understanding of the natural variation in C3h genes and their associations to wood properties is required to effectively improve wood quality. We used a candidate gene-based association mapping approach to identify CfC3h allelic variants associated with traits that affect the wood properties of Catalpa fargesii. We first isolated the full-length CfC3h cDNA (1825 bp), which was expressed at relatively high levels in xylem according to real time-polymerase chain reaction. In totally, 17 common single-nucleotide polymorphisms (minor allele frequency > 5%) were identified through cloning and sequencing the CfC3h locus from a mapping population (including 88 unrelated natural C. fargesii individuals collected from main distribution area). Nucleotide diversity and linkage disequilibrium (LD) in CfC3h indicate that CfC3h has low nucleotide diversity (πt = 0.0031 and θw = 0.0103) and relatively low LD (within 1800 bp; r2 ≥ 0.1). An association analysis identified eight common single-nucleotide polymorphisms (SNPs) (false discovery rate, Q < 0.10) and ten haplotypes (Q < 0.10) associated with wood properties, explaining 4.92–12.09% of the phenotypic variance in an association population consisted of 125 unrelated natural individuals (The 88 individuals from the mapping population were comprised in the association population). Our study would provide new insight into C3h gene affecting wood quality, and the SNP markers identified would have potential applications in marker-assisted breeding in the future.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Trees are important as sustainable and renewable sources of lumber, pulp, and biofuels. Wood formation is actually a major form of carbon sink, involves deposition of secondary cell walls that mainly composed of lignin, hemicellulose, cellulose, and others. The secondary cell wall biosynthesis is a complex and dynamic process that cooperatively regulated by various metabolic pathways involving lignin and polysaccharides according to previous studies (Carocha et al. 2015).
Lignin, a complex racemic mixture of aromatic heteropolymers mainly present in secondary thickened plant cell walls, is the second most abundant organic polymer next to cellulose in trees (Lisperguer et al. 2009). Lignins are essential for plant structure formation and defence. Lignins also support the stem mechanical properties and the cell wall structural integrity (Jones et al. 2001). Downregulation of lignin in poplar significantly decreases the elastic modulus, yield stress and modifies other wood properties (Özparpucu et al. 2017). Wood types vary in their properties, composition and structural or physical features, making wood suitable for different applications (Du et al. 2013). For example, wood with good mechanical properties, such as stiffness and ultimate stress, is useful as furniture or construction materials. Therefore, forest tree breeding programs should select woods according to the required applications. Catalpa fargesii Bur. (2n = 2X = 40) is a popular timber tree species native to China with straight stem, high density and excellent mechanical characters that make C. fargesii valuable materials for the production of furniture and other upmarket woodware (Zhao et al. 2012). Identifying the genes and allelic variations associated with wood quality in C. fargesii would yield important information for breeding programs, and will be of practical importance to production (Li et al. 2013). The most important wood traits are complex, quantitative traits, showing phenotypic variation that is typically influenced by multiple quantitative trait loci (QTLs) and environmental factors (Resende et al. 2012). Because perennial forest trees have a long lifespan, which make it different to get an advanced population and usually high heterozygosity, traditional QTL mapping using F1 individuals has low mapping resolution such that few alleles are detected (Dillon et al. 2012). Indeed, phenotypic variation can sometimes only be detected after years of growth. Linkage disequilibrium (LD)-based association is an effective way to examine the associations between natural allelic variation and target traits, and has a higher mapping resolution. Single-nucleotide polymorphism (SNP) markers are usually used in association studies given their wide distribution in the genome and potential to be in LD with the polymorphism (Rafalskia 2002). SNP markers associated with wood properties have been identified in several forest trees, such as Populus (Du et al. 2013; Tian et al. 2014; Wang et al. 2017), Eucalyptus (Thavamanikumar et al. 2014; Resende et al.2017), and spruce (Lamara et al. 2016; Lenz et al. 2017), among others.
Within the lignin pathway, coumarate 3-hydroxylase (C3h) catalyzes the coumaroyl shikimic acid to caffeoyl shikimic acid, which is a key step to synthesis of guaiacyl and syringyl lignin subunits in dicotyledonous plants (Poovaiah et al. 2014). Down regulation of C3h reduces lignin content in several plants (Fornalé et al. 2015; Sykes et al. 2015). Although functional studies of C3h have been carried out, we still know little about their allelic effects on the wood properties of trees, which is the foundation for marker-assisted breeding in forest. To study the allelic variation in the coumarate 3-hydroxylase gene and associated with wood properties and we first cloned a gene encoding the C3h homolog CfC3h from C. fargesii, and measured its expression in specific tissues. Single-marker and haplotype-based association methods were combined to identify factors underlying natural variations in wood properties in a C. fargesii population. This is the first association study about the allelic variations in the C3h and wood property. Molecular markers identified in our study would lay a foundation for improving wood quality through molecular breeding of C. fargesii in the future.
Materials and methods
Plant materials and DNA extraction
The C. fargesii population in this study consisted of 125 unrelated individuals growing in the Xiaolongshan conservation area, Gansu Province, China (33°40′ N, 106°23′ E) (Zhao et al. 2012). Branch segments of the 125 individuals were collected from eight cities in four provinces covering the main natural distribution range of C. fargesii and grafted to establish a clonal plantation in 2009 using a randomised complete block design with two plants per clone in each block and six replications (“row spacing” is 2 m and “plant spacing” is 2 m). These individuals were divided into four groups by geolocation: Fenhe River valley, Jinghe River valley, Jialingjiang River valley and Yellow River valley. The 88 unrelated individuals were selected from among these groups to identify SNPs associated with wood properties via polymerase chain reaction (PCR) amplification and sequencing. (At least one individual was selected for each location).
Fresh leaves were collected from each individual and total genomic DNA was extracted using the DNAeasy Plant kit (Qiagen, Shanghai, China) following the manufacturer’s protocol.
Phenotypic data
Nine phenotypic traits associated with wood property, including wood basic density and eight microstructural characteristics were measured: pore rate, cell wall percentages (the percentage of cell wall in whole cells), cell wall thickness, radial lumen diameter, chordwise lumen diameter, radial fibre central cavity diameter, chordwise fibre central cavity diameter, and average fibre central cavity diameter. The nine traits were selected for their possible influences on the final mechanical properties of timber according the other studies (Li et al. 2015). The 125 individuals were sampled in 2012. Cores containing bark and pith were collected at breast height (1.3 m above the ground) from stems of trees in the south-facing direction to evaluate wood basic density and other properties using an increment borer (7 mm). Wood basic density was measured as follows (Eq. 1):
where, W1, W2, and ρcw represent water-saturated weight, oven dry weight and cell wall density (here we used the constant 1.53 g cm−3 for ρcw), respectively (Zheng et al.2015; Duan et al.2016).
The anatomical parameters (pore rate, cell wall percentage, cell wall thickness, radial lumen diameter, chordwise lumen diameter, radial fibre central cavity diameter, chordwise fibre central cavity diameter and average fibre central cavity diameter) were determined according to Li et al. (2015): the cores were split into 3-cm-long pieces and cross-Sects. (10–15 μm thick) were prepared using a sliding microtome (Leica, Heidelberg, Germany), stained with Safranin-O (1% in distilled water) and permanently fixed with Eukitt (BiOptica, Milan, Italy) (Li et al. 2015). A digital image processing system, including a light microscope (80i; Nikon, Tokyo, Japan), video camera sensor (Penguin 600CL; Pixera, Santa Clara, CA, USA) and TDY-5.2 colour image analysis system (Beijing Tiandiyu Science and Technology Co., Ltd., Beijing, China) were used to measure the wood microstructural characteristics (Wang et al. 2005).
The frequency distributions for each trait are calculated using Excel (ver. 2013; Microsoft, Redmond, WA, USA) and shown in Fig. S1. The phenotypic data are listed in Table S1. SPSS software (ver. 18.0; SPSS Inc., Chicago, IL, USA) was used to evaluate the nine phenotypic traits, including in terms of mean values, ranges and coefficients of variation (Table S2). The proportion of the phenotypic variance explained by population structure (Table S2) was evaluated by SAS software (ver. 9.1.3; SAS Institute Inc., Cary, NC, USA) using generalized linear model (GLM). The variance components and narrow-sense heritability (h2) were evaluated using R/ASReml (ver. 4.0; VSN International Ltd., Hemel Hempstead, UK).
Isolation of the whole coding sequence (CDS) and genomic DNA amplification of the C3h homologue in Catalpa fargesii
Total RNA was extracted from the young branches of a 1-year-old “Xianhuiqiu” (C. fargesii) clone using the Plant Qiagen RNAeasy kit (Qiagen) according to the manufacturer’s instructions. First-strand cDNA was synthesised from 2 g of DNase I-treated RNA using the PrimeScript™ 1st Strand cDNA Synthesis Kit (TaKaRa Bio, Shiga, Japan). The entire open reading frame (ORF) of the C3h homologue from C. fargesii was isolated in the following way: We first obtained the partial C3h homologue sequence from previous RNA-seq data, i.e. an internal coding region of a C3h homologue sequence according to the National Center for Biotechnology Information (NCBI) database. The 3′ end was isolated by 3′ rapid amplification of cDNA ends (RACE) using the 3′-full RACE Core Set (ver. 2.0; TaKaRa Bio) and designed primers (C3h-3′ RACE; Table S3), and a 3′ RACE adaptor primer (C3h-3′ RACE adaptor primer; Table S3). To isolate the 5′end, 5′ RACE was carried out using the 5′-full RACE Core Set (ver. 2.0; TaKaRa Bio) according to the manufacturer’s instructions using specific primers, i.e. a designed 5′RACE primer (C3h-5′ RACE; Table S3) and a 5′ RACE adaptor primer (C3h-5′ RACE adaptor primer; Table S3). PCR was carried out using the C3h-CDS primers to verify the integrity of the C3h homologue CDS sequence (Table S3).
Total genomic DNA was extracted from young leaves of a 1-year-old “Xianhuiqiu” clone with the DNAeasy Plant Mini kit (Qiagen). The intron sequences were cloned using specific primers designed based on the cDNA sequences C3h-a, C3h-b and C3h-c, and were then sequenced (Table S3). After PCR amplification, three fragments were cloned into pMD 19-T Vector (Takara Bio) and sequenced. The entire CfC3h DNA sequence was obtained according to the assembly result of sequenced fragments using DNAman software (Lynnon BioSoft, Vaudreuil, Quebec, Canada). The entire CfC3h DNA sequence was identified using C3h-d primers (Table S3).
Sequence alignment and phylogenetic analyses
The CfC3h amino acid sequences were subjected to BLAST analysis via the GenBank database (http://www.ncbi.nlm.nih.gov/sites/entrez?db=nucleotide). During the BLAST searches, multiple C3h proteins from various species were selected for alignment using DNAMan software. To analyse the phylogenetic relationship of CfC3h to the C3h genes from other species, the amino acid sequences of the C3h gene from Sesamum indicum (AAL47545.1), Scutellaria baicalensis (BAJ09387.1), Salvia miltiorrhiza (ACA64048.1), Coffea Arabica (AFP49812.1), Populus tomentosa (AFZ78540.1), Platycodon grandifloras (AEM63674.1), Caragana korshinskii (AEV93473.1), Neosinocalamus affinis (AFD29885.1), Panicum virgatum (BAO20879.1), Ginkgo biloba (AAY54293.1), Cunninghamia lanceolate (AFX98060.1), Narcissus tazetta (AGI97941.1), Pinus taeda (AAL47685.1) and Isatis tinctori (AEH20527.1) were downloaded from NCBI (http://www.ncbi.nlm.nih.gov) and aligned with the ClustalW program using the default settings. The phylogenetic tree was constructed using MEGA 5.0 software. The phylogenetic tree was constructed based on the maximum likelihood method with the following parameters: bootstrap (1000 replicates), Jones–Taylor–Thornton substitution model, uniform rates, partial deletion of gaps/missing data and nearest neighbour interchange.
Expression of CfC3h in different C. fargesii organs
We collected tissue and organ samples of tree bark, phloem, xylem, leaves, flowers and juvenile branch meristem from three 11-year-old Xianhuiqiu (C. fargesii) trees planted in Luoyang, Henan in the middle of April for RNA extraction. Each tree was considered as one repetition. All tissues were immediately frozen in liquid nitrogen and stored at − 80 °C. The RNA extraction and cDNA synthesis procedures were performed as described above. Tissue-specific expression analysis of CfC3h was done using real-time quantitative polymerase chain reaction (RT-qPCR).
RT-qPCR was performed using a LightCycler 480 System (Roche, Basel, Switzerland) and the SYBR Premix Ex Taq Kit (TaKaRa Bio), under the amplification conditions recommended by TaKaRa Bio. The CfC3h-specific primers (C3h-q) were designed using Primer Express 5.0 software (Applied Biosystems, Foster City, CA, USA) and the actin gene was selected as the internal control according to Jing et al. (2015). The PCR program included initial denaturation at 95 °C for 30 s, and 40 cycles for 5 s at 95 °C and 30 s at 60 °C. All reactions were carried out four times, and the tissue-specific expression levels were calculated using the 2−ΔΔCT formula.
SNP identification and genotyping
To identify SNPs within the CfC3h gene, the 44 bp 5′-untranslated region (UTR), the entire coding region and the 128 bp 3′ UTR were sequenced and analysed in 88 unrelated individuals from the mapping population, to identify SNPs without consideration of insertions/deletions. To ensure the accuracy of sequencing, three pairs of primers (C3h-1, C3h-2 and C3h-3) were used to amplify three fragments of the entire sequence (Table S3). Primer pairs for amplification were designed using Primer Express 5.0 software. DNAMAN and ClustalX2 (Larkin et al. 2007) were used for sequence alignment, and manual editing was performed to confirm sequence quality. Eight clones of each individual were used to identify putative SNP variants, and fragments were randomly selected for initial allele sequencing via the ABI3730XL instrument (Applied Biosystems). The 88 genomic clones were aligned and compared using MEGA5.0 (Tamura et al. 2011) and DnaSP v5 software (UB Web, Barcelona, Spain, 2010) to identify SNPs and analyse the nucleotide polymorphisms. Common SNPs (SNPs with minor allele frequencies more than 5%) were genotyped across all 125 DNA samples from the overall population. The genotypic data of CfC3h identified in this population were shown in Table S4.
Nucleotide diversity and linkage disequilibrium analysis
Summary statistics for the SNP polymorphisms were generated by linear regression analysis using DnaSP v5. Nucleotide diversity was estimated according to the average number of pairwise differences per site between sequences, π (Nei 1987), and the average number of segregating sites, θw (Watterson 1975). The HAPLOVIEW software package (http://www.broad.mit.edu/mpg/haploview.html) was used to assess LD among the common SNPs. The squared allelic correlation coefficient (r2) was used to estimate LD (Hill and Robertson 1968). The significance (P value) of r2 for each SNP locus was calculated using 100,000 permutations.
SNP‑based associations and modes of gene action
Single-marker models were created for all SNP–trait combinations. A mixed linear model (MLM) was fitted to each trait-SNP combination using TASSEL v5.0 software. The MLM, from the Q + K model, uses the values of estimated membership probability (Q) to evaluate the effects of population structure, and those of pairwise kinship (K) to evaluate relatedness among individuals for marker-trait associations. The Q matrix was prepared, from the pattern of the population structure (K = 3) within the overall population (125 unrelated individuals), using STRUCTURE (ver. 2.3.1). The K matrix was obtained via the method proposed by Ritland (1996) using the SPAGeDi program (ver. 1.2). Corrections for multiple testing were performed using the positive false discovery rate (FDR) method in QVALUE software (Storey and Tibshirani 2003). The percentage of phenotypic variation (R2) explained by each SNP was calculated by using the below formula:
where, SSt and SST represented the variance between genotypes and the total variance, respectively. The detailed information can be found in Lu et al. (2018).
The ratios of dominant (d) to additive (a) effects calculated from least square means for each genotypic class were used to quantify the modes of gene action according to Wegrzyn et al. (2010). Values of |d/a| in the range of 0.50 to 1.25 were considered to indicate partial or complete dominance, and those in the range of |d/a| no more than 0.5 were considered additive effects. In addition, values of |d/a| more than 1.25 were considered to indicate under- or over-dominance. The detailed algorithm and formulas for estimating the gene action were described by Eckert et al. (2009).
Haplotype analysis
Within the genotypic data of 125 individuals, we analysed the haplotypes based on information of the contiguous common SNPs. Haplotype frequencies were estimated, and haplotype association tests were performed using a three-marker sliding window via haplotype trend regression software (Zaykin et al. 2002). We used a 1000 permutation tests to evaluate the significance of haplotype-based associations and haplotypes with a frequency ≥ 1% were selected for further analysis. The multiple testing was corrected using a positive FDR (Q ≤ 0.1) in QVALUE.
Results
Cloning of Catalpa fargesii C3h
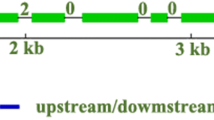
The full-length CfC3h cDNA isolated using RACE was 1825 bp, including a 1530 bp ORF encoding 510 amino acids and a 69 bp 5′ UTR sequence, as well as 226 bp 3′ UTR sequence. The full-length CfC3h DNA sequence was 3511 bp and contained a 3104 bp coding region, flanked by a 125 bp 5′ UTR sequence and a 282 bp 3′ UTR sequence (Fig. 1). Alignment of the cDNA sequence to the full-length DNA sequences showed that CfC3h has three exons and two introns.
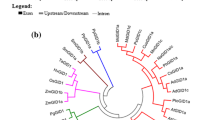
C3h genes were divided into four groups according to the molecular phylogeny analysis. CfC3h belongs to group IV, the same group as C3h genes of other three Tubiflorae species, namely Sesamum indicum, Salvia miltiorrhiza, and Scutellaria baicalensis. It is interesting that two dicotyledonous groups (group I and IV) belonged to two different branches, and the genetic evolutionary relationships of the C3h genes from group I were closer to genes from monocotyledons and gymnosperms than those from group IV. The phylogeny analysis suggested that separation of C3h genes may have occurred before gymnosperms and angiosperms diverged (Fig. 2). The sequence alignment showed that CfC3h in C. fargesii had close similarity at the amino acid level to C3h from other species (Fig. 3). C3h belonged to the P450 superfamily, and a cytochrome P450 cysteine heme–iron ligand signature (FGXGRRXCPG) was also found in the C terminal region of CfC3h from F (432) to G (441).
An unrooted phylogenetic tree of C3h members from different species. Sesamum indicum (AAL47545.1): SiC3H; Scutellaria baicalensis (BAJ09387.1): SbC3H; Salvia miltiorrhiza (ACA64048.1): SmC3H; Coffea Arabica (AFP49812.1): CaC3H; Populus tomentosa (AFZ78540.1): PtoC3H; Platycodon grandiflora (AEM63674.1): PgC3H; Caragana korshinskii (AEV93473.1): CkC3H; Neosinocalamus affinis (AFD29885.1): NaC3H; Panicum virgatum (BAO20879.1): PvC3H1; Ginkgo biloba (AAY54293.1): GbC3H; Cunninghamia lanceolate (AFX98060.1): ClC3H; Narcissus tazetta (AGI97941.1): NtC3H; Pinus taeda (AAL47685.1): PtaC3H; Isatis tinctori (AEH20527.1): ItC3H; Catalpa fargesii Bur.: CfC3H
Sequence comparison of CfC3h with other C3h proteins. The sequence of the cytochrome P450 cysteine heme-iron ligand signature is shown by the red box. Sesamum indicum (AAL47545.1): SiC3H; Scutellaria baicalensis (BAJ09387.1): SbC3H; Salvia miltiorrhiza (ACA64048.1): SmC3H; CaC3H; Populus tomentosa (AFZ78540.1): PtoC3H; Platycodon grandiflora (AEM63674.1): PgC3H; Caragana korshinskii (AEV93473.1):CkC3H; Catalpa fargesii Bur.: CfC3H
Expression of CfC3h in different organs
We used RT-qPCR to determine the tissue-specific expression of CfC3h in C. fargesii. As shown in Fig. 4, the expression of CfC3h was highest in xylem (0.406 ± 0.048), followed by phloem (0.229 ± 0.056) and leaves (0.188 ± 0.056). It was the lowest in flowers (0.022 ± 0.006). This result shows that CfC3h is mainly expressed in xylem.
Phenotypic variations in the Catalpa fargesii population
Phenotypic variations in all nine traits were evaluated in the overall C. fargesii population (125 individuals) to test quantitative traits for association mapping. All traits varied significantly within the overall population; for example, pore rate, cell wall percentages and radial lumen diameter ranged from 6 to 14% (mean, 9.94%), 22.95% to 41.38% (mean, 35.03%), and 7.31 to 27.74 μm (mean of 14.96 μm), respectively. To estimate the phenotypic variance within the overall population, we computed the coefficient of variation (CV) for all nine traits (Table S2). The results demonstrated that pore rates had the highest CV (14.45%), followed by cell wall thickness (12.98%) and radial lumen diameter (9.82%). All nine traits followed an approximately normal distribution (Fig. S1).
Nucleotide diversity and linkage disequilibrium in CfC3h
We amplified and sequenced a 3276 bp genomic region of CfC3h from 88 unrelated individuals within the overall population, including the 44 bp 5′ UTR, the entire coding region and the 151 bp 3′ UTR sequence, to determine SNP diversity. Alignment of the 88 samples revealed a total of 163 SNPs in CfC3h, with a polymorphism of 4.94% (Table 1). Of the 163 SNPs, only 17 (10.43%) were considered common SNPs (Fig. S2). The highest level of nucleotide polymorphism in the coding regions occurred in intron 2 (7.22%), and the lowest in exon 2 (2.76%). The CfC3h locus had low nucleotide diversity, with πt = 0.0031 and θw = 0.0103 (Table 1). Specifically, nucleotide diversity ranged from 0.0024 (exon 2) to 0.0094 (5′ UTR), and θw varied between 0.0060 (exon 2) and 0.0270 (5′ UTR). The coding region had more nonsynonymous changes (40) than synonymous changes (15).
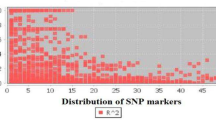
The SNPs identified in the 88 unrelated individuals were used to calculate r2 and the LD level was assessed according to the pattern of r2 with base-pair distance within the CfC3h. The r2 value decreased to 0.1 within 1800 bp (Fig. 5), indicating that LD may not extend over the entire region that we sequenced. We then genotyped 17 SNPs common across 125 individuals, and LD analysis using genotype data revealed four distinct haplotype blocks within the CfC3h locus: from SNP 6 to 7, 9 to 10, 11 to 12 and 14 to 15 (Fig. 6). LD between the SNPs was relatively high within each block (r2 > 0.75).
Decay of LD within CfC3h based on sequences of the CfC3h region from 88 unrelated individuals. We sequenced the CfC3h regions of 88 unrelated individuals. Pairwise correlations between single-nucleotide polymorphisms (SNPs) are plotted against the physical distance between the SNPs in base pairs. The curves show the linear regression of r2 according to the physical distance in the base pair
SNP-trait associations
MLM was used to detect associations between phenotypes and genotypes for the different SNPs, after correcting for multiple testing using the FDR method (Q ≤ 0.1). We identified eight significant associations at a threshold of P ≤ 0.05, encompassing seven unique SNPs (SNP 1, SNP 2, SNP 3, SNP 5, SNP 9, SNP 10 and SNP 17) significantly associated with wood basic density, pore rate, cell wall percentage, cell wall thickness and chordwise lumen diameter (Table 2), explaining 4.92−7.99% of the phenotypic variance in these traits. Five of these eight associations were consistent with over-dominance modes of gene action (|d/a|> 1.25) and one association was partial or complete dominance (Table 3). Of the seven significant SNPs, five were located in exons, including four nonsynonymous and one synonymous SNP. The nonsynonymous marker, SNP 1, showed an amino acid change from Val to Leu in exon 1, and was significantly associated with cell wall thickness, explaining 6.85% of the phenotypic variance therein. Heterozygous trees (CG) exhibited higher cell wall thickness (2.93 μm) than trees with the CC and GG genotypes (2.83 and 2.82 μm, respectively). The SNP 5 was significantly associated with pore rate, explaining 4.92% of the variance and exhibiting over-dominance for this trait (|d/a|> 1.25). The GG genotype of SNP 3 exhibited a lower cell wall percentage (32.61%) compared with the CG and CC genotypes (36.58% and 35.15%, respectively) (Fig. S3), thus exerting an over-dominance effect on cell wall percentage (|d/a|> 1.25). In the 3′ UTR, SNP 17 was associated with wood basic density (explaining 6.08% of the variance therein) and the mean values of two main genotypic groups: TT and TC were 0.417 and 0.429 g cm−3, respectively. SNP 10 was significantly associated with wood basic density (explaining 7.99% of the variance therein) and chordwise lumen diameter (6.39% of the variance).
Haplotype-based association tests were performed to identify haplotypes significantly associated with the nine phenotypic traits (Table 4). This analysis identified 10 associations between 11 common haplotypes (frequency ≥ 1%) in six blocks. Eight of the traits (i.e. all except average central diameter) reached the significance threshold of P ≤ 0.05 and FDR ≤ 0.1 among the entire region. Of these, three haplotypes from SNPs 5–7 were associated with pore rate, cell wall percentage and cell wall thickness, and three haplotypes from SNPs. 15–17 were associated with wood basic density, radial lumen diameter and radial fibre central cavity diameter. The proportion of phenotypic variation explained by these haplotypes ranged from 6.32 to 12.30%.
Discussion
The putative function of CfC3h
C3h is an important enzyme in lignin synthesis, where lignins are a major component of plant secondary cell walls. C3h mutants have been studied in Arabidopsis thaliana in the context of recovery of the function of the C3h gene (Kim et al. 2014). Defects in coumarate 3′-hydroxylase cause dwarfism and reduce cell wall lignin content. Wang et al. (2018) reported that downregulation of C3h in poplar not only reduces lignin levels, but also markedly increase the proportion of G and S-type lignin, and finally influence the wood properties.
We cloned a CfC3h gene from C. fargesii, which shared 69% and 71% identity at the nucleotide level with Arabidopsis C3h (AT2G40890) and Populus alba × grandidentata C3h (GenBank accession no. EU391631), respectively. We further analysed the expression of CfC3h in different organs and observed the highest expression in xylem, which may due to the higher degree of lignification in xylem.
This study identified an association between allelic variation in CfC3h and several wood quality traits, including cell wall percentage and cell wall thickness (Table 2). These results are consistent with previous studies and confirmed the importance of C3h within the structure of the secondary cell wall (Ralph et al. 2006; Fornalé et al. 2015).
Nucleotide diversity and LD in CfC3h
An understand of the extent of LD and nucleotide diversity level in a natural population could evaluate the precision and effectiveness of association mapping, as well as reflecting the forces in charge of the evolutionary change (Zhang et al. 2010). So, a comprehensive study of the patterns of SNP distribution and frequency within the CfC3h locus of from the C. fargesii population is necessary before SNP-based association mapping. The SNP frequency in exons regions, intron regions and the genomic sequence was 3.59%, 5.97%, and 6.62%, respectively. The exons showed substantially lower levels of nucleotide diversity compared with introns in the coding region (Table 1), which is consistent with previous studies (Du et al. 2013; Wang et al.2017) and indicates that the exon regions may have undergone strong purifying selection and thus remained relatively conserved. The sequence that codes the cytochrome P450 conserved domain (FGXGRRXCPG) was located in exon 3, which had a low level of nucleotide diversity (πt = 0.0027) thus indicating that CfC3h is extremely conserved due to its crucial role in the synthesis of monolignols and other 3,4-hydroxylated phenylpropanoid secondary metabolites (Bate et al. 1998; Kim et al. 1998). Compared to our previous study, the nucleotide diversity of CfC3h was similar to that of CfSUS (πt = 0.0031) (Lu et al. 2018), which indicated that the two genes may have a similar pattern of genetic variance in the natural population. However, and nucleotide diversity detected in a population may influenced by the population size, sampling strategy and other factors (Tian et al. 2014), so in the future study, larger population and more reasonable sampling strategy should be used to evaluate the nucleotide diversity level of C. fargesii.
Understanding the level of LD can help to determine whether candidate gene-based association studies are appropriate for understanding the molecular basis underlying quantitative variation, and whether a genome-wide approach is feasible (Du et al. 2013). In our study, CfC3h showed a relatively low level of LD and a rapid decline, indicating that candidate gene-based association studies may be appropriate in this instance to identify SNPs responsible for the detected traits. In fact, a low and rapidly declining LD has been reported in other studies (Guerra et al. 2013; Chu et al. 2014), which may due to the outcrossing habit, long history of recombination and large population size of these species (Abdurakhmonov and Abdukarimov 2008). The LD level of CfC3h was similar to that of CfSUS (r2 < 0.1, within 1600 bp) in the same population (Lu et al. 2018). Additionally, we detected four distinct haplotype blocks within the CfC3h gene and the distances between adjacent SNPs in the blocks were small (20 to 79 bp). Low LD observed in CfC3h gene may suggested high resolution of marker-trait associations.
Determining the allelic polymorphisms underlying wood properties
In our study, a gene-based association analysis has been used to identify alleles associated with wood properties in several tree species including Populus, Eucalyptus and some Pinus plants. However, SNP association studies have not been reported for C. fargesii. Therefore, we employed single-marker and haplotype-based association studies of a candidate gene in C. fargesii. The results showed that several single SNP markers and haplotypes were associated with wood properties in our C. fargesii population, indicating that these markers may be in close proximity to, or in fact are, the functional variant.
The eight single-SNP associations identified in our study only explain a small proportion of the variance in wood traits, which is in accordance with previous studies of other tree species (Porth et al. 2013; Wang et al. 2017). This may be because wood traits are usually quantitatively characterised and controlled by multiple genes. In addition, most of the SNPs (five of seven) were located in exon regions and the mode of gene action of SNP1 and SNP3 was over-domination. Mutations in coding regions, particularly nonsynonymous mutations, can affect gene function. Vanholme et al. (2013) identified a stop codon mutant in the hydroxycinnamoyl-CoA: shikimate hydroxycinnamoyl transferase gene, resulting in modified lignin composition in Populus nigra. The four SNPs (SNP1, SNP3, SNP9 and SNP10) were located in exons and identified as nonsynonymous mutations. It remains unclear how amino acid changes in these four locations influenced the function of CfC3h. However, Wang et al. reported that transfer between amino acids with similar polarities, charges or sizes, such as Cys and Ser, may not affect the function of genes (Wang et al. 2017). Whether amino acid changes of these four locations influence the function of CfC3h gene, need to be further studied.
Wood basic density is one of the most important factors associated with wood mechanical strength. Our study showed that SNPs 9 and 10 explained 5.50% and 7.99% of the variance in wood basic density, while a haplotype from SNPs 9 to 11 explained 11.59% of the phenotypic variation, slightly higher than single-marker association (5.50% and 7.99%), indicating that markers surrounding SNPs 9 and 10 may interact with the two loci and contribute to phenotypic effects; however it need be further investigated. SNP 5 (located in intron 1) was associated with 4.92% of the variation in pore rate, which was lower than that of a haplotype from SNPs 5 to 7 (8.51%). SNP 5 may interact with loci nearby or loci that influence RNA splicing, and thus influence the pore rate; however, further investigation is required to reveal the detailed mechanisms. Notably, SNP 17 in the 3′ UTR region was significantly associated with wood basic density. Although the polymorphism in the 3′ UTR region did not alter the amino acids, 3′ UTRs participate in the regulation of gene expression by affecting mRNA deadenylation and degradation (Fang et al. 2010). In addition, SNPs in 5′ UTR region can affect gene regulation by influencing transcriptional binding (Beaulieu et al. 2011; Tian et al. 2014), particularly SNPs in some important motifs of the promoter part of a gene (Wang et al. 2017). However, this study focused on the CfC3h coding region, and only a small part of the non-coding region was detected; thus, SNPs in 5′ UTR and 3′ UTR = regions will be sought in a further study.
Association analysis has been used to study the genetic architecture of important traits in forest. For example, Du et al. identified 202 significant SNPs in 63 candidate genes selected by transcriptome and QTL mapping that associated to plant growth (Du et al. 2016). In addition, dynamic association studies have been used to integrally identify the genetic basis of complex traits (Du et al. 2019). In future study, more association strategies will be undertaken to previously obtain important molecular markers to serve the C. fargesii breeding.
Conclusion
In our study, we first cloned a putative C3h homologous gene in C. fargesii and totally 163 SNPs were identified according to the alignment result from a mapping population including 88 natural C. fargesii individuals. The LD decay distance short within the CfC3h (r2 < 0.1 within 1800 bp). In additional, 8 SNPs and 10 haplotypes were identified significantly associated with 5 and 8 detected traits, respectively, using association a(h2) analysis. Our study implies allelic variations within CfC3h may influence wood properties of C. fargesii and the SNP markers identified in this study may be useful for marker-assisted selection, to improve wood traits in C. fargesii in the future.
Availability of data and material
Sequence data of the CfC3h from 125 Catalpa fargesii individuals have been deposited with the GenBank Data Library under the accession numbers: MH394548-MH394635.
Abbreviations
- CDS:
-
Coding sequence
- C3h:
-
Coumarate 3-hydroxylase
- LD:
-
Linkage disequilibrium
- MLM:
-
Mixed linear model
- ORF:
-
Open reading frame
- QTL:
-
Quantitative trait loci
- RACE:
-
Rapid amplification of cDNA ends
- SNP:
-
Single-nucleotide polymorphism
- UTR:
-
Untranslated region
- WBD:
-
Wood basic density
References
Abdurakhmonov I, Abdukarimov A (2008) Application of association mapping to understanding the genetic diversity of plan germplasm resources. Int J Plant Genomics 2008(2):574927
Bate N, Sivasankar S, Moxon C, Riley J, Thompson J, Rothstein S (1998) Molecular characterization of an Arabidopsis gene encoding hydroperoxide lyase, a cytochrome P-450 that is wound inducible. Plant Physiol 117(4):1393–1400
Beaulieu J, Doerksen T, Boyle B, Clément S, Deslauriers M (2011) Association genetics of wood physical traits in the conifer white spruce and relationships with gene expression. Genetics 188:197–214
Carocha V, Soler M, Hefer C, Cassan-Wang H, Fevereiro P, Myburg A, Paiva J, Grima-pettenati J (2015) Genome-wide analysis of the lignin toolbox of Eucalyptus grandis. New Phytol 206(4):1297–1313
Chu Y, Huang Q, Zhang B, Ding C, Su X (2014) Expression and molecular evolution of two DREB1 genes in black poplar (Populus nigra). PLoS ONE 9:e98334
Dillon S, Brawner J, Meder R, Lee D, Southerton S (2012) Association genetics in Corymbia citriodora subsp variegata identifies single nucleotide polymorphisms affecting wood growth and cellulosic pulp yield. New Phytol 195(3):596–608
Du Q, Xu B, Pan W, Gong C, Wang Q, Tian J, Li B, Zhang D (2013) Allelic variation in a cellulose synthase gene (PtoCesA4) associated with growth and wood properties in Populus tomentosa. G3 Genes Genomes Genet 3(11):2069–2084
Du Q, Gong C, Wang Q, Zhou D, Yang H, Pan W, Li B, Zhang D (2016) Genetic architecture of growth traits in Populus revealed by integrated quantitative trait locus (QTL) analysis and association studies. New Phytol 209(3):1067–1082
Du Q, Yang X, Xie J, Quan M, Xiao L, Lu W, Tian J, Gong C, Chen J, Li B, Zhang D (2019) Time-specific and pleiotropic quantitative trait loci coordinately modulate stem growth in Populus. Plant Biotechnol J 17(3):608–624
Duan H, Cao S, Zheng H, Hu D, Lin J, Lin H, Hu R, Sun Y, Li Y (2016) Variation in the growth traits and wood properties of Chinese Fir from six provinces of southern China. Forests 7(9):192
Eckert A, Bower A, Wegrzyn J, Pande B, Jermstad K, Krutovsky K, Clair J, Neale D (2009) Association genetics of coastal Douglas fir (Pseudotsuga menziesii var. menziesii, Pinaceae) I Cold-hardiness related traits. Genetics 182:1289–1302
Fang X, Xu H, Zhang C, Zhang J, Lan X, Gu C, Chen H (2010) Polymorphisms in BMP-2 gene and their associations with growth traits in goats. Genes Genom 32:29–35
Fornalé S, Rencoret J, Garcia-Calvo L, Capellades M, Encina A, Santiago R, Rigau J, Gutierrez A, del Rio J, Caparros-Ruiz D (2015) Cell wall modifications triggered by the down-regulation of Coumarate 3-hydroxylase-1 in maize. Plant Sci 236:272–282
Guerra F, Wegrzyn J, Sykes R, Davis M, Stanton B, Neale D (2013) Association genetics of chemical wood properties in black poplar (Populus nigra). New Phytol 197:162–176
Hill W, Robertson A (1968) Linkage disequilibrium in finite populations. Theor Appl Genet 38:226–231
Jing D, Xia Y, Chen F, Wang Z, Zhang S, Wang J (2015) Ectopic expression of a Catalpa bungei (Bignoniaceae) PISTILLATA homologue rescues the petal and stamen identities in Arabidopsis pi-1 mutant. Plant Sci 231:40–51
Jones L, Ennos AR, Turner S (2001) Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis. Plant J 26(2):205–216
Kim G, Tsukaya H, Uchimiya H (1998) The ROTUNDIFOLIA3 gene of Arabidopsis thalianaen codes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev 12(15):2381–2391
Kim J, Ciesielski P, Donohoe B, Chapple C, Li X (2014) Chemically induced conditional rescue of the reduced epidermal fluorescence8 mutant of Arabidopsis reveals rapid restoration of growth and selective turnover of secondary metabolite pools. Plant Physiol 164(2):584–595
Lamara M, Raherison E, Lenz P, Beaulieu J, Bousquet J, Mackay J (2016) Genetic architecture of wood properties based on association analysis and co-expression networks in white spruce. New Phytol 210(1):240–255
Larkin M, Blackshields G, Brown N, Chenna R, McGettigan P, McWillam H, Valentin F, Wallace I, Wilm A, Lopez R, Thompon J, Gibson T, Higgins D (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Lenz P, Beaulieu J, Mansfield SD, Clément S, Desponts M, Bousquet J (2017) Factors affecting the accuracy of genomic selection for growth and wood quality traits in an advanced-breeding population of black spruce (Picea mariana). BMC Genomics 18(1):335
Li X, Jia J, Wang J, Ma W, Ma J, Zhao Q (2013) Morphological diversity analysis and preliminary construction of core collection of Catalpa fargesii Bureau. J Plant Genet Resour 14(2):243–248 (in Chinese)
Li Q, Wang J, Li D, Hu C, Qi S (2015) Wood traits of Picea crassifolia clones. J Northeast For Univ 43(12):12–16 (in Chinese)
Lisperguer J, Perez P, Urizar S (2009) Structure and thermal properties of lignins: characterization by infrared spectroscopy and differential scanning calorimetry. J Chil Chem Soc 54(4):460–463
Lu N, Mei F, Wang Z, Wang N, Xiao Y, Kong L, Qu G, Ma W, Wang J (2018) Single-nucleotide polymorphisms (SNPs) in a sucrose synthase gene are associated with wood properties in Catalpa fargesii Bur. BMC Genet 19(1):99
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Özparpucu M, Rüggeberg M, Gierlinger N, Cesarino I, Vanholme R, Boerjan W, Burgert I (2017) Unravelling the impact of lignin on cell wall mechanics–a comprehensive study on young poplar trees downregulated for Cinnamyl Alcohol Dehyrogenase (CAD). Plant J 91:480–490
Poovaiah C, Nageswara-Rao M, Soneji J, Baxter H, Stewart C Jr (2014) Altered lignin biosynthesis using biotechnology to improve lignocellulosic biofuel feedstocks. Plant Biotechnol J 12(9):1163–1173
Porth I, Klapšte J, Skyba O, Hannemann J, McKown AD, Guy RD, DiFazio SP, Muchero W, Ranjan P, Tuskan GA, Friedmann MC, Ehlting J, Cronk Q, EI-Kassaby YA, Douglas CJ, Mansfied SD (2013) Genome wide association mapping for wood characteristics in populus identifies an array of candidate single nucleotide polymorphisms. New Phytol 200:710–726
Rafalskia A (2002) Applications of single nucleotide polymorphisms in crop genetics. Curr Opin Plant Biol 5:94–100
Ralph J, Akiyama T, Kim H, Lu F, Schatz P, Marita J, Ralph S, Reddy M, Chen F, Dixon R (2006) Effects of coumarate 3-hydroxylase down-regulation on lignin structure. J Biol Chem 281(13):8843–8853
Resende M, Resende M, Sansaloni C, Petroli C, Missiaggia A, Aguiar A, Abad J, Takahashi E, Rosado A, Faria D, Pappas G Jr, Kilian A, Grattapaglia D (2012) Genomic selection for growth and wood quality in Eucalyptus: capturing the missing heritability and accelerating breeding for complex traits in forest trees. New Phytol 194(1):116–128
Resende R, Resende M, Silva F, Azevedo C, Takahashi E, Silva-Junior O, Grattapagila D (2017) Regional heritability mapping and genome-wide association identify loci for complex growth, wood and disease resistance traits in Eucalyptus. New Phytol 213(3):1287–1300
Ritland K (1996) Estimators for pairwise relatedness and individual inbreeding coefficients. Genet Res 67:175–185
Storey J, Tibshirani R (2003) Statistical significance for genome wide studies. Proc Natl Acad Sci 100:9440–9445
Sykes R, Gjersing E, Foutz K, Rottmann W, Kuhn S, Foster C, Ziebell A, Turner G, Decker S, Hinchee M, Davis M (2015) Down-regulation of p-coumaroyl quinate/shikimate 3′-hydroxylase (C3′H) and cinnamate 4-hydroxylase (C4H) genes in the lignin biosynthetic pathway of Eucalyptus urophylla × E. grandis leads to improved sugar release. Biotechnol Biofuels 8(1):128
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thavamanikumar S, McManus L, Ades P, Bossinger G, Stackpole D, Kerr R, Hadjigol S, Freeman J, Vaillancourt R, Zhu P, Tibbits J (2014) Association mapping for wood quality and growth traits in Eucalyptus globulus ssp. globulus Labill identifies nine stable marker-trait associations for seven traits. Tree Genet Genomes 10(6):1661–1678
Tian J, Chang M, Du Q, Xu B, Zhang D (2014) Single-nucleotide polymorphisms in PtoCesA7 and their association with growth and wood properties in Populus tomentosa. Mol Genet Genomics 289(3):439–455
Vanholme B, Cesarino I, Goeminne G, Kim H, Marroni F, Acker RV, Vanholme R, Morreel K, Ivens B, Pinosio S, Morgante M, Ralph J, Bastien C, Mansfield S (2013) Breeding with rare defective alleles (BRDA): a natural Populus nigra HCT mutant with modified lignin as a case study. New Phytol 198:765–776
Wang X, Liu Z, Liu Y (2005) Principal component analysis on micrograph character parameter of wood transverse section. J Northeast For Univ 33(5):30–32 ((in Chinese))
Wang L, Wang B, Du Q, Chen J, Tian J, Yang X, Zhang D (2017) Allelic variation in PtoPsbW associated with photosynthesis, growth, and wood properties in Populus tomentosa. Mol Genet Genomics 292(1):77–91
Wang JP, Matthews ML, Williams CM, Shi R, Yang C, Tunlaya-Anukit S, Chen HC, Li Q, Liu J, Lin CY, Naik P, Sun YH, Loziuk PL, Yeh TF, Kim H, Gjersing E, Shollenberger T, Shuford CM, Song J, Miller Z, Huang YY, Edmunds CW, Liu B, Sun Y, Lin YJ, Li W, Chen H, Peszlen I, Ducoste JJ, Ralph J, Chang HM, Muddiman DC, Davis MF, Smith C, Isik F, Sederoff R, Chiang VL (2018) Improving wood properties for wood utilization through multi-omics integration in lignin biosynthesis. Nat Commun 9(1):1579. https://doi.org/10.1038/s41467-018-03863-z.PMID:29679008;PMCID:PMC5910405
Watterson G (1975) On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7:256–276
Wegrzyn J, Eckert A, Choi M, Lee J, Stanton B, Sykes R, Davis M, Tsai C, Neale D (2010) Association genetics of traits controlling lignin and cellulose biosynthesis in black cottonwood (Populus trichocarpa, Salicaceae) secondary xylem. New Phytol 188(2):515–532
Zaykin D, Westfall P, Young S, Karnoub M, Wagner M, Ehm M (2002) Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum Hered 53:79–91
Zhang D, Yang X, Zhang Z, Li B (2010) Expression and nucleotide diversity of the poplar COBL gene. Tree Genet Genomes 6:331–344
Zhao Q, Ma J, Wang J, Feng X, Ma L, Chen J (2012) Blastation and diversities of Catalpa Fargesii genetic resources among basions. J Plant Genet Resour 13(5):803–809 (in Chinese)
Zheng H, Hu D, Wang R, Wei R, Yan S (2015) Assessing 62 Chinese Fir (Cunninghamia lanceolata) breeding parents in a 12-year grafted clone test. Forests 6(10):3799–3808
Acknowledgements
We would like to appreciate Dr. Longxing Wang and Dr. Chenrui Gong for guiding the data analysis in this work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Project funding: This work was supported by the National Key Research and Development Program of China [No.2017YFD0600201] and Forestry Industry Research Special Funds for Public Welfare Projects [No. 201404101].
The online version is available at http://www.springerlink.com.
Corresponding editor: Yu Lei.
Rights and permissions
About this article
Cite this article
Lu, N., Mei, F., Wang, Z. et al. Allelic variation in the coumarate 3-hydroxylase gene associated with wood properties of Catalpa fargesii Bur.. J. For. Res. 32, 2517–2529 (2021). https://doi.org/10.1007/s11676-020-01276-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11676-020-01276-7