Abstract
Biomass estimation using allometric models is a nondestructive and popular method. Selection of an allometric model can influence the accuracy of biomass estimation. Bangladesh Forest Department initiated a nationwide forest inventory to assess biomass and carbon stocks in trees and forests. The relationship between carbon storage and sequestration in a forest has implications for climate change mitigation in terms of the carbon sink in Bangladesh. As part of the national forest inventory, we aimed to derive multi-species biomass models for the hill zone of Bangladesh and to determine the carbon concentration in tree components (leaves, branches, bark and stem). In total, 175 trees of 14 species were sampled and a semi-destructive method was used to develop a biomass model, which included development of smaller branch (base dia < 7 cm) biomass allometry and volume estimation of bigger branches and stems. The best model of leaf, branches, and bark showed lower values for adjusted R2 (0.3152–0.8043) and model efficiency (0.436–0.643), hence these models were not recommended to estimate biomass. The best fit model of stem and total aboveground biomass (TAGB) showed higher model efficiency 0.948 and 0.837, respectively, and this model was recommended for estimation of tree biomass for the hill zone of Bangladesh. The best fit allometric biomass model for stem was Ln (Stem) = − 10.7248 + 1.6094*Ln (D) + 1.323*Ln (H) + 1.1469*Ln (W); the best fit model for TAGB was Ln (TAGB) = − 6.6937 + 0.809*Ln (D^2*H*W), where DBH = Diameter at Breast Height, H = Total Height, W = Wood density. The two most frequently used pan-tropical biomass models showed lower model efficiency (0.667 to 0.697) compared to our derived TAGB model. The best fit TAGB model proved applicable for accurate estimation of TAGB for the hill zone of Bangladesh. Carbon concentration varied significantly (p < 0.05) by species and tree components. Higher concentration (48–49%) of carbon was recorded in the tree stem.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Forests of Bangladesh have been classified as tropical evergreen or semi evergreen forest, tropical moist or dry deciduous forests and tidal mangrove forest, all of which have been managed scientifically for over 100 years. Twenty-six inventories have been conducted for the forests of Bangladesh at regional and national scales. The objectives of these inventories shifted from volume estimation to biomass and carbon stock assessment (FD 2017a). Timber volume stock was assessed using species specific volume tables or volume equations (Mahmood et al. 2016a). Biomass estimation during the first National Forest and Tree Resources Assessment; and carbon inventory for the Sundarbans and eight protected areas were highly dependent on generalized and species-specific volume equations, and pantropical biomass models (FD 2007; Donato et al. 2011; Latif et al. 2015). The use of such allometric models can reduce accuracy in biomass and carbon stock assessment (Maulana et al. 2016; Nam et al. 2016).
The second National Forest Inventory was underway in Bangladesh in 2018 with the objectives of accurate assessment of biomass and carbon stock in trees and forests (FD 2017b). This inventory has stratified the trees and forests into hill, sal, sundarbans, coastal and village zone based on climatic and geophysical properties (Akhter et al. 2016). Localized, multi-species allometric biomass models are expected to be developed for each of these zones. Allometric technique is a semi- or non-destructive method of tree biomass estimation (Golley et al. 1975; Picard et al. 2012), where mathematical functions are derived to relate tree biomass with easily measurable biometrical predictors such as diameter at breast height (DBH), height (H) and wood density (W) (Sileshi 2014). Allometric equations can be developed for individual species or to represent a community at local, regional or biospheric (pan-tropical) scales. Species-specific and site-specific multi-species biomass allometric models can ensure desired accuracy in biomass estimation (Ketterings et al. 2001; Manuri et al. 2014; Maulana et al. 2016). Bangladesh has 222 validated allometric volume and biomass allometric models. But, the hill zone has only 10 allometric biomass models for three species (Senna siamea (Lam.) Irwin et Barneby, Artocarpus chaplasha Roxb. and Tectona grandis L.f. (Mahmood et al. 2016a), while this zone contains at least 47 tree species both in natural stands and in plantations (Nur et al. 2016). Development of species-specific allometric biomass models for tree species of this zone would be time consuming and labour intensive if based on destructive sampling (felling of trees). As part of the current National Forest Inventory, the aim of this study was to derive multi-species allometric biomass models for the frequently occurring tree species of the hill zone of Bangladesh.
Materials and methods
Study area
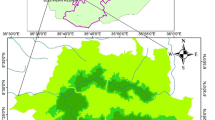
The hill zone of Bangladesh is located in the northeast and southeast corners of Bangladesh and covers about 17,174 Km2 of land area (Fig. 1). This zone consists of tropical evergreen and semi-evergreen vegetation with natural and plantation stands. The major naturally occurring tree species are A. chaplasha, Albizzia spp., Dipterocarpus spp., Duabanga grandiflora (Roxb. ex DC.) Walpers, Gmelina arborea Roxb., Hopea odorata Roxb., Lagerstroemia speciosa (L.) Pers., Mangifera sylvatica Roxb., Swintonia floribunda Griff., Syzigium spp., Toona ciliata M. Roem. (FD 2017c). The soil of this zone is yellowish brown to reddish brown in colour, loam to clay loam and sandy clay loam. Soil is acidic and organic matter content of the topsoil varies from 0.15 to 3.32%. While, total nitrogen concentration varies from 0.03 to 0.24%. The ranges of available form of soil phosphorus, potassium, calcium, and magnesium are 1.38 to 7.32 mg kg−1, 62 to 206 mg kg−1, 279 to 928 mg kg−1, and 74 to 371 mg kg−1, respectively (Hossain et al. 2014).
Sampling of tree species
A total of 175 individuals of 14 tree species representing 11 families (Table 1) were collected during two sessions from 50 sample plots of 19 m radius that covered a wide range of DBH and total height classes. All sample trees were selected to avoid specimens with broken top, hollow trunk, damage caused by natural calamities or animals, and evidence of suppression or disease. We collected 136 individuals during the first event (Table S1 Biomass Data Set A, Supplementary data), which was used to derive the allometric models. While, the remaining 39 individuals were collected during the second event (Table S2 Biomass Data Set B, Supplementary data) and that was used for validation and evaluation of the derived allometric models.
Field measurement and laboratory analysis
Total height and DBH of sampled trees were measured. A non-destructive method was used to estimate the volume of stems and larger branches (diameter > 7 cm). Base diameter of all small branches (diameter < 7 cm) of the sampled trees were measured and 2 to 3 smaller branches were trimmed from each sampled tree. These trimmed branches were separated into components (leaves, leaf containing smaller branch, and woody parts of branch) to measure their fresh biomass in the field.
Ten sub-samples (about 0.25 kg) of each component (leaves, leaf containing smaller branch, and woody parts of branch) of trimmed branches of each species were collected from the field. These were oven-dried at 105 °C to constant weight to derive a fresh to oven-dried weight conversion ratio (Mahmood et al. 2015). Wood samples of bigger branches and stems of sampled species were collected using battery powered hand drill. Carbon concentration of components (leaf, smaller branches, bigger branches and stem) were measured by use of the ignition loss method (Allen 1989). Carbon concentration were compared by two-way analysis of variance using R (3.2.3) statistical software.
Allometric biomass models for the smaller branches
Base diameter of branches and biomass of branch components (leaf, leaf containing smaller branch (LCB), and woody parts of branch) were considered as predictor and output variables respectively. These variables were Ln (natural logarithm) transformed to derived the species-specific branch allometric biomass models (Ln (biomass) = a + b Ln (base diameter of branch)) using R (3.2.3) statistical software. A correction factor (CF) was calculated for each equation to minimize the systematic bias during the back transformation of Ln values to biomass values (Sprugel 1983).
Biomass of sampled tree
Oven-dry biomass of stem and larger branches was estimated from their calculated volume (m3) and the mean wood density (Kg m−3) of the respective tree species. Mean wood densities were used from the data base of Zanne et al. (2009) (Table 1) and these values were compared by one-way analysis of variance using R (3.2.3) statistical software. Oven-dry biomass of smaller branches of the individual sampled trees was estimated from the derived species-specific branch allometric model and oven-dry biomass of trimmed branches of the sampled trees (Picard et al. 2012).
Allometric models for tree components and total above-ground biomass (TAGB)
DBH, tree height, and wood density were designated as predictor variables while biomass of leaf, branch, bark, stem and TAGB were designated as output variables. All variables were Ln (natural logarithm) transformed to improve linearity and homoscedasticity (Zar 1996). A total of eight allometric models (Table 2) were tested to derive the allometric model for the output variables using R (3.2.3) statistical software. A correction factor (CF) was calculated for each equation to minimize the systematic bias during the back transformation of Ln values to biomass values (Sprugel 1983).
Selection, validiation and comparisn of allometric model
The best-fit allometric model was selected based on lowest Akaike Information Criterion (AIC) and Residual Standard Error (RSE); and the highest Akaike Information Criterion Weight (AICw) and coefficient of determination (R2) (Sileshi 2014; Mahmood et al. 2015, 2016b). But, models having RSE value greater than 0.30 may not be selected as best fit (Sileshi 2014). A variance influence factor (VIF) test was performed to check the multicollineary among the identical predictor variables of each model. Models having VIF > 5 indicate the existence of multicollineary among the identical independent variables (Sileshi 2014). The selected models of leaf, branch, bark and stem biomass were validated using sample trees of Data Set B in terms of model efficiency (ME). Modes having ME value less than zero cannot be recommended, while models with ME value close to 1 are considered to be “near-perfect” (Mayer and Butler 1993).
where Yp = Predicted biomass from model, Yo = Observed biomass in field measurement, and \({\bar{{\text{Y}}}}\) = Mean of the observed biomass.
Data Set B was used to compare the ME of the most frequently used pan-tropical biomass models of Brawn (1997) and Chave et al. (2005). Regression between Yp (in the X-axis) and Yo (in the Y-axis) was calculated for the best fit and compared pan-tropical models using the Data Set B (Table 8). Significance of slope (b = 1) and intercept (a = 1) were also tested according to Piñeiro et al. (2008). This analysis helped to understand graphically the over- or under-estimation of the predicted biomass using each model from 1:1 line (Sileshi 2014). Unless otherwise noted in the following text, the alpha level for all statistical analyses was 0.05.
Results
Carbon in different components of samples tree species and wood density
Carbon concentration varied significantly by tree component as well as species. Among tree components, leaf contained lower concentration (43–48%) of carbon, while significantly higher concentration (48–49%) of carbon was recorded in stem (Table 3). Wood density values of the sampled tree species were found to vary significantly with species. Higher (822 kg m−3) density was observed for T. arjuna followed by T. bellirica and lowest density (320 kg m−3) was recorded for A. malaccensis (Table 1).
Allometric models for the smaller branches
The adjusted R2 values of the allometric model for leaf, leaf containing smaller branches and woody parts of smaller branches varied by species. Lower values (0.2487 to 0.8608) of adjusted R2 were observed for leaves, while higher values (0.7270 to 0.9767) were recorded for woody parts of smaller branches (Tables 4, 5, 6).
Allometric biomass models and their validiation
The ranges of DBH and H were 10.9 to 62 cm and 6 to 24 m, respectively. Model 6 (Ln(Leaf) = − 11.0054 + 1.2476*Ln(D) + 1.355*Ln(W)) was selected as the best allometric model for leaf with adjusted R2 value of 0.3152. In case of branch, Model 4 (Ln (Branch) = − 31.244 + 29.4233*Ln (D) − 9.2811*(Ln (D))^2 + 1.0181*(Ln (D))^3 + 0.4927*Ln (W)) showed lowest AIC and RSE, and highest AICw, adjusted R2, and variance influence factor (VIF) for multicollineary test than the reference value (5) compared to other tested models. While, Model 3 (Ln (Branch) = − 4.0022 + 2.0826*Ln (D) − 0.5103*Ln (H) + 0.4863*Ln (W)) with second lowest AIC (224.494) and RSE (0.5364), and second highest AICw (0.0544) and adjusted R2 (0.6458) was selected as the best model. In case of bark, Model 3 (Ln (Bark) = − 13.8954 + 1.0877*Ln (D) + 1.5993*Ln (H) + 1.4478*Ln (W)) showed better selection criteria with lowest AIC (181.326) and RSE (0.4576), and highest adjusted R2 (0.8043). However, RSE for these allometric models (leaf, branch and bark) exceeded the reference level (Table 7). In other ways, the best fit allometric models of stem without bark and TAGB were Model 3 (Ln (Stem) = − 10.7248 + 1.6094*Ln (D) + 1.323*Ln (H) + 1.1469*Ln (W)) and Model 5 (Ln (TAGB) = − 6.6937 + 0.809*Ln (D^2*H*W)), respectively, where DBH = Diameter at Breast Height in cm, H = Total Height in m, and W = Wood Density (kg m−3) (Table 7). The allometric models for leaves, branch and bark showed very low model efficiency (0.436 − 0.643) compared to the best-fit allometric models of stem without bark (0.948) and TAGB (0.837) (Table 8).
TAGB model comparison
The pan-tropical biomass model of Brown (1997) and Chave et al. (2005) showed lowest model efficiency (0.667 to 0.697) compared to the best-fit TAGB model of this study (Table 8). Visualization of the observed and predicted biomass demonstrated deviation in biomass estimation from the line of significance of slope (b = 1) and intercept (a = 1), which indicated that compared pan-tropical biomass models of Brown (1997) and derived best-fit TAGB models underestimated TAGB, while model of Chave et al. (2005) overestimated TAGB (Fig. 2).
Discussion
This study showed differences in carbon concentration among the tree components and species. Similarly, Thomas et al. (2012) reported wide variation in carbon concentration (41.9–51.6%) in stem tissue of tropical tree species
Accuracy in biomass estimation is largely depends on appropriate selection of allometric models (Nam et al. 2016). Model selection involves careful investigation of model parameters as described by Sileshi (2014), Birigazzi et al. (2015) and Mahmood et al. (2016a). This study included tree species having wide range of wood density and the best model for all components of trees included DBH, H and W as predictor variables (Table 7).
Most of the multi-species biomass models include wood density along with DBH and H as predictor variables to account for species-specific variability (Nelson et al. 1999; Chave et al. 2005, 2014; Djomo et al. 2010). The fitted model of leaf, branch and bark of this study showed lower adjusted R2 as 0.3152, 0.6458 and 0.8043, respectively, compared to the recommended value (0.85) by UNFCCC (2011), hence these models might be less statistically reliable for future use. RSE of these models were higher than the reference level (Sileshi 2014), which could result in unrealistic biomass estimation for leaf, branch and bark by using these derived models (Mugasha et al. 2016). Thus, we failed to identify any best-fit model for leaf, branch and bark. A pool of tree species with wide range of DBH (10.9 to 62 cm) and H (6 to 24 m) were used to derive the allometric models. Each tree species has inherent architecture that varies with stage of growth, morphological characteristics, adaptation ability, physiological characteristics and site quality (Tomlinson and Zimmermann 1978; White 1979; Hibbs 1981). Tree architecture influences the form and shape of crown and stem growth, which shows identical morphological differences among tree species. Tree crown is highly variable with species and stages of growth (sapling, pole and tree), which can directly affect the branching pattern as well as biomass of leaves and branches (Jack et al. 1982; Echereme et al. 2015; Mugasha et al. 2016). Finally, DBH, H and W as predictor variables may not able to address the variability among the tree crown biomass of the sampled species. In such case, inclusion of some more predictor variables like crown dimensions (crown length and crown diameter) may increase the efficiency of multi species biomass models for leaf and branch. The selected models for stem and TABG were linear and/or interactive (Model 3) and compound derivatives (Model 5), respectively, and both showed statistical credibility to be selected as best fit model (Sileshi 2014; Birigazzi et al. 2015)
Accurate estimation of biomass stock is needed, but some error is always associated with biomass estimation using allometric models. Our best fit TAGB models showed higher efficiency compared to the frequently used pan-tropical models of Brown (1997) and Chave et al. (2005). Such comparison is essential to assess the uncertainty and suitability of the derived model and frequently used pan-tropical biomass models at local scale (Nam et al. 2016). Numerous studies have demonstrated that biomass estimation using pan-tropical models generated higher uncertainty than did locally developed models, for instance the biomass study of Kalimantan (Basuki et al. 2009), Sarawak (Kenzo et al. 2009), Columbia (Alvarez et al. 2012; Ngomanda et al. 2014), Indonesia (Manuri et al. 2014; Maulana et al. 2016), and Vietnam (Nam et al. 2016). However, the context provided by this study and the results presented herein demonstrate that our derived best-fit model will able to accurately estimate TAGB for the hill zone of Bangladesh.
Conclusions
Best fit biomas models for leaf, branch and bark were not identified due to unacceptably low model efficiency. Diameter at breast height, height and wood density as predictor variables might not be enough to address the variabliity of leaf and branch biomass for a pool of tree species having different crown architecture. The best fit total aboveground biomass model of this study showed higher model efficiency compared to some frequently used pan-tropical models. The results of this study demonstrate that the development of local models derived from an appropriate sample of representative species with appropriate predictor variables can greatly improve the estimation of total aboveground biomass. The best-fit models presented here can provide greater confidence when estimating biomass, carbon stock, and monitoring of forest productivity. This might help to guide new management initiatives for the Hill zone of Bangladesh.
References
Akhter M, Jalal R, Costello L, Rahman L, Tasnuva U (2016) Zoning for tree and forest assessment in Bangladesh. Bangladesh Forest Department and Food and Agricultural Organization of the United Nations, Dhaka, Bangladesh, p 36
Allen SE (1989) Chemical analysis of ecological materials. Blackwell Scientific Publications, Oxford, p 565
Alvarez E, Rodríguez L, Duque A, Saldarriaga J, Cabrera K, de las Salas G, del Valle ILA, Moreno F, Orrego S (2012) Tree above-ground biomass allometries for carbon stocks estimation in the natural forests of Colombia. For Ecol Manag 267:297–308
Basuki TM, van Laake PE, Skidmore AK, Hussin YA (2009) Allometric equations for estimating the above-ground biomass in tropical lowland Dipterocarp forests. For Ecol Manag 257:1684–1694
Birigazzi L, Javier GP, Gamarra Gael S, Stefano G, Emily D, Julian M, Matieu H, Nicolas P (2015) Toward a transparent and consistent quality control procedure for tree biomass allometric equations. In: XIV World Forestry Congress, vol 7, no 11. Durban, South Africa
Brown S (1997) Estimating biomass and biomass change of tropical forests: a primer. FAO Forestry Paper, vol 134. Rome, p 55
Brown S, Gillespie AJR, Lugo AE (1989) Biomass estimation method for tropical forests with applications to forest inventory data. For Sci 35:881–902
Chave J, Andalo C, Brown S, Cairns MA, Chambers JQ, Eamus D, Folster H, Fromard F, Higuchi N, Kira T, Lescure JP, Nelson BW, Ogawa H, Puig H, Riera B, Yamakura T (2005) Tree allometry and improved estimation of carbon stocks and balance in tropical forests. Oecologia 145:87–99
Chave J, Réjou-Méchain M, Búrquez A, Chidumayo E, Colgan MS, Delitti WB, Duque A, Eid T, Fearnside PM, Goodman RC, Henry M, Martínez-Yrízar A, Mugasha WA, Muller-Landau HC, Mencuccini M, Nelson BW, Ngomanda A, Nogueira EM, Ortiz-Malavassi E, Pélissier R, Ploton P, Ryan CM, Saldarriaga JG, Vieilledent G (2014) Improved allometric models to estimate the aboveground biomass of tropical trees. Global Change Biol 10(10):3177–3190
Djomo AN, Ibrahimab A, Saborowskic J, Gravenhorst G (2010) Allometric equations for biomass estimations in Cameroon and pan moist tropical equations including biomass data from Africa. For Ecol Manag 260:1873–1885
Donato DC, Ahmed I, Iqbal Z (2011) Carbon assessment report 2009–2010 inventory of the sundarbans reserve forest. Bangladesh Forest Department, Dhaka, p 22
Echereme CB, Mbaekwe EI, Ekwealor KU (2015) Tree crown architecture: approach to tree form, structure and performance: a review. Int J Sci Res Pub 5(9):2250–3153
FD (2007) National forest and tree resources assessment 2005–2007, Bangladesh. Ministry of Environment and Forest (MoEF), Dhaka, p 118
FD (2017a) Forest Type. http://www.bforest.gov.bd/site/page/28615def-835e-4b3a-a412-da954f55a943/. Accessed 30 July 2018
FD (2017b) Bangladesh Tree & Forest Inventory 2016. http://www.bforest.gov.bd/site/page/8fce3a01-7119-4083-9448-489a6a38a1a5/Bangladesh-Forest-Inventory. Accessed 21 July 2018
FD (2017c) Hill Forest. http://www.bforest.gov.bd/site/page/28615def-835e-4b3a-a412-da954f55a943/Tropical-Evergreen-and-Semi-evergreen-Forest-(Hill-Forest). Accessed 21 July 2018
Golley BF, Mc Ginnis TJ, Clements GR, Child IG, Duever JM (1975) Mineral cycling in a tropical moist forest ecosystem. University of Georgia Press, Athens, p 248
Hibbs DE (1981) Leader growth and the architecture of three North American hemlocks. Can J Bot 59:476–480
Hossain MdI, Kashem MdA, Osman KT (2014) Fertility status of some forested soil of Chittagong Hill Tract, Bangladesh. Int J Latest Res Sci Technol 3(1):82–87
Jack B, Fisher David EH (1982) Plasticity of tree architecture: specific and ecological variation found in aubreville’s model. Am J Bot 69(5):690–702
Kenzo T, Furutani R, Hattori D, Kendawang JJ, Tanaka S, Sakurai K, Ninomiya I (2009) Allometric equations for accurate estimation of above-ground biomass in logged-over tropical rainforests in Sarawak, Malaysia. J For Res 14:365–372
Ketterings QM, Coe R, Noordwijk MV, Amagau Y, Palm CA (2001) Reducing uncertainty in the use of allometric biomass equations for predicting above-ground tree biomass in mixed secondary forest. For Ecol Manag 146:199–209
Latif MAF, Netzer M, Banik H, Chowdhury RM (2015) Forest carbon inventory 2014 at eight protected areas in Bangladesh. Bangladesh Forest Department and Winrock International, Dhaka, p 58
Mahmood H, Siddique MRH, Saha S, Abdullah SMR (2015) Allometric models for biomass, nutrients and carbon stock in Excoecaria agallocha of the Sundarbans, Bangladesh. Wetl Ecol Manag 23:765–774
Mahmood H, Saha C, Abdullah SMR, Saha S, Siddique MRH (2016a) Allometric biomass, nutrient and carbon stock models for Kandelia candel of the Sundarbans, Bangladesh. Trees 30(3):709–717
Mahmood H, Siddique MRH, Akhter M (2016b) A critical review and database of biomass and volume allometric equation for trees and shrubs of Bangladesh. IOP Conf Ser, Earth Environ Sci 39:012057
Manuri S, Brack C, Nugroho NP, Hergoualc’h K, Novita N, Dotzauer H, Verchot L, Putra CAS, Widyasari E (2014) Tree biomass equations for tropical peat swamp forest ecosystems in Indonesia. For Ecol Manag 334:241–253
Maulana SI, Wibisono Y, Utomo S (2016) Development of local allometric equation to estimate total aboveground biomass in Papua tropical forest. Indonesian J For Res 3(2):107–118
Mayer D, Butler D (1993) Statistical validation. Ecol Model 68(1):21–32
Mugasha WA, Mwakalukwa EE, Luoga E, Malimbwi RE, Zahabu E, Silayo DS, Sola G, Crete P, Henry M, Kashindye A (2016) Allometric models for estimating tree volume and aboveground biomass in lowland forest of Tanzania. Int J For Res. https://doi.org/10.1155/2016/8076271
Nam VT, van Kuijk M, Anten NPR (2016) Allometric equations for aboveground and belowground biomass estimations in an evergreen forest in Vietnam. PLoS ONE 11(6):e0156827
Nelson BW, Mesquita R, Pereira JLG, Souza SGAD, Batista GT, Couto LB (1999) Allometric regressions for improved estimate of secondary forest biomass in the central Amazon. For Ecol Manag 117:149–167
Ngomanda A, Engone Obiang NL, Lebamba J, Moundounga Mavouroulou Q, Gomat H, Mankou GS, Loumeto J, Midoko Iponga D, Kossi Ditsouga F, Zinga Koumba R (2014) Site-specific versus pantropical allometric equations: Which option to estimate the biomass of a moist central African forest? For Ecol Manag 312:1–9
Nur A, Nandi R, Jashimuddin M, Hossain MA (2016) Tree species composition and regeneration status of Shitalpur Forest Beat under Chittagong North Forest Division, Bangladesh. Adv Ecol 2016:5947874
Picard N, Saint-André L, Henry M (2012) Manual for building tree volume and biomass allometric equations: from field measurement to prediction. In: Food and agricultural organization of the United Nations, Rome, and Centre de Coopération Internationale en Recherche Agronomique pour le Développement, Montpellier, p 213
Piñeiro G, Perelman S, Guerschman JP, Paruelo JM (2008) How to evaluate models: observed vs predicted or predicted vs observed. Ecol Model 216:316–322
Sileshi GW (2014) A critical review of forest biomass estimation models, common mistakes and corrective measures. For Ecol Manag 329:237–254
Sprugel DG (1983) Correcting for bias in log-transformed allometric equations. Ecology 64(1):209–210
Thomas SC, Martin AR (2012) Carbon content of tree tissues: a synthesis. Forests 3:332–352
Tomlinson PB, Zimmermann MH (1978) Tropical trees as living systems. Cambridge University Press, Cambridge, p 202
UNFCCC (2011) Report of the Conference of the Parties on Its Sixteenth Session, Held in Cancun from 29 November to 10 December 2010. In: Addendum. Part Two: Action Taken by the Conference of the Parties at Its Sixteenth Session. Geneva: (United Nations Framework Convention on Climate Change)
White J (1979) The plant as a metapopulation. Annu Rev Syst Ecol 10:109–145
Zanne AE, Lopez-Gonzalez G, Coomes DA, Ilic J, Jansen S, Lewis SL, Miller RB, Swenson NG, Wiemann MC, Chave J (2009) Global wood density database. Dryad. Identifier: http://hdl.handle.net/10255/dryad.235. Accessed 10 June 2018
Zar JH (1996) Biostatistical analysis, 3rd edn. Prentice Hall, Englewood Cliffs, p 662
Acknowledgements
We greatly acknowledge the financial support of FAO through GCP/BGD/058/USA (LOA Code: FAOBGDLOA 2017-008) to accomplish the field and laboratory work. We would like to Bangladesh Forest Department and Forestry and Wood Technology Discipline, Khulna University for their logistic support during the field and laboratory analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
Project funding: The work was supported by FAO through GCP/BGD/058/USA (LOA Code: FAOBGDLOA 2017-008).
The online version is available at http://www.springerlink.com
Corresponding editor: Tao Xu.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mahmood, H., Siddique, M.R.H., Islam, S.M.Z. et al. Applicability of semi-destructive method to derive allometric model for estimating aboveground biomass and carbon stock in the Hill zone of Bangladesh. J. For. Res. 31, 1235–1245 (2020). https://doi.org/10.1007/s11676-019-00881-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11676-019-00881-5