Abstract
Genome editing is a valuable tool to target specific DNA sequences for mutagenesis in the genomes of microbes, plants, and animals. Although different genome editing technologies are available, the clustered regularly interspaced short palindromic repeats/Cas9 (CRISPR/Cas9) system, which utilizes engineered endonucleases to generate a double-stranded DNA break (DSB) in the target DNA region and subsequently stimulates site-specific mutagenesis through DNA repair machineries, is emerging as a powerful genome editing tool for elucidating mechanisms of protection from plant viruses, plant disease resistance, and gene functions in basic and applied research. In this review, we provide an overview of recent advances in the CRISPR system associated genome editing in plants by focusing on application of this technology in model plants, crop plants, fruit plants, woody plants and grasses and discuss how genome editing associated with the CRISPR system can provide insights into genome modifications and functional genomics in plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genome editing that targets specific genome sequences has emerged as a powerful technology for improving crop yield, enhancing crop quality, and increasing plant disease and virus resistance (Nagamangala Kanchiswamy et al. 2015; Nejat et al. 2016; Ricroch and Henard-Damave 2015; Schaart et al. 2015; Teotia et al. 2016; Zlotorynski 2015). Targeted genome engineering is expected to contribute significantly to future plant improvement and holds great promise in plant molecular breeding, biotechnology, agriculture, environmental protection, and basic plant biology research because of its highly efficient modification and the ability to precisely modify genome sequence in a site-specific manner (Baltes and Voytas 2015; Belhaj et al. 2015; Bogdanove 2014; Bortesi and Fischer 2015; Lozano-Juste and Cutler 2014). Although plant genome editing based on Agrobacterium and ionizing radiations is efficient, these methods led to randomly distributed genome modifications (Quetier 2016). Unlike Agrobacterium and ionizing-radiation-based methods, zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) provide customizable DNA-binding modules designed to bind to any sequence of interest and have been used with much success across eukaryotic species to edit genomes because these technologies introduce site-specific double strand DNA breaks and improve genome editing that led to a dramatic decrease in off-target events (Belhaj et al. 2013; Bortesi and Fischer 2015; Butler et al. 2015; Chandrasegaran and Carroll 2015; Jia and Wang 2014a; Kumar and Jain 2015; Liang et al. 2014; Osakabe and Osakabe 2015; Puchta and Fauser 2013; Scott et al. 2014; Shan et al. 2014; Wang et al. 2015; Weeks et al. 2015). However, these technologies have not been widely applied because of the complicated design and laborious assembly of specific DNA-binding proteins for each target gene.
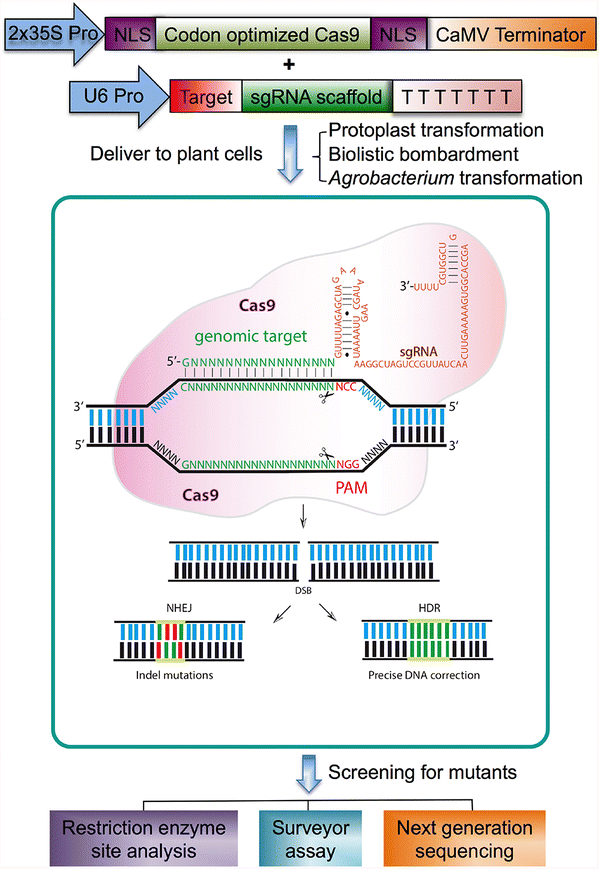
Recently, an easier method based on the bacterial type II CRISPR (clustered regularly interspaced short palindromic repeats)/Cas (CRISPR-associated) immune system has emerged as a powerful technology for genome editing (Belhaj et al. 2013; Teotia et al. 2016). CRISPRs are used as guide RNAs for Cas9 nuclease-specific digestion (Chaparro-Garcia et al. 2015; Quetier 2016; Zlotorynski 2015). The CRISPR/Cas system has been a highly efficient DNA-targeting platform for genome editing since it allows targeted cleavage of genomic DNA guided by a customizable small, noncoding RNA, resulting in gene modifications by both nonhomologous end joining (NHEJ) and homology-directed repair (HDR) mechanisms (Baltes and Voytas 2015; Bortesi and Fischer 2015; Shariat and Dudley 2014; Trevino and Zhang 2014). The CRISPR/Cas9 system is technically easy to use (Fig. 1), and scientists can use commercially available kits for targeted editing of genomes after carefully choosing the location of the DNA double strand breaks to be induced (Mahfouz et al. 2014). The CRISPR/Cas9 system represents dramatic scientific progress that allows the development of more complex useful agronomic traits through synthetic oligonucleotides (Belhaj et al. 2013; Quetier 2016). Genome editing based on the CRISPR/Cas9 system has been successfully applied in dozens of diverse plant species, including the sweet orange (Jia and Wang 2014a), arabidopsis (Gao et al. 2015b; Jiang et al. 2013; Li et al. 2014), tobacco (Gao et al. 2015a, b; Jiang et al. 2013; Li et al. 2014; Lowder et al. 2015), maize (Basak and Nithin 2015; Liang et al. 2014; Puchta and Fauser 2013; Schuster et al. 2015; Svitashev et al. 2015; Xing et al. 2014), rice (Mikami et al. 2015a, b; Wang et al. 2015; Xu et al. 2014; Zhang et al. 2016b), and wheat (Basak and Nithin 2015; Shan et al. 2014; Sugano et al. 2014; Upadhyay et al. 2013; Wang et al. 2014).
Overview of the mechanism for genome editing by the CRISPR/Cas9 system and subsequent targeted mutagenesis. After Cas9 and the targeted genomic sequence guided by the sgRNA are delivered into the plant cell by protoplast transformation, biolistic bombardment, or Agrobacterium-mediated transformation, Cas9 stimulates DNA strand separation and allows a sgRNA to hybridize with a specific sequence in the targeted gene for targeted DNA cleavage. The CRISPR/Cas9 system positions the target DNA into the active site of Cas9 in proper orientation in relation to a PAM binding site and allows separate nuclease domains of Cas9 to independently cleave each strand of the target DNA sequence 3-nt upstream of the PAM site. The double-strand DNA break then undergoes error-prone nonhomologous end joining (NHEJ) and homology-directed repair (HDR) DNA repair when deletions or insertions of a few nucleotides often occur. Approximately one in three deletions or insertions that restore the proper reading frame in the gene’s coding region thus restore gene activity. Mutants are examined by different approaches, including restriction enzyme site analysis, surveyor assay, and next generation sequencing. The cassette expressing Cas9 is driven by the cauliflower mosaic virus 35S promoter and the target and scaffold sequence of the gRNA is driven by the U6 promoter. The PAM sequence (NGG; red) is essential for the Cas9 nuclease activity. Green letters represent the targeted genomic DNA molecule; scissors indicate cleavage sites; Cas9, (CRISPR)-associated protein 9; PAM, protospacer adjacent motifs; sgRNA, single guide RNA; NLS, nuclear localization signal
The use of ZFP/ZFNs, TALE/TALENs, and CRISPR/Cas has produced unprecedented advances in gene targeting and genome editing in plants (Bortesi and Fischer 2015; Fichtner et al. 2014; Liang et al. 2014; Osakabe and Osakabe 2015). These genome editing techniques allow researchers to specifically alter genes, reprogram epigenetic marks, generate site-specific deletions and potentially cure diseases (Chandrasegaran and Carroll 2015; Fichtner et al. 2014; Kumar and Jain 2015; Puchta and Fauser 2013). Principles and advances of these technologies in plant physiology and metabolism (Bogdanove 2014; Kumar and Jain 2015), gene expression and regulation (Basak and Nithin 2015; Belhaj et al. 2015), knockout of host susceptibility genes (Ilardi and Tavazza 2015), generation of disease resistant plants (Nejat et al. 2016), protection from plant viruses (Zlotorynski 2015), mutagenesis in plants (Belhaj et al. 2013), and probe gene function and modify epigenetic marks (Fichtner et al. 2014) have been well reviewed.
Here we review the capabilities of the CRISPR/Cas9 system, the improvement of plants by increasing yield, quality, and tolerance to limiting biotic and abiotic stress and highlight the exciting applications of the CRISPR/Cas9 system in plant molecular biology and crop genetic engineering. In this review, we provide an overview of recent advances in CRISPR-system-associated genome editing in plants by focusing on application of this technology in model plants, crop plants, fruit plants, woody plants and grasses (Table 1) and discuss how it can provide insights into genome modifications and functional genomics research in plants.
CRISPR/Cas9 system-based genome editing in plants
Genome editing is a technique that adds, removes or replaces a specific target DNA sequence of the genome using synthetic DNA bases. The CRISPR/Cas system is an invaluable genome editing technology that is very efficient and preferred (Jacobs et al. 2015; Liang et al. 2014; Wang et al. 2015). The CRISPR/Cas system employs an RNA-guided nuclease, Cas9, to induce double-strand breaks and allows targeted cleavage of genomic DNA guided by a customizable small noncoding RNA, resulting in gene modifications (Belhaj et al. 2013; Osakabe and Osakabe 2015; Quetier 2016). The CRISPR/Cas9 system has been used to precisely mutate DNA sequences in a number of plants, and the Cas9-mediated breaks are repaired by cellular DNA repair mechanisms (Kumar and Jain 2015; Nagamangala Kanchiswamy et al. 2015; Nejat et al. 2016; Zlotorynski 2015). A novel cloning strategy and vector system based on In-Fusion(R) cloning was developed to simplify the production of CRISPR/Cas9 targeting vectors, which should be applicable for targeting any gene in any plant species (Jacobs et al. 2015).
The CRISPR/Cas system-based genomic manipulations are revolutionizing virtually all areas of molecular biosciences, including functional genomics, genetics, applied biomedical research, and agricultural biotechnology (Bortesi and Fischer 2015; Nagamangala Kanchiswamy et al. 2015; Teotia et al. 2016; Zlotorynski 2015). The CRISPR/Cas system is a rapidly developing genome editing technology that has been successfully applied in many plants for targeted mutagenesis, and the heritable, CRISPR/Cas9-mediated modifications to the plant genome have made gene targeting in plants promising and provided a foundation for efficient genome editing of crop genomes without the random integration of foreign DNA (Baltes and Voytas 2015; Bogdanove 2014; Cermak et al. 2015; Lozano-Juste and Cutler 2014). Compared with the two well-established genome editing platforms ZFNs and TALENs, the CRISPR/Cas9 is simple, efficient, and highly specific (Bortesi and Fischer 2015; Liang et al. 2014; Wang et al. 2015).
Cas9, an RNA-guided DNA endonuclease, can be targeted to specific genomic sequences and induce heritable mutations by engineering a separately encoded guide RNA with which it forms a complex (Belhaj et al. 2013, 2015). CRISPR/Cas9 has proven to be extremely versatile because only a short synthetic RNA sequence is needed to recognize a new target (Belhaj et al. 2013, 2015; Bortesi and Fischer 2015; Liang et al. 2014; Wang et al. 2015). In some plant species, homozygous knockout mutants can be produced in a single generation, indicating its importance as a breakthrough technology for genome editing in basic research and plant breeding (Belhaj et al. 2015; Chandrasekaran et al. 2016; Quetier 2016; Teotia et al. 2016). CRISPR loci are transcribed into ncRNA and eventually form a functional complex with Cas9 and further guide the complex to cleave complementary invading DNA (Basak and Nithin 2015). Applications of the CRISPR/Cas technologies in plants demonstrated that these systems were powerful tools for genome editing in conventional biological model systems, functional genome screening, cell-based human hereditary disease modeling, epigenomic studies, and visualizing cellular processes (Belhaj et al. 2013; Nemudryi et al. 2014).
Delivery of the CRISPR/Cas9 system in plants
The CRISPR/Cas9 system is delivered into plant cells for genome editing by a number of approaches including biolistic delivery, virus-based guide RNA (gRNA) delivery, and Agrobacterium-mediated delivery. Biolistic delivery for CRISPR/Cas9 mediated plant genome editing has been used in maize (Zea mays) for targeted mutagenesis of endogenous genes and site-specific insertion of a trait gene (Sauer et al. 2015; Svitashev et al. 2015). DNA vectors expressing maize codon-optimized S. pyogenes Cas9 endonuclease and single guide RNAs were co-introduced with or without DNA repair templates into maize immature embryos by biolistic transformation, targeting upstream of the liguleless1 (LIG1) gene, male fertility genes (Ms26 and Ms45), and acetolactate synthase (ALS) genes (Frampton et al. 2012; Svitashev et al. 2015). Mutations were subsequently identified at all sites targeted, and plants containing biallelic multiplex mutations at LIG1, Ms26, and Ms45 were recovered, indicating the utility of Cas9-guide RNA technology as a plant genome editing tool for plant breeding (Svitashev et al. 2015). Oligonucleotide-mediated technology to target different genes in Arabidopsis thaliana (hereafter, Arabidopsis) protoplasts showed that the selection of traits in plant varieties may improve crop performance by genome editing technologies (Sauer et al. 2015).
A virus-based guide RNA (gRNA) delivery system for CRISPR/Cas9-mediated plant genome editing (VIGE) has been used in Nicotiana benthamiana, fruit trees, and crop plants (Ali et al. 2015; Ilardi and Tavazza 2015; Yin et al. 2015). Using a modified cabbage leaf curl virus (CaLCuV) vector to express gRNAs in stable transgenic plants expressing Cas9, mutations in NbPDS3 and NbIspH genes were confirmed by DNA sequencing, indicating that geminivirus-based VIGE could be a powerful tool in plant genome editing (Yin et al. 2015). Plum pox virus (PPV) is the etiological agent of sharka, the most devastating and economically important viral disease affecting Prunus species. Robust and predictable resistance can be achieved using CRISPR/Cas9-engineered nucleases to target conserved regions of the PPV genome and knockout host susceptibility genes in stone fruit trees for efficient and fast breeding programs (Ali et al. 2015; Cermak et al. 2015; Ilardi and Tavazza 2015; Yin et al. 2015). The results of tobacco rattle virus (TRV)-mediated genome editing in Nicotiana benthamiana demonstrated the persistence of the TRV-mediated Cas9 activity for up to 30 d after agroinfection, indicating that virus-mediated CRISPR/Cas9 is a useful tool for targeted engineering of plant genomes (Ali et al. 2015).
Agrobacterium has been used as a tool to deliver the CRISPR/Cas9 system, consists of a short guide RNA (gRNA) and the gene encoding Cas9, into a wide variety of plant species including Arabidopsis, tobacco, rice, sorghum, wheat, and the liverwort Marchantia polymorpha for targeted mutagenesis (Hyun et al. 2015; Jiang et al. 2013; Sugano et al. 2014). For example, using Agrobacterium-mediated transformation, Sugano et al. (2014) isolated stable mutants in the gametophyte generation of M. polymorpha (Sugano et al. 2014). In A. thaliana, after Agrobacterium-mediated introduction of T-DNAs encoding RGENs that targets FLOWERING LOCUS T (FT) and SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 4 genes, somatic mutagenesis at the targeted loci was detected in transgenic plants (Hyun et al. 2015). Rice protoplast cells transformed with Cas9/sgRNA constructs targeting the promoter region of the bacterial blight susceptibility genes OsSWEET14 and OsSWEET11 were confirmed by DNA sequencing to contain mutated DNA sequences at the target sites (Jiang et al. 2013).
The CRISPR/Cas9-associated genome editing in model plants
Arabidopsis thaliana
The CRISPR system has also emerged as a powerful tool for targeted gene editing in Arabidopsis, and the Cas9 nuclease is able to induce heritable mutations (Schiml et al. 2014). Agrobacterium tumefaciens was used for delivery of genes encoding Cas9, sgRNA and a nonfunctional, mutant green fluorescence protein (GFP) to Arabidopsis, and the results demonstrated that the Cas9/sgRNA system could be a powerful means of plant genetic engineering for scientific and agricultural applications (Jiang et al. 2013). Feng et al. (2014) examined several plant generations with seven genes at 12 different target sites to determine the patterns, efficiency, specificity, and heritability of CRISPR/Cas-induced gene mutations or corrections in Arabidopsis (Feng et al. 2014). They reported that the rate of mutations (chimeric, heterozygous, biallelic, or homozygous) was 71.2 % in the T1 generation, 58.3 % in the T2, and 79.4 % in the T3, and most mutations were 1-bp insertions or short deletions (Feng et al. 2014). Mutations in T1 plants are mostly in somatic cells. In contrast, approximately 22 % of T2 plants were found to be homozygous for a modified gene (Feng et al. 2014).
The efficiency of Cas9/sgRNA induced modification of a target reporter gene in T1 transgenic plants of A. thaliana and the inheritance of the modified gene in T2 and T3 progeny was examined, and efficient conversion of an out-of-frame GFP gene into a functional GFP gene was confirmed in T1 plants by green fluorescent signals in leaf tissues and by the presence of mutagenized DNA sequences at the sgRNA target site within the GFP gene (Jiang et al. 2014). Analyses of 42 individual T2 generation plants derived from six T1 progenitor plants showed that 50 % of the T2 plants inherited a single T-DNA insert, indicating the promise of this system for facile editing of plant genes. Gene targeting (GT) by homologous recombination can also be induced by DSBs. Schiml et al. (2014) demonstrated that this Cas9 paired nickase strategy has a mutagenic potential comparable with that of the nuclease, while the resulting mutations are mostly deletions that are stably inherited in A. thaliana (Schiml et al. 2014).
A CRISPR/Cas9 binary vector set based on the pGreen or pCAMBIA backbone, as well as a gRNA (guide RNA) module vector set for multiplex genome editing in plants has been developed (Xing et al. 2014). This toolkit was validated using transgenic Arabidopsis lines and was shown to exhibit high efficiency and targeted mutations of three Arabidopsis genes were detected in transgenic seedlings of the T1 generation (Xing et al. 2014). Recent work showed that CRISPR/Cas9-induced gene mutations in Arabidopsis were mostly somatic mutations in the early generation, although some mutations could be stably inherited in later generations (Zhang et al. 2014).
The Cas9/sgRNA-mediated small deletions/insertions at single cleavage sites have been reported in transient and stable transformations, although genetic transmission of edits has been reported only in Arabidopsis (Basak and Nithin 2015; Zhou et al. 2014). Auxin binding protein 1 (ABP1) functions as an auxin receptor and has an essential role in many developmental processes. Using ribozyme-based CRISPR technology to generate an Arabidopsis abp1 mutant that contains a 5-bp deletion in the first exon of ABP1, Gao et al. (2015b) identified the T-DNA insertion abp1 allele that harbors a T-DNA insertion located 27 bp downstream of the ATG start codon in the first exon and that the two new abp1 mutants are null alleles (Gao et al. 2015b). Furthermore, the abp1 plants are not resistant to exogenous auxin, and the induction of known auxin-regulated genes is similar in both wild-type and abp1 plants in response to auxin treatments, suggesting that ABP1 is not a key component in auxin signaling or Arabidopsis development.
To induce mutagenesis in proliferating tissues during embryogenesis, sgRNAs and Cas9 DNA endonuclease were expressed from the U6 snRNA and INCURVATA2 promoters to target FLOWERING LOCUS T (FT) and SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 4 genes (Hyun et al. 2015). Late-flowering plants homozygous for heritable null alleles of FT including a 1-bp insertion or short deletions were recovered in the T2 and T3 generations, demonstrating that dividing tissue-targeted mutagenesis provides an efficient heritable genome engineering method in A. thaliana (Hyun et al. 2015). It has been reported that a germinally transmitted deletion in a repeat array has been induced in Arabidopsis, and this work emphasizes the efficiency of the CRISPR/Cas system in plants, suggesting that optimal design of sgRNAs or Cas9 variants is important (Johnson et al. 2015).
To streamline and facilitate rapid and wide-scale use of CRISPR/Cas9-based technologies for plant research, a comprehensive molecular toolbox for multifaceted CRISPR/Cas9 applications was developed for Arabidopsis for multiplexed gene editing and transcriptional activation or repression of plant endogenous genes (Ali et al. 2015; Lowder et al. 2015). With this system, uniform biallelic, heterozygous, homozygous, and chimeric mutations in Arabidopsis T1 plants have been obtained, and the targeted mutations in Arabidopsis were heritable; thus, a versatile toolbox is available for studying functions of multiple genes and gene families in plants for basic research and genetic improvement (Ma et al. 2015; Sauer et al. 2015). Editing plant genomes without introducing foreign DNA into cells may alleviate regulatory concerns related to genetically modified plants. Transformed preassembled complexes of purified Cas9 protein and guide RNA into plant protoplasts of A. thaliana have achieved targeted mutagenesis in regenerated plants at frequencies of up to 46 % with the targeted sites contained germline-transmissible small insertions or deletions (Steinert et al. 2015; Woo et al. 2015).
Tobacco
The CRISPR/Cas system also served as an efficient tool to target phytoene desaturase (pds) genes in leaves of Nicotiana benthamiana (Upadhyay et al. 2013). The expression of chimeric guide RNAs (cgRNA) at two sites in the same gene resulted in deletion of DNA fragment between the targeted sequences, and target specificity analysis of the cgRNAs showed that mismatches at the 3′ end of the target site abolished the cleavage activity completely, suggesting that this approach provides a powerful method for genome engineering in N. benthamiana (Upadhyay et al. 2013). Using mesophyll protoplasts of N. benthamiana as a model cell system, targeted genome modifications and gene regulations have been generated with plant codon-optimized Cas9-mediated genome editing (Li et al. 2014). Tobacco rattle virus (TRV)-mediated CRISPR/Cas9 system has been used for targeted genome-editing application in the persistent activity and specificity in N. benthamiana. Experimental results demonstrated the persistence of TRV-mediated Cas9 activity for up to 30 d post-agroinfection and that TRV-mediated genome editing resulted in no off-target activities at potential off-targets, indicating the feasibility and exciting possibilities of using virus-mediated CRISPR/Cas9 for targeted engineering of plant genomes (Ali et al. 2015).
The CRISPR/Cas9 system has also been used for targeted mutagenesis of NtPDS and NtPDR6 in tobacco (Nicotiana tabacum) (Gao et al. 2015a; Johnson et al. 2015). Transient genome editing activity in tobacco protoplasts resulted in insertion and deletion mutations in the NtPDS locus in 16.2–20.3 % of the protoplasts after transfection with guide RNA (gRNA) and nuclease Cas9 (Gao et al. 2015a). The mutation percentage in transgenic tobacco plants with NtPDS and NtPDR6 mutations induced by Cas9/gRNA was 81.8 % for NtPDS gRNA4 and 87.5 % for NtPDR6 gRNA2 (Gao et al. 2015a). A comparative study of the CRISPR/Cas system, TALENs, and zinc-finger nucleases focused on different types of custom-designed nucleases in N. benthamiana leaves demonstrated that cleavage by CRISPR/Cas was more efficient than cleavage of the same target by TALENs. Cleavage efficiency of the S. pyogenes Cas9 protein differed significantly from both TALENs and zinc-finger nucleases (Johnson et al. 2015).
Protoplasts of N. benthamiana have been used as a model system in basic and applied plant research for targeted modification of the plant genome to elucidate and manipulate gene functions (Ali et al. 2015; Lowder et al. 2015). Dual gRNAs were designed, constructed, and evaluated for plant codon-optimized Cas9-mediated genome editing to generate targeted genome modifications in whole plants (Ali et al. 2015). A comprehensive molecular toolbox for multifaceted CRISPR/Cas9 applications in plants provides researchers with a protocol to quickly and efficiently assemble functional CRISPR/Cas9 transfer DNA constructs for plants using Golden Gate and Gateway cloning method, and the functionality and effectiveness of this toolbox in tobacco (N. benthamiana) demonstrated its utility for basic and applied plant research (Lowder et al. 2015).
CRISPR/Cas9-associated genome editing in crop plants
Rice
The type II CRISPR/Cas system has emerged as a potent new tool for targeted gene knockout in rice. A. tumefaciens was used for delivery of genes encoding Cas9 and sgRNA into rice protoplasts. Cells transformed with Cas9/sgRNA constructs targeting the promoter region of the bacterial blight susceptibility genes OsSWEET14 and OsSWEET11 were confirmed by DNA sequencing to contain mutated DNA sequences at the target sites (Jiang et al. 2013). Targeted gene mutation in rice plants via the CRISPR/Cas9 system has also been reported (Xie and Yang 2013). Three gRNAs with a 20–22-nt seed region were designed to pair with distinct rice genomic sites that are followed by the protospacer-adjacent motif. The engineered gRNAs were shown to direct the Cas9 nuclease for precise cleavage at the desired sites and introduce a mutation by nonhomologous end joining. The mutation efficiency at these target sites was 3–8 % (Xie and Yang 2013). In addition, an off-target effect of an engineered gRNA-Cas9 was found on an imperfectly paired genomic site, but it had lower genome-editing efficiency than the perfectly matched site (Xie and Yang 2013). Shan et al. (2014) established a protocol for the selection of target sites and the use of sgRNAs for sequence-specific CRISPR/Cas-mediated mutagenesis and gene targeting in rice. This CRISPR/Cas system provides a straightforward method for rapid gene targeting in protoplasts within 1–2 weeks, and mutated rice plants can be generated within 13–17 weeks (Schiml et al. 2014; Shan et al. 2014).
The CRISPR/Cas9 system was also introduced into rice for gene targeting by A. tumefaciens-mediated transformation. Three 20-nt CRISPR RNAs were designed to pair with diverse sites followed by the protospacer adjacent motif of the rice herbicide resistance gene BEL (Xu et al. 2014). After integrating the single-guide RNA (sgRNA) and Cas9 cassette in a single binary vector, transgenic rice plants harboring sgRNA:Cas9 were stably generated by A. tumefaciens-mediated transformation; the mutagenesis efficiency varied from ~2 to 16 % (Xu et al. 2014). Phenotypic analysis revealed that the biallelic-mutated transgenic plant was sensitive to bentazon, indicating that the agricultural trait could be purposely modified by sgRNA:Cas9-induced gene targeting (Xu et al. 2014). Zhang et al. (2014) tested 11 target genes in two rice subspecies for their amenability to CRISPR/Cas9-induced editing and determined the patterns, specificity and heritability of the gene modifications. They showed that the CRISPR/Cas9 system was highly efficient in rice, with target genes edited in nearly half of the transformed embryogenic cells before their first cell division. The gene mutations were passed to the next generation (T1) following classic Mendelian law, without any detectable new mutation or reversion, suggesting the CRISPR/Cas9 system is a powerful tool for crop genome engineering (Zhang et al. 2014). Zhou et al. (2014) reported large Cas9/sgRNA-induced chromosomal segment deletions in rice, inheritance of the genome edits in multiple generations, and construction of a set of facile vectors for high-efficiency, multiplex gene targeting. Four sugar efflux transporter genes were modified in rice at high efficiency; the most efficient system yielding 87–100 % editing in T0 transgenic plants, all with diallelic edits. They also demonstrated proof of efficiency of Cas9/sgRNAs in producing large chromosomal deletions (115–245 kb) involving three different clusters of genes in rice protoplasts and verified deletions in two clusters in regenerated T0 generation plants (Zhou et al. 2014).
The transcriptional response of rice RAV genes (OsRAV) to salt stress was characterized by examining the expression of all five OsRAV genes. Expression profile analyses indicated that OsRAV2 is transcriptionally regulated by salt, but not KCl, osmotic stress, cold or ABA (abscisic acid) treatment. Serial 5′ deletions and site-specific mutations in the promoter region of OsRAV2 revealed that a GT-1 element located at position -664 relative to the putative translation start site is essential for induction of the salt response. The regulatory function of the GT-1 element in the salt induction of OsRAV2 was verified in situ in plants with targeted mutations generated using the CRISPR/Cas9 system, indicating that the GT-1 element directly controls the salt response of OsRAV2 (Basak and Nithin 2015; Duan et al. 2015). Thus, a comprehensive molecular toolbox for multifaceted CRISPR/Cas9 applications in rice (Oryza sativa) now provides researchers with a protocol and reagents to quickly and efficiently assemble functional CRISPR/Cas9 transfer DNA constructs for rice using Golden Gate and Gateway cloning methods. Its full suite of capabilities includes multiplexed gene editing and transcriptional activation or repression of plant endogenous genes (Lowder et al. 2015).
Ma et al. (2015) reported a robust CRISPR/Cas9 vector system, utilizing a plant codon optimized Cas9 gene, for convenient and high-efficiency multiplex genome editing in rice. They designed PCR-based procedures to rapidly generate multiple sgRNA expression cassettes, which can be assembled into the binary CRISPR/Cas9 vectors in one round of cloning by Golden Gate ligation or Gibson Assembly. With this system, they edited 46 target sites in rice with an average 85.4 % rate of mutation, mostly in biallelic and homozygous states. They provided examples of loss-of-function gene mutations in T0 rice by simultaneous targeting multiple (up to eight) members of a gene family, multiple genes in a biosynthetic pathway, or multiple sites in a single gene (Ma et al. 2015). Frequency of CRISPR/Cas9-mediated targeted mutagenesis varies depending on Cas9 expression level and the culture period of the rice callus. Mikami et al. (2015b) examined factors affecting mutation frequency in rice calli. After sequential transformation of the calli with Cas9- and gRNA-expression constructs, the mutation frequency in independent Cas9 transgenic lines was positively correlated with the Cas9 expression level. By extending the culture period, they increased the proportion of mutated cells and the variety of mutations obtained, suggesting that a prolonged tissue culture period increases the chance of inducing de novo mutations in nonmutated cells. They also evaluated the mutation frequency in rice using different Cas9 and/or gRNA expression cassettes under standardized experimental conditions. Mutation frequencies differed significantly depending on the Cas9 expression cassette used. In addition, a gRNA driven by the OsU6 promoter was superior to one driven by the OsU3 promoter. Using an all-in-one expression vector harboring the best combined Cas9/gRNA expression cassette improved the frequency of targeted mutagenesis in rice calli, and biallelic mutant plants were produced in the T0 generation.
Rice-pathogenic bacteria Burkholderia glumae, Burkholderia gladioli, and Burkholderia plantarii, which primarily cause grain rot, sheath rot, and seedling blight, respectively, can severely reduce rice yields in all rice-growing countries, yet comprehensive studies to control the diseases are only in the early stages. Although the genome of B. plantarii ATCC 43733T has many common features with those of B. glumae and B. gladioli, this B. plantarii strain has several unique features, including quorum sensing and CRISPR/CRISPR-associated protein (Cas) systems, suggesting that B. glumae has evolved rapidly or has undergone rapid genome rearrangements or deletions in response to the hosts. Thus, this rice pathogenic Burkholderia species has unique features relative to other Burkholderia species of plants, animals and humans (Seo et al. 2015).
Wang et al. (2015) attempted to substitute a single base in situ on the rice OsEPSPS gene, which encodes enolpyruvylshikimate phosphate synthase, a key enzyme in the shikimate pathway, by co-transformation of TALEN with chimeric RNA/DNA oligonucleotides (COs), including different strand composition such as RNA/DNA (C1) or DNA/RNA (C2) but containing the same target base to be substituted. They obtained one mutant with a target base substitution but it was accompanied by an undesired deletion of 12 bases downstream of the target site from the co-transformation of TALEN and C1. Phenotypic assessments in T1 generation showed that the heterozygous plants with either one or three bases in target sequence deleted were more sensitive to glyphosate and the heterozygous d1 plants had significantly lower seed-set than the wild type did (Wang et al. 2015). They achieved targeted mutagenesis at frequencies up to 46 % in the regenerated plants. The targeted sites contained germline-transmissible small insertions or deletions that are indistinguishable from naturally occurring genetic variation (Woo et al. 2015). As noted earlier, such editing of plant genomes without introducing foreign DNA may alleviate regulatory concerns related to genetically modified rice (Wang et al. 2015).
Xie et al. (2015) developed a general strategy to produce numerous gRNAs from a single polycistronic gene. The endogenous tRNA-processing system, which precisely cleaves both ends of the tRNA precursor, was engineered as a simple, robust platform to boost the targeting and multiplex editing capability of the CRISPR/Cas9 system. They demonstrated that synthetic genes with tandemly arrayed tRNA-gRNA architecture were efficiently and precisely processed into gRNAs with thedesired 5′ targeted sequences in vivo, which directed Cas9 to edit multiple chromosomal targets. Using this strategy, multiplex genome editing and chromosomal-fragment deletion were readily achieved in stable transgenic rice plants with a high efficiency (up to 100 %) (Xie et al. 2015). Zhang et al. (2016a, b) studied the efficiency of the TALEN system in rice and the nature and inheritability of TALEN-induced mutations and described important features of this technology. The N287C230 TALEN backbone resulted in low mutation rates (0–6.6 %), but truncations in its C-terminal domain greatly increased the efficiency to 25 %. The TALEN-induced mutations were also stably transmitted to the T1 and T2 populations by normal Mendelian inheritance. Their results contrast with other published report for the CRISPR/Cas9 system in rice, in which the predominant mutations affected single bases and deletions accounted for only 3.3 % of the overall mutations (Zhang et al. 2016b).
Maize
Since a sequence-specific endonuclease was first used to induce a double-strand break (DSB) at a target locus in 1996, site-specific endonucleases, meganucleases, and the CRISPR/Cas system have been used for genome editing in maize (Liang et al. 2014; Puchta and Fauser 2013). Targeted genome modifications have become routine in crop plants using these nucleases along with newly developed techniques (Puchta and Fauser 2013). Targeted mutagenesis in Zea mays using the CRISPR/Cas system at a targeting efficiency for ZmPDS, ZmIPK1A, ZmIPK, and ZmMRP4 up to 23.1 %, and about 13.3 to 39.1 % of the transgenic plants had somatic mutations (Liang et al. 2014; Puchta and Fauser 2013). The efficiency of gRNAs targeting the ZmIPK gene in maize protoplasts was 16.4–19.1 %. In addition, the CRISPR/Cas system induced targeted mutations in 13.1 % of the Z. mays protoplasts (Liang et al. 2014).
A CRISPR/Cas9 binary vector set based on the pGreen or pCAMBIA backbone and a gRNA module vector set, have been validated as a toolkit for multiplex genome editing in plants using maize protoplasts and transgenic maize lines and demonstrated high efficiency and specificity of multiple gene mutations (Xing et al. 2014). Although the CRISPR/Cas technology has been successfully used in maize, all these studies have focused on protein-coding genes, and information on targeting non-coding genes is scarce. Several strategies to overcome the challenges of applying the CRISPR/Cas technology in editing ncRNAs in plants have been suggested (Basak and Nithin 2015).
The type II bacterial CRISPR/Cas9 system has been used to efficiently disrupt target genes in the fungal maize pathogen Ustilago maydis (Schuster et al. 2015). Transformation of a self-replicating plasmid constitutively expressing the U. maydis codon-optimized cas9 gene and a suitable sgRNA under control of the U. maydis U6 snRNA promoter was sufficient to induce genome editing and an average of 70 % of the progeny of a single transformant were disrupted within the gene, indicating the importance of this technology for the simultaneous disruption of functionally redundant genes and gene families to investigate their contribution to virulence of U. maydis (Schuster et al. 2015). DNA vectors expressing maize codon-optimized S. pyogenes Cas9 endonuclease and sgRNAs were co-introduced biolistically into immature maize embryos to target five different genomic regions in maize: upstream of the liguleless1 (LIG1) gene, male fertility genes (Ms26 and Ms45), and acetolactate synthase (ALS) genes (ALS1 and ALS2) (Svitashev et al. 2015). Mutations were subsequently identified at all sites targeted, and plants containing biallelic multiplex mutations at LIG1, Ms26, and Ms45 were recovered (Svitashev et al. 2015). Progeny showed the expected Mendelian segregation of mutations, edits, and targeted gene insertions, indicating the utility of Cas9-guide RNA technology as a plant genome editing tool to enhance plant breeding and crop research (Svitashev et al. 2015).
Wheat
CRISPR/Cas-mediated genome editing has also been used in wheat (Triticum aestivum), which has a very large, complex genome (Shan et al. 2014; Upadhyay et al. 2013). Mutations were targeted in the inositol oxygenase (inox) and phytoene desaturase (pds) genes using a cell suspension culture, and the expression of duplex cgRNA with Cas9 targeting two sites in the same gene resulted in deletion of a DNA fragment between the targeted sequences (Shan et al. 2014; Upadhyay et al. 2013). Target specificity analysis of cgRNA showed that mismatches at the 3′ end of the target site abolished the cleavage activity and the mismatches at the 5′ end reduced cleavage, suggesting that the off-target effects can be abolished in vivo by selecting target sites with unique sequences at the 3′ end (Upadhyay et al. 2013). A stepwise protocol for the selection of target sites, as well as the design, construction, verification and use of sgRNAs for sequence-specific CRISPR/Cas-mediated mutagenesis and gene targeting in wheat demonstrated that the CRISPR/Cas system provides a straightforward method for rapid gene targeting within 1-2 weeks in protoplasts (Shan et al. 2014).
The CRISPR/Cas9 system has been used to introduce targeted mutations in the three homoeoalleles that encode MILDEW-RESISTANCE LOCUS (MLO) proteins in hexaploid bread wheat (Sugano et al. 2014). Mutation of all three TaMLO homoeologs in the same plant conferred heritable broad-spectrum resistance to powdery mildew, showing the feasibility of engineering targeted DNA insertion in bread wheat through nonhomologous end joining of the double-strand breaks and providing a methodological framework to improve polyploid crops (Sugano et al. 2014; Wang et al. 2014). CRISPR loci are transcribed into ncRNA and eventually form a functional complex with Cas9 and further guide the complex to cleave complementary invading DNA. However, strategies to overcome the challenges of applying the CRISPR/Cas technology in editing ncRNAs in plants are needed (Basak and Nithin 2015).
Sorghum
The type II CRISPR/Cas system from S. pyogenes and its simplified derivative, the Cas9/single guide RNA (sgRNA) system, provide tools for targeted gene knockout in sorghum (Jiang et al. 2013; Sugano et al. 2014). A. tumefaciens was used to deliver genes encoding Cas9, sgRNA and a nonfunctional, mutant green fluorescence protein (GFP) to sorghum. The mutant GFP gene contained target sites in its 5′ coding regions that were successfully cleaved by a CAS9/sgRNA complex that, along with error-prone DNA repair, resulted in the creation of functional GFP genes. DNA sequencing confirmed Cas9/sgRNA-mediated mutagenesis at the target site. Successful application of the Cas9/sgRNA system in sorghum demonstrated that this technology is a powerful means of plant genetic engineering for scientific and agricultural applications (Jiang et al. 2013).
Soybean
The CRISPR/Cas9 system was effective for modifying nine endogenous loci in soybean by knocking-out a green fluorescent protein (GFP) transgene, and targeted DNA mutations were detected in 95 % of 88 hairy-root transgenic events analyzed (Jacobs et al. 2015). Homoeologous genes were successfully targeted singly and together, demonstrating that CRISPR/Cas9 can both selectively, and generally, target members of gene families to enable the production of plants with heritable mutations. The frequency of the DNA modifications increased with increasing culture time, suggesting that the CRISPR/Cas9 is a simple, efficient, and highly specific genome editing system for soybean. Based on an online web tool for fast identification of CRISPR/Cas9 target loci within soybean gene models and generic DNA sequences, a soybean codon-optimized CRISPR/Cas9 platform was designed to direct double-stranded breaks to the targeted loci in hairy root transformed cells, and the modified Cas9 enzyme successfully mutated target genes in somatic cells of soybean (Michno et al. 2015).
The precision gene editing tools, including the CRISPR/Cas9 system, developed in basic research will speed the development of transgenic soybeans (Ricroch and Henard-Damave 2015). In a comparative analysis of soybean genome editing targets GmPDS11 and GmPDS18 using CRISPR/Cas9, the efficiency of single gene targeting in soybean hairy roots ranged from 11.7 to 18.1 % with the AtU6-26 promoter, and 43.4 to 48.1 % with the GmU6-16 g-1 promoter, suggesting that the CRISPR/Cas9 and the GmU6-16 g-1 promoter is an efficient tool compared with the other technologies (Du et al. 2016). In a double mutation gene targeting efficiency experiment with CRISPR/Cas9 technology, a targeting efficiency of 12.5 % was achieved with the AtU6-26 promoter and 43.4 to 48.1 % with the GmU6-16 g-1 promoter; thus CRISPR/Cas9 is also a good choice for simultaneous editing of multiple homoeoalleles in soybean (Du et al. 2016).
Alfalfa
A web-based server was developed for the CRISPR/Cas9 system has been used on alfalfa (Medicago truncatula) to identify specific promoters and terminators for optimal expression of the Cas9 enzyme, promoters for expression of the CRISPR gRNA, and potential CRISPR/Cas9 target sites, including restriction enzyme sites that can facilitate the detection of new mutations (Michno et al. 2015). Michno et al. (2015) designed codon-optimized CRISPR/Cas9 platform to direct double-stranded breaks to the targeted loci in hairy root cells; the modified Cas9 enzyme successfully mutated target genes in somatic cells of M. truncatula, indicating that these new optimized tools may help facilitate targeted mutagenesis in legume and other plant species.
Potato
Defense mechanisms acquired by a CRISPR/Cas system, which enables sequence-specific targeting of foreign nucleic acids in potato, for example, is distinct from those generated by other means because the defense is heritable, (Richter et al. 2012). Pectobacterium atrosepticum, a plant pathogen that causes soft-rot and blackleg disease in potato, has been used to investigate protein–protein interactions and complex formation in the subtype I-F CRISPR/Cas system including Cas1, Cas3, and the four subtype specific proteins Csy1, Csy2, Csy3 and Cas6f (Richter et al. 2012). The results show that the subtype I-F Cas1 and Cas3 (a Cas2-Cas3 hybrid) proteins interact, suggesting the formation of a protein complex for adaptation and a role for subtype I-F Cas3 proteins in both the adaptation and interference steps of the CRISPR/Cas mechanism (Richter et al. 2012). The CRISPR/Cas system has been also used in potato (Solanum tuberosum L. group Tuberosum) for targeted mutagenesis via nonhomologous end joining (Schaart et al. 2015). In this study, CRISPR/Cas reagents expressing one of two single-guide RNA (sgRNA) targeting the potato ACETOLACTATE SYNTHASE1 (StALS1) gene were tested for inducing targeted mutations in callus and stable events of diploid and tetraploid potato using Agrobacterium-mediated transformation with either a conventional T-DNA or a modified geminivirus T-DNA (Butler et al. 2015; Schaart et al. 2015). The percentage of primary events with targeted mutations ranged from 3 to 60 % per transformation. Single targeted mutations were inherited through the germline of both diploid and tetraploid primary events with transmission percentages ranging from 87 to 100 %, demonstrating the application of CRISPR/Cas in potato and providing a framework for future studies (Butler et al. 2015). Genome-editing techniques can produce improved varieties for crops that are difficult to obtain through traditional breeding methods. The CRISPR/Cas system has been successfully used to create new disease resistancees in potato (Schaart et al. 2015).
Tomato
Although the use of homologous recombination to precisely modify plant genomes has been challenging from the lack of efficient methods to deliver DNA repair templates to plant cells, Cermak et al. (2015) used geminivirus replicons to modify the tomato genome at frequencies 10-fold higher than Agrobacterium-mediated DNA delivery. They used CRISPR/Cas9 to target a gene controlling anythocyanin biosynthesis to obtain overexpression and ectopic accumulation of anythocyanin in tissues. The modification, insertion of a strong promoter upstream of the target gene, was also carried to progeny by Mendelian inheritance. Thus, the gemini vector seems to be able to overcome the efficiency barrier that has made gene targeting in plants challenging. No extra-chromosomal replicons or off-target insertion of replicon sequences were found, so this efficient editing of the genome will be avoid the controversy of foreign DNA in genetically improved crops (Cermak et al. 2015). The CRISPR/Cas9 also holds potential for generating mlo-type resistance in tomato against powdery mildew and other pathogens (Acevedo-Garcia et al. 2014).
Cucumber
The CRISPR/Cas9 technology has been utilized to develop virus resistance in cucumber (Cucumis sativus) by disrupting the function of the recessive gene eIF4E. Cas9/sgRNA constructs were targeted to the N’ and C’ terminus of the eIF4E gene, and small deletions and SNPs were observed in the eIF4E gene targeted sites of T1 generation transformed cucumber plants, but not in putative off-target sites (Chandrasekaran et al. 2016). Nontransgenic heterozygous eIF4E mutant plants were selected for production of nontransgenic homozygous T3 generation plants. Homozygous T3 progeny after Cas9/sgRNA that had been targeted to both eIF4E sites exhibited immunity to cucumber vein yellowing virus (ipomovirus) infection and resistance to the potyviruses zucchini yellow mosaic virus and papaya ring spot mosaic virus-W (Chandrasekaran et al. 2016). In contrast, heterozygous-mutant and nonmutant plants were highly susceptible to these viruses, indicating that virus resistance was generated nontransgenically in the cucumber crop, without visibly affecting plant development or long-term backcrossing. This technology can be expected to be applicable to a wide range of crop plants (Chandrasekaran et al. 2016).
The CRISPR/Cas9-associated genome editing in fruit and woody plants
Orange
The CRISPR/Cas9 system and a synthetic sgRNA targeting the CsPDS gene were delivered into sweet orange leaves via agroinfiltration facilitated by pretreatment with Xanthomonas citri subsp. citri (Xcc) (Jia and Wang 2014a). DNA sequencing confirmed that the CsPDS gene was mutated at the target site at an efficiency of approximately 3.2–3.9 % without any off-target mutagenesis detected, suggesting targeted genome modification in citrus using the Cas9/sgRNA system-a system holds significant promise for the study of citrus gene function and for targeted genetic modification. In addition, the Xcc pretreatment before agroinfiltration significantly promoted transient protein expression in citrus leaves and in Valencia sweet orange leaves, which are recalcitrant to traditional agroinfiltration. Expression of beta-glucuronidase (GUS) expression in five citrus varieties and GFP in six citrus varieties tested was also enhanced (Jia and Wang 2014b).
Populus
When RNA-guided genome editing and targeted gene mutation via the CRISPR/Cas9 system was used to edit the genome of Populus tomentosa, four gRNAs were designed to target distinct poplar genomic sites of the phytoene desaturase gene 8 (PtoPDS) that are followed by the protospacer-adjacent motif (Fan et al. 2015; Tsai and Xue 2015). After Agrobacterium-mediated transformation, an obvious albino phenotype was observed in transgenic poplar plants. Analysis of the RNA-guided genome-editing events demonstrated that 30 of 59 PCR clones were homozygous mutants, 2 of 59 were heterozygous mutants, and the mutation efficiency at these target sites was estimated to be 51.7 % (Fan et al. 2015). The CRISPR/Cas9 system was better than previous approaches for determining functional redundancy of paralogous genes that are prevalent in plant genomes, indicating that CRISPR/Cas9 will accelerate not only basic research but also crop improvement progress (Tsai and Xue 2015).
CRISPR/Cas9-associated genome editing in liverwort
The CRISPR/Cas9 system has been applied to the liverwort M. polymorpha, a model species for studying land plant evolution, by Agrobacterium-mediated transformation for targeted mutagenesis (Sugano et al. 2014). The U6 promoter of M. polymorpha was identified and cloned to express the gRNA and the target sequence of the gRNA was designed to disrupt the gene encoding auxin response factor 1 (ARF1) in M. polymorpha (Sugano et al. 2014). CRISPR/Cas9-based site-directed mutagenesis was achieved using either the cauliflower mosaic virus 35S or M. polymorpha EF1alpha promoter to express Cas9 (Sugano et al. 2014). Stable mutants were produced by asexual reproduction of T1 plants, and multiple arf1 alleles were established using CRIPSR/Cas9-based targeted mutagenesis; thus, the CRISPR/Cas9 system is a fast, simple method for targeted genome modification and molecular genetics in M. polymorpha.
Conclusions
The CRISPR/Cas system is an extremely promising tool for genome editing in plants due to its simplicity, efficiency, minimal off-target effects, and high specificity for targeted mutagenesis. It provides a novel platform for recruitment of activation or repression domains to specific genomic loci to regulate endogenous gene expression and to identify regulatory proteins that bind with specific DNA sequences to control gene expression. The system promises antibiotic-selection-free genomic engineering with high precision in a large number of plant species and provides exciting opportunities for increasing disease and virus resistance, improving crop yield, enhancing crop quality, and increasing stress tolerance in plants.
Despite its increasing use in many plant species, the precise molecular mechanism, the length of sgRNA needed to achieve higher efficiency, off-target effects of a given sgRNA, and efficient delivery methods still need to be addressed in plants. Germline transmission and mutation heritability also need to evaluated further. Undoubtedly, the CRISPR/Cas9 system can facilitate research on genome modifications and functional genomics in plants. Ongoing efforts and future advances in this technology will accelerate both basic and applied research in improving a wide variety of agronomic traits in crop plants, fruit plants, woody plants, and grass.
References
Acevedo-Garcia J, Kusch S, Panstruga R (2014) Magical mystery tour: mLO proteins in plant immunity and beyond. New Phytol 204:273–281
Ali Z, Abul-Faraj A, Piatek M, Mahfouz MM (2015) Activity and specificity of TRV-mediated gene editing in plants. Plant Signal Behav 10:e1044191
Baltes NJ, Voytas DF (2015) Enabling plant synthetic biology through genome engineering. Trends Biotechnol 33:120–131
Basak J, Nithin C (2015) Targeting Non-Coding RNAs in plants with the CRISPR-Cas technology is a challenge yet worth accepting. Front Plant Sci 6:1001
Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V (2013) Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9:39
Belhaj K, Chaparro-Garcia A, Kamoun S, Patron NJ, Nekrasov V (2015) Editing plant genomes with CRISPR/Cas9. Curr Opin Biotechnol 32:76–84
Bogdanove AJ (2014) Principles and applications of TAL effectors for plant physiology and metabolism. Curr Opin Plant Biol 19:99–104
Bortesi L, Fischer R (2015) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv 33:41–52
Butler NM, Atkins PA, Voytas DF, Douches DS (2015) Generation and inheritance of targeted mutations in potato (Solanum tuberosum L.) Using the CRISPR/Cas system. PLoS ONE 10:e0144591
Cermak T, Baltes NJ, Cegan R, Zhang Y, Voytas DF (2015) High-frequency, precise modification of the tomato genome. Genome Biol 16:232
Chandrasegaran S, Carroll D (2015) Origins of programmable nucleases for genome engineering. J Mol Biol 33:543–548
Chandrasekaran J, Brumin M, Wolf D, Leibman D, Klap C, Pearlsman M, Sherman A, Arazi T, Gal-On A (2016) Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol Plant Pathol. doi:10.1111/mpp.12375
Chaparro-Garcia A, Kamoun S, Nekrasov V (2015) Boosting plant immunity with CRISPR/Cas. Genome Biol 16:254
Du H, Zeng X, Zhao M, Cui X, Wang Q, Yang H, Cheng H, Yu D (2016) Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J Biotechnol 217:90–97
Duan YB, Li J, Qin RY, Xu RF, Li H, Yang YC, Ma H, Li L, Wei PC, Yang JB (2015) Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Mol Biol 90(1–2):49–62
Fan D, Liu T, Li C, Jiao B, Li S, Hou Y, Luo K (2015) Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the first generation. Sci Rep 5:12217
Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL, Wang Z, Zhang Z, Zheng R, Yang L, Zeng L, Liu X, Zhu JK (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci U S A 111:4632–4637
Fichtner F, Urrea Castellanos R, Ulker B (2014) Precision genetic modifications: a new era in molecular biology and crop improvement. Planta 239:921–939
Frampton RA, Pitman AR, Fineran PC (2012) Advances in bacteriophage-mediated control of plant pathogens. Int J Microbiol 2012:326452
Gao J, Wang G, Ma S, Xie X, Wu X, Zhang X, Wu Y, Zhao P, Xia Q (2015a) CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol Biol 87:99–110
Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y (2015b) Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc Natl Acad Sci USA 112:2275–2280
Hyun Y, Kim J, Cho SW, Choi Y, Kim JS, Coupland G (2015) Site-directed mutagenesis in Arabidopsis thaliana using dividing tissue-targeted RGEN of the CRISPR/Cas system to generate heritable null alleles. Planta 241:271–284
Ilardi V, Tavazza M (2015) Biotechnological strategies and tools for Plum pox virus resistance: trans-, intra-, cis-genesis, and beyond. Front Plant Sci 6:379
Jacobs TB, LaFayette PR, Schmitz RJ, Parrott WA (2015) Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol 15:16
Jameson PE, Song J (2015) Cytokinin: a key driver of seed yield. J Exp Bot 67:593–606
Jia H, Wang N (2014a) Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS ONE 9:e93806
Jia H, Wang N (2014b) Xcc-facilitated agroinfiltration of citrus leaves: a tool for rapid functional analysis of transgenes in citrus leaves. Plant Cell Rep 33:1993–2001
Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 41:e188
Jiang W, Yang B, Weeks DP (2014) Efficient CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana and inheritance of modified genes in the T2 and T3 generations. PLoS ONE 9:e99225
Johnson RA, Gurevich V, Filler S, Samach A, Levy AA (2015) Comparative assessments of CRISPR-Cas nucleases’ cleavage efficiency in planta. Plant Mol Biol 87:143–156
Kumar V, Jain M (2015) The CRISPR-Cas system for plant genome editing: advances and opportunities. J Exp Bot 66:47–57
Li JF, Zhang D, Sheen J (2014) Cas9-based genome editing in Arabidopsis and tobacco. Methods Enzymol 546:459–472
Liang Z, Zhang K, Chen K, Gao C (2014) Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J Genet Genomics 41:63–68
Lowder LG, Zhang D, Baltes NJ, Paul JW 3rd, Tang X, Zheng X, Voytas DF, Hsieh TF, Zhang Y, Qi Y (2015) A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol 169:971–985
Lozano-Juste J, Cutler SR (2014) Plant genome engineering in full bloom. Trends Plant Sci 19:284–287
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu YG (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8:1274–1284
Mahfouz MM, Piatek A, Stewart CN Jr (2014) Genome engineering via TALENs and CRISPR/Cas9 systems: challenges and perspectives. Plant Biotechnol J 12:1006–1014
Michno JM, Wang X, Liu J, Curtin SJ, Kono TJ, Stupar RM (2015) CRISPR/Cas mutagenesis of soybean and Medicago truncatula using a new web-tool and a modified Cas9 enzyme. GM Crops Food 6:243–252
Mikami M, Toki S, Endo M (2015a) Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol Biol 88:561–572
Mikami M, Toki S, Endo M (2015b) Parameters affecting frequency of CRISPR/Cas9 mediated targeted mutagenesis in rice. Plant Cell Rep 34:1807–1815
Nagamangala Kanchiswamy C, Sargent DJ, Velasco R, Maffei ME, Malnoy M (2015) Looking forward to genetically edited fruit crops. Trends Biotechnol 33:62–64
Nejat N, Rookes J, Mantri NL, Cahill DM (2016) Plant-pathogen interactions: toward development of next-generation disease-resistant plants. Crit Rev Biotechnol. doi:10.3109/07388551.2015.1134437
Nekrasov V, Staskawicz B, Weigel D, Jones JD, Kamoun S (2013) Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol 31:691–693
Nemudryi AA, Valetdinova KR, Medvedev SP, Zakian SM (2014) TALEN and CRISPR/Cas genome editing systems: tools of discovery. Acta Naturae 6:19–40
Osakabe Y, Osakabe K (2015) Genome editing with engineered nucleases in plants. Plant Cell Physiol 56:389–400
Puchta H, Fauser F (2013) Gene targeting in plants: 25 years later. Int J Dev Biol 57:629–637
Quetier F (2016) The CRISPR-Cas9 technology: closer to the ultimate toolkit for targeted genome editing. Plant Sci 242:65–76
Richter C, Gristwood T, Clulow JS, Fineran PC (2012) In vivo protein interactions and complex formation in the Pectobacterium atrosepticum subtype I-F CRISPR/Cas system. PLoS ONE 7:e49549
Ricroch AE, Henard-Damave MC (2015) Next biotech plants: new traits, crops, developers and technologies for addressing global challenges. Crit Rev Biotechnol 35:1–16
Sauer NJ, Mozoruk J, Miller RB, Warburg ZJ, Walker KA, Beetham PR, Schopke CR, Gocal GF (2015) Oligonucleotide-directed mutagenesis for precision gene editing. Plant Biotechnol J 14(2):448–462
Schaart JG, van de Wiel CC, Lotz LA, Smulders MJ (2015) Opportunities for products of new plant breeding techniques. Trends Plant Sci 21(5):438–449
Schiml S, Fauser F, Puchta H (2014) The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J 80:1139–1150
Schuster M, Schweizer G, Reissmann S, Kahmann R (2015) Genome editing in ustilago maydis using the crispr-cas system. Fungal Genet Biol 89:3–9
Scott JN, Kupinski AP, Boyes J (2014) Targeted genome regulation and modification using transcription activator-like effectors. FEBS J 281:4583–4597
Seo YS, Lim JY, Park J, Kim S, Lee HH, Cheong H, Kim SM, Moon JS, Hwang I (2015) Comparative genome analysis of rice-pathogenic Burkholderia provides insight into capacity to adapt to different environments and hosts. BMC Genom 16:349
Shan Q, Wang Y, Li J, Gao C (2014) Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc 9:2395–2410
Shariat N, Dudley EG (2014) CRISPRs: molecular signatures used for pathogen subtyping. Appl Environ Microbiol 80:430–439
Steinert J, Schiml S, Fauser F, Puchta H (2015) Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J 84:1295–1305
Sugano SS, Shirakawa M, Takagi J, Matsuda Y, Shimada T, Hara-Nishimura I, Kohchi T (2014) CRISPR/Cas9-mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol 55:475–481
Svitashev S, Young JK, Schwartz C, Gao H, Falco SC, Cigan AM (2015) Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and Guide RNA. Plant Physiol 169:931–945
Teotia S, Singh D, Tang X, Tang G (2016) Essential RNA-based technologies and their applications in plant functional genomics. Trends Biotechnol 34:106–123
Trevino AE, Zhang F (2014) Genome editing using Cas9 nickases. Methods Enzymol 546:161–174
Tsai CJ, Xue LJ (2015) CRISPRing into the woods. GM Crops Food 6:206–215
Upadhyay SK, Kumar J, Alok A, Tuli R (2013) RNA-guided genome editing for target gene mutations in wheat. G3 (Bethesda) 3(12):2233–2238
Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32:947–951
Wang M, Liu Y, Zhang C, Liu J, Liu X, Wang L, Wang W, Chen H, Wei C, Ye X, Li X, Tu J (2015) Gene editing by co-transformation of TALEN and chimeric RNA/DNA oligonucleotides on the rice OsEPSPS gene and the inheritance of mutations. PLoS ONE 10:e0122755
Weeks DP, Spalding MH, Yang B (2015) Use of designer nucleases for targeted gene and genome editing in plants. Plant Biotechnol J 14(2):483–495
Woo JW, Kim J, Kwon SI, Corvalan C, Cho SW, Kim H, Kim SG, Kim ST, Choe S, Kim JS (2015) DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33:1162–1164
Xie K, Yang Y (2013) RNA-guided genome editing in plants using a CRISPR-Cas system. Mol Plant 6:1975–1983
Xie K, Minkenberg B, Yang Y (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA 112:3570–3575
Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14:327
Xu R, Li H, Qin R, Wang L, Li L, Wei P, Yang J (2014) Gene targeting using the Agrobacterium tumefaciens-mediated CRISPR-Cas system in rice. Rice (N Y) 7:5
Yin K, Han T, Liu G, Chen T, Wang Y, Yu AY, Liu Y (2015) A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci Rep 5:14926
Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N, Zhu JK (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 12:797–807
Zhang B, Yang X, Yang C, Li M, Guo Y (2016a) Exploiting the CRISPR/Cas9 system for targeted genome mutagenesis in petunia. Sci Rep 6:20315
Zhang H, Gou F, Zhang J, Liu W, Li Q, Mao Y, Botella JR, Zhu JK (2016b) TALEN-mediated targeted mutagenesis produces a large variety of heritable mutations in rice. Plant Biotechnol J 14:186–194
Zhou H, Liu B, Weeks DP, Spalding MH, Yang B (2014) Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res 42:10903–10914
Zlotorynski E (2015) Plant cell biology: cRISPR-Cas protection from plant viruses. Nat Rev Mol Cell Biol 16:642
Acknowledgments
The authors are grateful to Dr. Luo, Dr. Whitley, Dr. Lauressergues, Dr. Omidbakhshfard, and Dr. Page for their critical reading and valuable suggestions during the preparation of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
The online version is available at http://www.springerlink.com
Corresponding editor: Chai Ruihai
Rights and permissions
About this article
Cite this article
Tang, W., Tang, A.Y. Applications and roles of the CRISPR system in genome editing of plants. J. For. Res. 28, 15–28 (2017). https://doi.org/10.1007/s11676-016-0281-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11676-016-0281-7