Abstract
Common bottlenose dolphin (Tursiops truncatus) is a well-known cetacean species that inhabits temperate and tropical seas worldwide. Limited supply and poor quality of samples hinder the investigation of the effects of various pathogens and environmental pollutants on this cetacean species. Cultured cells are useful for experimental studies; however, no cell lines derived from cetaceans are generally available. Therefore, in this study, we established a novel kidney cell line, TK-ST, derived from T. truncatus. Primary cells exhibited the morphological characteristics of epithelial and fibroblast cells, but their immortalization and passaging resulted in a predominantly epithelial cell morphology. TK-ST was immortalized using the large T SV40 antigen and human telomerase reverse transcriptase and exhibited long-term stable cell growth. TK-ST cells are generally cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum at 37°C and 5% CO2 but can also be cultured in 5–20% fetal bovine serum and several other classical media commonly used for common animal cell culture. TK-ST cells were found to be susceptible to several viruses, including the dolphin morbillivirus (most important virus in cetaceans), and exhibited cytopathic effects, facilitating the replication of the dolphin morbillivirus. Furthermore, mRNA expression levels of cytokine genes were increased in TK-ST cells after stimulation with lipopolysaccharides and poly(I:C). Therefore, the novel TK-ST cell line derived in this study can potentially be used for further in vitro studies on cetaceans.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Common bottlenose dolphin (Tursiops truncatus; Cetacea, Delphinidae) is one of the most popular cetacean species, inhabiting temperate and tropical sea worldwide. They are long-term coastal residents and apex predators who are affected by human activities. Common bottlenose dolphins are considered useful sentinel species for monitoring marine ecosystems (Wells et al. 2004; Bossart 2011). Various pathogens, such as the Morbillivirus members, and environmental pollutants, such as pollutants of concern, adversely affect this species. Therefore, it is essential to understand their physiological status and pathological mechanisms to accurately assess the effects of external factors on this species. Although biological samples are necessary for these investigations, it is difficult to obtain suitable samples from cetaceans for various reasons, such as ethical constraints and their marine ecology. Moreover, the samples from stranded cetacean carcasses may be contaminated or poorly preserved, and hence, not suitable for analysis.
In vitro experiments using cultured cells can overcome these problems in cetacean research. However, the difficulty in obtaining fresh tissue limits the production of cultured cells from cetaceans. Although a few studies, including ours, have obtained primary cultured cells and cells derived from cetaceans that can be passaged for a long period, they do not exhibit complete immortalization (Carvan et al. 1994; Yu et al. 2005; Jin et al. 2013; Burkard et al. 2015; Suzuki et al. 2016; Yajing et al. 2018). To date, no cetacean-derived cell lines are available, even in global bioresource centers, such as the American Type Culture Collection.
In this study, we aimed to establish and characterize a novel cell line derived from the common bottlenose dolphin for application in cetacean research. Our established cell line, TK-ST, will aid in future cetacean research. Moreover, our findings can be used to generate cell lines for the conservation of genetic resources from endangered whale species.
Materials and Methods
Ethics Statement
A special note is required regarding any usage of killed animals in this study. Bottlenose dolphins off Wakayama are hunt by regional fishermen with the permission of the Wakayama Prefectural Government (Wakayama, Japan) and under the supervision of the Fisheries Agency of Japan (Tokyo, Japan). We collected samples and data from these catches with the cooperation of the biological surveys of the Fisheries Resources Institute (Kanagawa, Japan).
Materials
All media, phosphate-buffered saline (PBS), and antibodies for cell culture were purchased from FUJIFILM WAKO Pure Chemical (Osaka, Japan), unless otherwise stated. Cell culture flasks and multiwell plates were purchased from Violamo (As One Corporation, Osaka, Japan) or Greiner Bio-One (Frickenhausen, Germany), respectively. Fetal bovine serum (FBS) was obtained from Biosera (Nuaillé, France) or Biowest (Nuaillé, France).
Kidney Tissue Collection
Female bottlenose dolphin (T. truncatus; sample ID. 19TK036) was commercially hunted off the coast of Wakayama with the permission of the Wakayama Prefectural Government and with the cooperation of the Fisheries Agency of Japan in January 2019. Kidney tissue samples were collected immediately post-mortem from the dolphin, preserved in clean bags, and transported to our laboratory on ice (4°C) for approximately 12 h.
Primary Cell Culture and Subculture
Kidney tissue was aseptically cut into 1-mm3 sections with sterile surgical scissors and washed with PBS containing 200 units/mL penicillin, 200 µg/mL streptomycin, and 0.5 μg/mL amphotericin B. Then, kidney tissues were resuspended in 3 mL fresh DMEM (high-glucose; #044–29,765) with 10% FBS, 100 units/mL penicillin, and 100 µg/mL streptomycin. Tissue suspension was seeded into a 100-mm cell culture dish (Greiner Bio-One) and incubated at 37°C and 5% CO2. After 1 h, 13 mL of fresh medium was gently added to the culture medium. After a cell monolayer was formed in 1 week, the cells were rinsed twice with PBS and detached via trypsinization with 0.05% trypsin–EDTA solution. It was difficult to trypsinize the cells strongly adhered to the flask. Detached cells were resuspended in fresh DMEM with 10% FBS, 100 units/mL penicillin, and 100 µg/mL streptomycin and subcultured.
Cell Immortalization
Bottlenose dolphin kidney primary cells were cryopreserved, transported to Applied Biological Materials, Inc. (Richmond, BC, Canada), and immortalized via transduction with Lenti-SV40T lentivirus (#LV613; Applied Biological Materials, Inc.) and Lenti-human telomerase reverse transcriptase (hTERT) lentivirus (#LV615; Applied Biological Materials, Inc.). Briefly, cells were seeded in a 6-well plate, cultured for 24 h, infected with the two lentiviruses at a multiplicity of infection (MOI) of 5, and incubated for 72 h at 37 °C and 5% CO2. Transfected cells were cultured in the medium and continuously passaged. The resultant immortalized bottlenose dolphin kidney cell line was named the TK-ST cell line.
Growth Curve and Population Doubling Level
We calculated the population doubling level (PDL) to determine the long-term stable growth period for TK-ST cells. PDL refers to the total number of times the number of cells in the population doubled during culture. PDL was calculated using the formula: PDL = log2 (number of current cells/number of seeded cells) + PDL of the seeding culture. TK-ST cells were counted manually using a hemocytometer or an automated cell counter (Countess 3; Thermo Fisher Scientific, Waltham, MA) after trypan blue staining in successive subcultures. Subculture was performed at a 1:2 ratio, and a growth curve was generated based on the number of cells.
Sequence Analysis of Cytochrome b
We analyzed the partial sequence of cytochrome b (Cytb) to identify the origin of the species and determine the presence of any cross-contamination. DNA was extracted from TK-ST cells using NucleoSpin Tissue (Macherey–Nagel, Düren, Germany), according to the manufacturer’s instructions. Primer sequences are listed in Table 1. Bottlenose dolphin positive control used DNA derived from the blood of captive individuals in the Shinagawa Aquarium (Tokyo, Japan). Then, polymerase chain reaction (PCR) was performed using Blend Taq Plus (Toyobo, Osaka, Japan). PCR protocol was as previously described (Viricel and Rosel 2012): 95 °C for 30 s, followed by 35 cycles of 30 s at 94°C, 30 s at 45°C, 30 s at 72°C, and a final extension at 72°C for 7 min. PCR products were analyzed via 2% agarose gel electrophoresis, purified using the NucleoSpin Gel and PCR Clean-up Kit (Macherey–Nagel), and directly sequenced using an ABI PRISM 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA) with a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). The obtained sequences were compared with known sequences in the National Center for Biotechnology Information (NCBI) database using the Basic Local Align Search Tool (BLAST) (https://blast.ncbi.nlm.nih.gov).
Mycoplasma Detection Test
PCR was used to detect any Mycoplasma contamination in TK-ST cells. Briefly, cells were cultured in DMEM with 10% FBS without antibiotics for a week and the conditioned medium was collected for the test. DNA extraction and PCR were performed using the same reagents as those used in the Cytb experiment, according to the manufacturer’s instructions. Previously reported primer sequences (Nagasawa et al. 1992; Harasawa et al. 1993) (Table 1) were used to detect the Mycoplasma species commonly infecting cell lines: Mycoplasma arginini, M. fermentans, M. hominis, M. hyorhinis, M. orale, M. prium, and M. salivarum (Ayral et al. 2006). M. orale was used as a positive control. It was previously isolated from another cell line in our laboratory and was identified using 16S ribosomal RNA gene sequencing. PCR protocol consisted of an initial denaturation step at 94°C for 2 min, followed by 35 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s.
Growth Studies
To determine their growth characteristics, we cultured TK-ST cells in various medium compositions.
Serum Concentrations
TK-ST cells were suspended in DMEM (high-glucose) containing 0, 2.5, 5, 10, and 20% FBS (Biowest). Cells were seeded in a 6-well plate at a density of 1 × 105 cells/well. The plates were incubated at 37°C and 5% CO2, and observed for 8 d. Every 48 h post-subculture, the number of viable cells was counted using a Countess 3 Automated Cell Counter (Thermo Fisher Scientific). Three replicates were used for each group. Results are expressed as the mean (standard deviation [SD]) of replicates.
Classical Cell Culture Media
We determined the changes in TK-ST cell growth based on differences in the classical cell culture media. Six classical cell culture media were selected for this study: DMEM (high-glucose, with sodium pyruvate; #043–30,085), DMEM (high-glucose; #044–29,765), DMEM (low-glucose, with sodium pyruvate; #041–29,775), Eagle’s minimum essential medium (EMEM; #051–07,615), Roswell Park Memorial Institute (RPMI)-1640 medium (#189–02,025), and Ham’s F-12 medium (#087–08,335). TK-ST cells were cultured in a medium containing 10% FBS for eight days. Every 48 h post-subculture, the number of viable cells was counted using the same method as that used for the study of serum concentrations.
Gene Expression Analysis
Gene expression in TK-ST cells were determined using reverse transcription (RT)-PCR. Total RNA from TK-ST was extracted using NucleoSpin RNA (Macherey–Nagel), according to the manufacturer’s instructions. The quantity and purity (A260/A280 > 1.6) of total RNA were assessed using a spectrophotometer (NanoDrop One; Thermo Fisher Scientific). Total RNA (680 ng) was reverse transcribed using a high-capacity RNA-to-cDNA kit (Applied Biosystems). PCR was performed using Blend Taq Plus. PCR protocol was as follows: an initial denaturation step at 94°C for 2 min, followed by 35–40 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s. PCR amplicons were analyzed using 2% agarose gel electrophoresis. Expression of β-actin was used as a control for each cDNA sample.
Cell-Type Maker Gene Expression
Kidneys have many cell types. To characterize TK-ST cells and assess their composition, the expression profiles of cell-type marker genes were analyzed via RT-PCR. Targets of the cell-type marker genes were as follows: fibroblast cell marker, S100 calcium-binding protein A4 (S100A4); epithelial cell marker, cytokeratin18 (KRT18); myofibroblast cell marker, α-smooth muscle actin (α-SMA); mesenchymal cell marker, vimentin (VIM); and skeletal muscle cell marker, desmin (DES). All PCR primer sequences used in this study are listed in Table 1.
Virus Susceptibility Analysis
To analyze the viral susceptibility of TK-ST cells, we observed the cytopathic effect (CPE) of the cell line and evaluated the increase in the virus genome. Vero cells were used as controls. Vero cells were obtained from the RIKEN BRC Cell Bank (RCB0001) and cultured in EMEM (#051–07615) with 10% FBS.
CPE
Pseudorabies virus (PRV; YS-81 strain; Aujeszky’s disease virus) and Japanese encephalitis virus (JEV; AS-6 strain) kindly provided by the Livestock Hygiene Service Center (Kanagawa, Japan) and the National Institute of Animal Health, National Agriculture and Food Research Organization (Ibaraki, Japan), respectively, were used for CPE test. TK-ST and Vero cells were seeded in a 6-well plate at a density of 3.0 × 105 cells per well and cultured for approximately 24 h at 37°C and 5% CO2. Then, cells were infected at an MOI of 0.1. The virus was adsorbed for an hour, the virus inoculum was removed, and cells were washed twice with PBS. Next, 2 mL fresh growth medium was added to each well. Infected cells were observed at 0, 12, 24, and 48 h post-infection (hpi).
Susceptibility to Dolphin Morbillivirus
Dolphin morbillivirus (DMV) is the most important pathogen causing fatal infections in wild cetaceans. Replication of DMV (Muc strain) in TK-ST cells was analyzed via RT-qPCR to quantify the viral genome expression levels. DMV was kindly provided by Dr. Makoto Sugiyama (Gifu University, Gifu, Japan). Cells were seeded in a 6-well plate at a density of 3 × 105 cells/well, incubated for 24 h at 37°C, infected with 100 µL/well of DMV, and the virus was adsorbed for an hour. Inoculum was removed and the cells were washed twice with PBS. Infected cells were cultured in 2 mL fresh growth medium and 0, 1, 3, 5, and 7 d after DMV infection, the cells were freeze-thawed thrice. Suspension was then centrifuged at 2300 × g for 10 min, and the supernatant was collected for viral samples. Viral RNA was extracted using a QIAamp Viral RNA Mini kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions, and reverse transcription was performed in a manner similar to RT-PCR. PCR primer sequences used in this study are listed in Table 1. qPCR was performed using TB Green Premix Ex Taq II (Takara, Shiga, Japan) with a Thermal Cycler Dice Real-Time System III (Takara). The qPCR program was as follows: 95°C for 30 s, 40 cycles of 95°C for 5 s, and 60°C for 30 s, and dissociation curve analysis by gradual heating at 0.5°C intervals from 60 to 95°C. The standard template was constructed by Eurofins Genomics (standard gene synthesis service; Eurofins Genomics, Ebersberg, Germany) using synthetic DNA for absolute quantification of the DMV genome. Synthetic DNA standard (188 bp) was designed based on the amplified sequence (178 bp), and five bases were added to both ends of the primer-binding sites. qPCR standard was diluted by 10 to 106 copies/µL and used for absolute quantification. Signaling lymphocyte activation molecule (SLAM) and nectin cell adhesion molecule 4 (Nectin4) act as receptors for DMV in immune and epithelial cells, respectively (Tatsuo et al. 2000; Mühlebach et al. 2011; Noyce et al. 2011). RT-PCR was used to analyze the expression levels of these receptors in TK-ST cells.
Immune Responses to Viral and Bacterial Challenges
To assess the utility of TK-ST cells in immunological studies, we evaluated their immune responses. Toll-like receptors (TLRs) play important roles in the recognition of microbial pathogens during the innate immune response (Iwasaki and Medzhitov 2004; Takeda and Akira 2005; Kawasaki and Kawai 2014). TLR3 recognizes double-stranded RNA (dsRNA) synthesized by viruses, whereas TLR4 recognizes bacterial lipopolysaccharide (LPS) (Poltorak et al. 1998; Alexopoulou et al. 2001). We determined the expression of TLR3 and TLR4 in TK-ST cells using RT-PCR. Next, we determined the gene expression levels of cytokines and cyclooxygenase-2 (COX-2) in TK-ST cells stimulated with TLR3 and TLR4 agonists using RT-PCR and qPCR. TK-ST cells were seeded in a 12-well plate at a density of 4 × 104 cells per well and cultured for approximately 48 h at 37°C and 5% CO2. Cells were then stimulated with 50 μg of poly(I:C) (Tocris Bioscience, Bristol, England) in 1 mL medium or 50 μg LPS (#L3755; Sigma-Aldrich, St. Louis, MO) in 1 mL medium. Poly(I:C) is a synthetic dsRNA. Cells were stimulated only once at first and not washed. Cells were incubated for 0, 3, 6, 12, and 24 h at 37°C and 5% CO2. Then the samples were collected for subsequent analysis. RT-PCR and qPCR were performed as previously described. β-Actin was used as a housekeeping gene and its expression was measured in each sample as a positive control. Relative quantification was performed using the ΔΔCT method with the Thermal Cycler Dice Real Time System III Software version 6.01C (Takara). Three biological replicates were prepared for each condition, and the data are expressed as the mean (SD). Statistical analyses were conducted using the Mann–Whitney U test in each simulation vs. paired controls with Bell Curve for Excel (Social Survey Research Information Co., Ltd. Tokyo, Japan). All primer sequences used in this study are listed in Table 1.
Results
Establishment of the TK-ST Cell Line
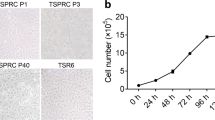
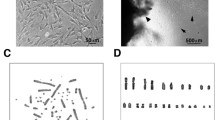
Primary cells were obtained by culturing the kidney tissues of the common bottlenose dolphin in DMEM (high-glucose) with 10% FBS. Radial growth was observed in the tissue pieces. Most cells exhibited an epithelial-like morphology, and some cells exhibited a fibroblast-like morphology (Fig. 1A, B). After reaching 90% confluency, the cells were subcultured in a fresh medium. In the second passage, epithelial-like and fibroblast-like cells appeared to be mixed (Fig. 1C). The fibroblast-like morphology increased as the cells became more confluent. Primary kidney cells stopped proliferating after approximately eight passages. We transfected SV40T and hTERT into kidney primary cells using a lentivirus vector and named this immortalized cell line as TK-ST cell line. The cells exhibited an epithelial-like morphology after immortalization and repeated passaging (Fig. 1D).
Phase contrast photomicrographs of cell cultures derived from the kidney tissues of the common bottlenose dolphin. (A, B) Primary culture on day 10. (C) Confluent monolayer of cells at the second passage. (D) Confluent monolayer of TK-ST cells at the 74th passage. Scale bar = 100 µm. *Kidney tissue piece.
To confirm the long-term proliferative ability of TK-ST cells, we calculated their PDL after immortalization. The total number of times the cells in the population doubled was more than 200 (Fig. 2). After 500 d of culturing, TK-ST cells continued to grow steadily.
PCR protocols targeting mitochondrial DNA are widely used for species identification in animal cells (Parodi et al. 2002; Cooper et al. 2007; Ono et al. 2007). In this study, the partial regions of Cytb gene were used for analysis. Amplified fragment of partial Cytb gene (704 bp) from TK-ST cells was obtained and sequenced (Fig. 3A, B). The obtained sequence was submitted to NCBI BLAST, which revealed 100% similarity to the T. truncatus Cytb gene (MW183360.1). This result indicates that the TK-ST cell line originates from T. truncatus.
Detection of Mycoplasma
PCR was used to detect Mycoplasma contamination in TK-ST cells. The tested sample showed no bands, whereas the Mycoplasma-positive control showed a single band (Fig. 4).
Growth Characteristics
To determine the optimal medium for TK-ST growth, the cells were cultured for 8 days in various medium compositions. Growth rate of TK-ST cells depended on the concentration of FBS (Fig. 5A). When cultured for 8 d with 20% FBS, TK-ST cells showed the maximum growth rate, and the number of cells was approximately 75.8-fold higher than the number of seeded cells. TK-ST cells also exhibited stable growth at FBS concentrations of 10% (approximately 53.9-fold cell growth) and 5% (approximately 22.9-fold cell growth), which are mainly used for mammalian cell cultures. Cells showed slight growth (approximately 6.9-fold cell growth) with 2.5% FBS. In a serum-free medium, TK-ST cells did not show any growth, and most cells died within a few days of culture.
Growth characteristics of TK-ST cells. (A) Growth of TK-ST cells in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) with different concentrations of fetal bovine serum (FBS; 0, 2.5, 5, 10, and 20%). (B) Growth of TK-ST cells in a six classical cell culture media with 10% FBS. Results are represented as the mean (standard deviation [SD]) of three replicates.
TK-ST growth in six different classical media for eight days is shown in Fig. 5B. Cells showed relatively good growth in the three types of DMEM and RPMI-1640 medium (approximately 57.6-fold cell growth), and the differences in cell numbers after 8 d of culture among these four media were almost the same, up to 1.2-fold. EMEM resulted in a cell growth of approximately 42.4-fold after 8 days of culture. Cells in Ham’s F-12 medium exhibited the lowest growth rate (approximately 21.7-fold).
Based on these results and costs, we selected DMEM with 10% concentration of FBS for subsequent experiments as its medium composition resulted in adequate cell growth.
Cell-Type Marker Gene Expression Levels in TK-ST Cells
To characterize TK-ST cells, expression levels of cell-type marker genes were determined via RT-PCR. TK-ST cell profiles were compared with those of kidney tissue and primary cells. Except for DES, all marker genes were similarly expressed in all samples (Fig. 6). DES, a skeletal muscle cell marker, was expressed in the kidney tissue and primary cells but not in TK-ST cells.
Virus Susceptibility and CPE
To analyze their viral susceptibility, TK-ST cells were infected with PRV and JEV at an MOI of 0.1, and CPEs were observed for 3 d. TK-ST cells exhibited CPEs with PRV and JEV infections, indicating that TK-ST cells are susceptible to these viruses (Fig. 7A). CPEs of PRV were clearly observed in TK-ST cells 24 hpi, and most cells detached by 48 hpi. CPEs of JEV were not as clear as those of PRV but were observed in the cells 48 hpi. The virus-uninfected negative control, MOCK, did not exhibit any CPEs throughout the experiment. Vero cells were used as a positive control as they are susceptible to PRV and JEV. Vero cells were also infected with PRV and JEV at an MOI of 0.1 and exhibited CPEs (Fig. 7B). CPEs of TK-ST cells are similar to those of Vero cells. The time of CPE appearance in TK-ST cells was almost the same as in Vero cells for PRV but later than that in Vero cells for JEV.
Cytopathic effects (CPEs) caused by the pseudorabies virus (PRV) and Japanese encephalitis virus (JEV). (A) CPEs observed in TK-ST cells infected with PRV and JEV 24, 48, and 72 h post-infection (hpi). MOCK did not exhibit any CPEs throughout the experiment. (B) CPEs in Vero cells infected with PRV (24 hpi) and JEV (72 hpi). Scale bar = 25 µm.
DMV Susceptibility of TK-ST Cells
DMV replication in TK-ST cells was analyzed using RT-qPCR. After 7 days of culture, the number of DMV genome copies increased by more than 2000-fold (Fig. 8A), but the CPEs of DMV were not observed throughout the experiment. Low expression of SLAM was observed in the kidney tissue, but not in TK-ST cells (Fig. 8B). Nectin4 expression was observed in all samples.
Dolphin morbillivirus (DMV) replication and expression of its receptors in TK-ST cells. (A) Data represent log10 genome copies per microliter. DMV genome copy number increased after the incubation of TK-ST cells for 7 d. Each point represents the mean (SD) of three independent experiments. (B) Expression of the signaling lymphocyte activation molecule (SLAM) was not detected in TK-ST cells but that of nectin cell adhesion molecule 4 (Nectin4) was observed in all samples.
Immune Responses in TK-ST Cells
Expression of both TLR3 and TLR4 was detected in all samples, including TK-ST cells (Fig. 9A). These receptors were stimulated by an agonist, LPS, and poly(I:C), for 0–24 h, and the expression levels of cytokines and COX-2 were determined via RT-PCR and qPCR (Fig. 9B, C).
Expression levels of immune-related genes in TK-ST cells. (A) Expression levels of toll-like receptors in TK-ST cells. Both TLR3 and TLR4 were expressed in all samples. (B) Expression levels of cytokines and cyclooxygenase-2 (COX-2) in TK-ST cells 0–24 h after stimulation with poly(I:C) or lipopolysaccharides (LPSs). β-actin was used as a control for the amount and the quality of cDNA. (C) qPCR analysis of the mRNA levels of cytokines and COX-2 in TK-ST cells 0–24 h after stimulation with poly(I:C) or LPS. Data are represented as the mean (SD) of three independent experiments. β-Actin was used as a control and housekeeping gene for analysis. * indicates statistically significant differences vs. control via Mann–Whitney’s U test (p < 0.05).
Interferon (IFN)-α and transforming growth factor (TGF)-β were highly expressed in the control cells, and their expression levels were almost the same before and after stimulation. Although IFN-α and TGF-β showed statistically significant differences compared to the control in some cases, they showed little fold changes and no biological differences after LPS and poly(I:C) stimulation (lower than twofold change). Expression levels of TNF-α and COX-2 were low in the control, but they showed statistically and biologically significant differences with the control after stimulation with LPS and poly(I:C) (more than tenfold change). Quantitative analysis of IFN-β and IFN-γ levels was not possible because their expression was not detected in the control or immediately after immunostimulation. However, their expression was detected via RT-PCR 3 h after poly (I:C) stimulation and 12 h after LPS stimulation.
Discussion
In this study, we established and characterized a novel cell line, TK-ST, derived from the common bottlenose dolphin (T. truncatus). Although several studies have attempted to establish cell lines in cetaceans, TK-ST cell line is superior to previously reported cetacean cell lines in terms of stable long-term growth and ease of in vitro culture.
Primary cells of the kidneys of the bottlenose dolphin were cultured in explant culture in DMEM with 10% FBS. Cells were cultured for eight passages but they stopped growing. Several methods are used to generate immortalized animal cells. Abrogation of p53 by the introduction of viral genes, such as SV40, is a popular method generate immortalized cells (Jha et al. 1998). Transduction of hTERT gene is also used for cell immortalization (Bodnar et al. 1998). In this study, we immortalized primary kidney cells by introducing SV40T and hTERT using lentiviral vectors. Stable and long-term growth of TK-ST cells after immortalization is shown by the growth curve based on the PDL. To date, the cells have maintained their growth rate (February, 2023), even after more than 200 cell divisions. This cell line is considered to have almost infinite growth potential.
Mycoplasma contamination is a major problem in cell culture. It causes various problems, such as cell death, and mycoplasmas are resistant to antibiotics commonly used in cell culture (Drexler and Uphoff 2002). These two methods are mainly used for the detection of Mycoplasma contamination: PCR-based methods and fluorescent dye-based methods (Uphoff and Drexler 2014). PCR was performed to examine Mycoplasma contamination in TK-ST. The results showed no evidence of Mycoplasma contamination as no Mycoplasma was detected.
Serum is added to a culture medium for cell culture because it contains factors that promote cell growth. TK-ST cells showed the maximum growth at an FBS concentration of 20%. TK-ST cells also showed stable growth at an FBS concentration of 5–10%, which is mainly used for mammalian cell culture. In contrast, the cells did not grow in DMEM without FBS, and most cell died a few days after culture. There are several advantages of using serum-free media for animal cells. For example, they reduce the risk of contamination by serum-derived pathogens. The serum contains unknown components, but does not facilitate the identification of substances released by cultured cells into the medium. Furthermore, it enables cell culture with consistent performance and is unaffected by serum quality. The results of this experiment showed that TK-ST cannot be cultured in serum-free DMEM. However, serum-free culture may become possible by examining the culture conditions using various commercially available serum-free media.
Classical cell culture medium consists of amino acids, vitamins, and inorganic salts. Classical media have traditional compositions and are widely used in research studies. TK-ST cells were cultured in six classical media of different compositions with 10% FBS: three types of medium based on DMEM (Dulbecco and Freeman 1959), EMEM (Eagle 1959), RPMI-1640 medium (Moore et al. 1966), and Ham’s F-12 medium (Ham 1965). We found that the three DMEM and RPMI-1640 cell lines showed almost the same growth rate in the TK-ST culture. The growth rate decreased in the order of EMEM > Ham’s F-12 medium. The original composition of DMEM was low glucose (1 g/L) and pyruvic acid. In this study, three DMEMs were prepared with different compositions, including the original composition according to glucose concentration (high-glucose [4.5 g/L] and low-glucose) and with or without pyruvic acid, but no notable differences were observed. This suggests that glucose concentration and the presence of pyruvic acid are ineffective in TK-ST growth. Previously reported cetacean-cultured cells required a special composition of medium with supplements for culture (Pine et al. 2004; Yu et al. 2005; Jin et al. 2013). However, TK-ST cells grew on all tested classical media without any supplements other than 10% FBS, indicating their easy of culture.
Except DES, S100A4, KRT18, α-SMA, and VIM genes were expressed in TK-ST cells. DES was expressed in the kidney tissue and primary cells. The results of this study suggested that TK-STs are a mixed population of various cell types. Furthermore, TK-ST cells were transfected with the viral oncogene SV40T to achieve immortalization, which is known to suppress tumor suppressor genes, such as p53 (Fridman and Tainsky 2008). This may result in the mRNA expression differing from the physiological state. Nevertheless, it is clear from the results of DES expression that the gene expression profile is altered during immortalization and passaging, and such changes in characteristics from the original tissue should be noted in experiments using TK-ST.
TK-ST was infected with several pathogenic viruses of mammalian origin, and CPEs were observed, indicating in vitro replication of the viruses. PRV is a member of the Herpesviridae family with a double-stranded DNA genome. PRV causes pseudorabies (Aujeszky’s disease) and severe neurological disorders in young piglets. JEV belongs to the family Flaviviridae and has a positive-sense single-stranded RNA genome. JEV causes Japanese encephalitis, inflammation of the brain in humans. The results obtained by CPE observations demonstrated that TK-ST is PRV- and JEV-sensitive. The CPE observed in TK-ST was similar to that observed in Vero. Thus, we showed that virus isolation is possible using TK-ST and that this cell line can be used for virus analysis.
Cetacean morbillivirus (CeMV) is a member of the genus Morbillivirus, subfamily Paramyxovirinae, family Paramyxoviridae, with several strains, including DMV. CeMV is a major pathogen in wild cetaceans (Van Bressem et al. 2014). The virus has caused several outbreaks of lethal diseases in cetaceans, including bottlenose dolphins, worldwide. Most previous studies detected gene fragments of CeMV using RT-PCR because virus isolation is difficult due to poorly preserved samples of stranded cetacean carcasses. In addition, the poor cellular resources of cetaceans limit virus isolation. However, viral isolation is an important step in the diagnosis of viral infections and subsequent viral analysis. Therefore, we analyzed the replication of DMV infected with TK-ST by evaluating the DMV phosphoprotein (P) gene copy number using qPCR. The results showed an increase in the number of copies, indicating the usefulness of TK-ST for DMV isolation and research.
Several morbilliviruses have been isolated from human and animals, such as CeMV, human meal virus, and canine distemper virus. SLAM and Nectin4 have been identified as receptors for these morbilliviruses (Tatsuo et al. 2000; Mühlebach et al. 2011; Noyce et al. 2011). The expression of Nectin4 was detected in TK-ST, but that of SLAM was only slightly detected in kidney tissue but not in TK-ST. SLAM is a receptor that is involved in viral entry and pathogenesis. In addition, SLAM plays a role in determining host specificity, depending on the significant sequence diversity among the host animals. Cell lines, such as Vero and CHO, expressing human and canine SLAM have been used in morbillivirus research (Ono et al. 2001; Seki et al. 2003). Morbillivirus replicates and CPE appears faster in the stably transfected Vero cell line expressing canine SLAM (Vero.DogSLAMtag) than in Vero (Nielsen et al. 2008). The CPE of DMV was not observed in the TK-ST during the infection experiments. Since qPCR has shown TK-ST susceptibility to DMV, these results alone do not lead us to point out the shortcomings of TK-ST for DMV studies. However, TK-ST could be improved by expressing cetacean SLAM, as shown in previous studies.
In summary, TK-ST was susceptible to several mammalian-derived viruses. Cellular susceptibility to viruses sometimes depends on the receptors that vary among species. Therefore, it is expected that this cell line will be useful for isolating cetacean-derived viruses that cannot be isolated by cell lines derived from other terrestrial mammalian species.
Foreign agents, such as environmental pollutants and viruses, have been reported to affect the immune system of cetaceans, and measurement methods have been established for each step of the immune response (Beineke et al. 2010). The detailed immune mechanisms remain unclear because they are difficult to verify using animal experiments.
The innate immune system recognizes infecting microbes using pattern recognition receptors that recognize microbe-specific molecular patterns. Toll-like receptors are one of them, activation of Toll-like receptor signaling induces cytokine production, and other factors that regulate the activation of adaptive immune responses (Iwasaki and Medzhitov 2004; Takeda and Akira 2005; Kawasaki and Kawai 2014). TLR3 and TLR4 are receptors of LPS and double-stranded RNA, respectively (Poltorak et al. 1998; Alexopoulou et al. 2001). Both TLR3 and TLR4 mRNA expression was detected in TK-ST. When TLR3 and TLR4 were stimulated by LPS and poly(I:C) in TK-ST, several significant changes were detected in the mRNA expression profiles of cytokines and COX-2. The results indicate that TK-ST has TLRs capable of recognizing LPS and poly(I:C) and also exhibits an immune reaction in response to these stimulations.
There are three types of IFN in mammals: type I IFNs include IFN-α and IFN-β, and type II include IFN-γ. IFN-α and IFN-β are induced following viral infection and play a role in the protection against viral infection. IFN-α and TGF-β are constitutively expressed in TK-ST. Constitutive expression of IFN-α in kidney tissue has been identified in humans and bats, and is thought to be a mechanism that helps maintain a rapid and robust innate and adaptive immune response to foreign bodies (Tovey et al. 1987; Gough et al. 2012; Zhou et al. 2016).
Some studies have reported positive selections of cetacean TLR associated with their adaptive evolution to the marine environment (Ishengoma and Agaba 2017; Xu et al. 2019). As the existence of large amounts of viruses in the ocean has been estimated (Suttle 2007), it is considered that the evolution of the cetacean immune system is important for marine adaptation. Cultured cells are useful experimental tools in immunology research that reflect biological responses to exposure to foreign agents. The functional TLRs possessed by TK-STs are useful for the study of immune function in cetaceans.
In this study, we successfully established a novel cell line, TK-ST, derived from the kidney tissues of the common bottlenose dolphin. TK-ST cells can easily stabilize long-term cultures under common culture conditions and can be used as in vitro systems for viral and immunological research in cetaceans.
Conclusions
In this study, we established a novel kidney cell line, TK-ST, derived from the common bottlenose dolphin. Our results may be used as an experimental basis for future studies in cetaceans. Moreover, our established cell line be used as a cell resource. TK-ST cells are easy to culture as they can grow stably over a long period under common culture conditions. Notably, their characteristics determined in this study indicate the suitability of TK-ST cells for viral and immunological research. Overall, TK-ST cells are useful tools for in vitro research in cetaceans.
References
Alexopoulou L, Holt AC, Medzhitov R, Flavell RA (2001) Recognition of double-stranded RNA and activation of NF-κB by toll-like receptor 3. Nature 413:732–738
Ayral AM, Clarkson S, Cheeseman M, Wells S, Dear TN (2006) A panel of optimized primers and positive-control DNAs for PCR detection of common biological contaminants in mouse cell lines and tissue samples. Lab Anim (NY) 35(8):31–36. https://doi.org/10.1038/laban0906-31
Barrett T, Visser IKG, Mamaev L, Goatley L, Van Bressem M-F, Osterhaus A (1993) Dolphin and porpoise morbilliviruses are genetically distinct from phocine distemper virus. Virology 193(2):1010–1012. https://doi.org/10.1006/viro.1993.1217
Beineke A, Siebert U, Wohlsein P, Baumgärtner W (2010) Immunology of whales and dolphins. Vet Immunol Immunopathol 133:81–94. https://doi.org/10.1016/j.vetimm.2009.06.019
Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE (1998) Extension of life-span by introduction of telomerase into normal human cells. Science (80- ) 279:349–352. https://doi.org/10.1126/science.279.5349.349
Bossart GD (2011) Marine mammals as sentinel species for oceans and human health. Vet Pathol 48:676–690. https://doi.org/10.1177/0300985810388525
Burkard M, Whitworth D, Schirmer K, Nash SB (2015) Establishment of the first humpback whale fibroblast cell lines and their application in chemical risk assessment. Aquat Toxicol 167:240–247. https://doi.org/10.1016/j.aquatox.2015.08.005
Carvan MJ, Santostefano M, Safe S, Busbee D (1994) Characterization of a bottlenose dolphin (Tursiops truncatus) kidney epithelial cell line. Mar Mam Sci 10(1):52–69. https://doi.org/10.1111/j.1748-7692.1994.tb00389.x
Cooper JK, Sykes G, King S, Cottrill K, Ivanova NV, Hanner R, Ikonomi P (2007) Species identification in cell culture: a two-pronged molecular approach. Vitr Cell Dev Biol - Anim 43:344–351. https://doi.org/10.1007/s11626-007-9060-2
Drexler HG, Uphoff CC (2002) Mycoplasma contamination of cell cultures: incidence, sources, effects, detection, elimination, prevention. Cytotechnology 39:75–90. https://doi.org/10.1023/A:1022913015916
Dulbecco R, Freeman G (1959) Plaque production by the polyoma virus. Virology 8(3):396–397. https://doi.org/10.1016/0042-6822(59)90043-1
Eagle H (1959) Amino acid metabolism in mammalian cell cultures. Science 130(3373):432–437. https://doi.org/10.1126/science.130.3373.432
Fridman AL, Tainsky MA (2008) Critical pathways in cellular senescence and immortalization revealed by gene expression profiling. Oncogene 27:5975–5987. https://doi.org/10.1038/onc.2008.213
Gough DJ, Messina NL, Clarke CJP, Johnstone RW, Levy DE (2012) Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity 36(2):166–174. https://doi.org/10.1016/j.immuni.2012.01.011
Ham RG (1965) Clonal growth of mammalian cells in a chemically defined, synthetic medium. Proc Natl Acad Sci U S A 53(2):288–293. https://doi.org/10.1073/pnas.53.2.288
Harasawa R, Mizusawa H, Nozawa K, Nakagawa T, Asada K, Kato I (1993) Detection and tentative identification of dominant mycoplasma species in cell cultures by restriction analysis of the 16S–23S rRNA intergenic spacer regions. Res Microbiol 144:489–493. https://doi.org/10.1016/0923-2508(93)90057-9
Ishengoma E, Agaba M (2017) Evolution of toll-like receptors in the context of terrestrial ungulates and cetaceans diversification. BMC Evol Biol 17:54. https://doi.org/10.1186/s12862-017-0901-7
Iwasaki A, Medzhitov R (2004) Toll-like receptor control of the adaptive immune responses. Nat Immunol 5:987–995. https://doi.org/10.1038/ni1112
Jha KK, Banga S, Palejwala V, Ozer HL (1998) SV40-mediated immortalization. Exp Cell Res 245(1):1–7. https://doi.org/10.1006/excr.1998.4272
Jin W, Jia K, Yang L, Chen J, Wu Y, Yi M (2013) Derivation and characterization of cell cultures from the skin of the Indo-Pacific humpback dolphin Sousa chinensis. Vitr Cell Dev Biol - Anim 49:449–457. https://doi.org/10.1007/s11626-013-9611-7
Kawasaki T, Kawai T (2014) Toll-like receptor signaling pathways. Front Immunol 5. https://doi.org/10.3389/fimmu.2014.00461
Moore GE, Ito E, Ulrich K, Sandberg AA (1966) Culture of human leukemia cells. Cancer 19(5):713–723. https://doi.org/10.1002/1097-0142(196605)19:5%3C713::aid-cncr2820190518%3E3.0.co;2-y
Mühlebach MD, Mateo M, Sinn PL, Prüfer S, Uhlig KM, Leonard VHJ, Navaratnarajah CK, Frenzke M, Wong XX, Sawatsky B, Ramachandran S, McCray PB, Cichutek K, Von Messling V, Lopez M, Cattaneo R (2011) Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature 480:530–533. https://doi.org/10.1038/nature10639
Nagasawa T, Uemori T, Asada K, Harasawa R (1992) Acholeplasma laidlawii has tRNA genes in the 16S–23S spacer of the rRNA operon. J Bacteriol 174(24):8163–8165. https://doi.org/10.1128/jb.174.24.8163-8165.1992
Nielsen O, Smith G, Weingartl H, Lair S, Measures L (2008) Use of a SLAM transfected Vero cell line to isolate and characterize marine mammal morbillivirus using an experimental ferret model. J Wildl Dis 44:600–611. https://doi.org/10.7589/0090-3558-44.3.600
Noyce RS, Bondre DG, Ha MN, Lin LT, Sisson G, Tsao MS, Richardson CD (2011) Tumor cell marker pvrl4 (nectin 4) is an epithelial cell receptor for measles virus. PLoS Pathog 7. https://doi.org/10.1371/journal.ppat.1002240
Ono K, Satoh M, Yoshida T, Ozawa Y, Kohara A, Takeuchi M, Mizusawa H, Sawada H (2007) Species identification of animal cells by nested PCR targeted to mitochondrial DNA. Vitr Cell Dev Biol - Anim 43:168–175. https://doi.org/10.1007/s11626-007-9033-5
Ono N, Tatsuo H, Hidaka Y, Aoki T, Minagawa H, Yanagi Y (2001) Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J Virol 75:4399–4401. https://doi.org/10.1128/JVI.75.9.4399-4401.2001
Palumbi S, Martin A, Romano S, McMillan WO, Stice L, Grabowski G (1991) The simples fool’s guide to PCR. University of Hawaii, Honolulu
Parodi B, Aresu O, Bini D, Lorenzini R, Schena F, Visconti P, Cesaro M, Ferrera D, Andreotti V, Ruzzon T (2002) Species identification and confirmation of human and animal cell lines: a PCR-based method. Biotechniques 32:432–440. https://doi.org/10.2144/02322rr05
Pine M, Schroeder M, Greer K, Hokanson R, Busbee D (2004) Generation and partial characterization of a transformed cetacean cell line. Aquat Toxicol 67(2):195–202. https://doi.org/10.1016/j.aquatox.2004.01.003
Poltorak A, He X, Smirnova I, Liu M-Y, Huffel C Van, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B (1998) Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science (80-. ) 282:2085–2088. https://doi.org/10.1126/science.282.5396.2085
Rosel PE, Dizon AE, Heyning JE (1994) Genetic analysis of sympatric morphotypes of common dolphins (genus Delphinus). Mar Biol 119:159–167. https://doi.org/10.1007/BF00349552
Seki F, Ono N, Yamaguchi R, Yanagi Y (2003) Efficient isolation of wild strains of canine distemper virus in Vero cells expressing canine SLAM (CD150) and their adaptability to Marmoset B95a cells. J Virol 77:9943–9950. https://doi.org/10.1128/JVI.77.18.9943-9950.2003
Sitt T, Bowen L, Blanchard MT, Smith BR, Gershwin LJ, Byrne BA, Stott JL (2008) Quantitation of leukocyte gene expression in cetaceans. Dev Comp Immunol 32:1253–1259. https://doi.org/10.1016/j.dci.2008.05.001
Suttle CA (2007) Marine viruses - major players in the global ecosystem. Nat Rev Microbiol 5:801–812. https://doi.org/10.1038/nrmicro1750
Suzuki M, Wakui H, Itou T, Segawa T, Inoshima Y, Maeda K, Kikuchi K (2016) Two isoforms of aquaporin 2 responsive to hypertonic stress in the bottlenose dolphin. J Exp Biol 219:1249–1258. https://doi.org/10.1242/jeb.132811
Takeda K, Akira S (2005) Toll-like receptors in innate immunity. Int Immunol 17:1–14. https://doi.org/10.1093/intimm/dxh186
Tatsuo H, Ono N, Tanaka K, Yanagi Y (2000) SLAM (CDw150) is a cellular receptor for measles virus. Nature 406:893–897. https://doi.org/10.1038/35022579
Tovey MG, Streulit M, Gressert I, Gugenheim J, Blanchardt B, Guymarhot J, Vignauxt F, Gigou M (1987) Interferon messenger RNA is produced constitutively in the organs of normal individuals. Proc Natl Acad Sci U S A 84:5038–5042. https://doi.org/10.1073/pnas.84.14.5038
Uphoff CC, Drexler HG (2014) Detection of mycoplasma contamination in cell cultures. Curr Protoc Mol Biol 106:1–14. https://doi.org/10.1002/0471142727.mb2804s106
Van Bressem M-F, Duignan P, Banyard A, Barbieri M, Colegrove K, De Guise S, Di Guardo G, Dobson A, Domingo M, Fauquier D, Fernandez A, Goldstein T, Grenfell B, Groch K, Gulland F, Jensen B, Jepson P, Hall A, Kuiken T, Mazzariol S, Morris S, Nielsen O, Raga J, Rowles T, Saliki J, Sierra E, Stephens N, Stone B, Tomo I, Wang J, Waltzek T, Wellehan J (2014) Cetacean morbillivirus: current knowledge and future directions. Viruses 6:5145–5181. https://doi.org/10.3390/v6125145
Viricel M, Rosel PE (2012) Evaluating the utility of cox1 for cetacean species identification. Mar Mamm Sci 28:37–62. https://doi.org/10.1111/j.1748-7692.2010.00460.x
Wells R, Rhinehart H, Hansen L, Sweeney J, Townsend F, Stone R, Casper DR, Scott M, Hohn A, Rowles T (2004) Bottlenose dolphins as marine ecosystem sentinels: developing a health monitoring system. EcoHealth 1:246–254. https://doi.org/10.1007/s10393-004-0094-6
Xu S, Tian R, Lin Y, Yu Z, Zhang Z, Niu X, Wang X, Yang G (2019) Widespread positive selection on cetacean TLR extracellular domain. Mol Immunol 106:135–142. https://doi.org/10.1016/j.molimm.2018.12.022
Yajing S, Rajput IR, Ying H, Fei Y, Sanganyado E, Ping L, Jingzhen W, Wenhua L (2018) Establishment and characterization of pygmy killer whale (Feresa attenuata) dermal fibroblast cell line. PLoS One 13:1–15. https://doi.org/10.1371/journal.pone.0195128
Yu J, Kindy MS, Ellis BC, Baatz JE, Peden-Adams M, Ellingham TJ, Wolff DJ, Fair PA, Gattoni-Celli S (2005) Establishment of epidermal cell lines derived from the skin of the Atlantic bottlenose dolphin (Tursiops truncatus). Anat Rec Part A Discov Mol Cell Evol Biol 287A:1246–1255. https://doi.org/10.1002/ar.a.20266
Zhou P, Tachedjian M, Wynne JW, Boyd V, Cui J, Smith I, Cowled C, Ng JHJ, Mok L, Michalski WP, Mendenhall IH, Tachedjian G, Wang LF, Baker ML (2016) Contraction of the type i IFN locus and unusual constitutive expression of IFN-α in bats. Proc Natl Acad Sci U S A 113:2696–2701. https://doi.org/10.1073/PNAS.1518240113/-/DCSUPPLEMENTAL/PNAS.201518240SI.PDF
Acknowledgements
We would like to thank Dr. Makoto Sugiyama (Gifu University, Gifu, Japan) for providing the dolphin morbillivirus Muc strain. We are also grateful to the cooperation of the Taiji Fisheries Cooperative Union (Wakayama, Japan) and the Fisheries Agency of Japan for providing the specimens used in this study. We would also like to thank the staff of the Shinagawa Aquarium for their assistance in collecting the blood samples used in this study.
Funding
This research was funded by a Grant-in-Aid for Scientific Research C (18K05831) from the Japan Society for the Promotion of Science (KAKENHI).
Author information
Authors and Affiliations
Contributions
Kaede Tashiro, Takao Segawa, and Takuya Itou conceptualized the study. Kaede Tashiro and Takuya Itou conducted the experiments. Kaede Tashiro, Taketo Futami, Miwa Suzuki, and Takuya Itou processed samples. Kaede Tashiro, Takao Segawa, Miwa Suzuki, and Takuya Itou performed the data analysis; Kaede Tashiro drafted, revised, and edited the manuscript. All authors read the manuscript and approved the final submitted manuscript for publication.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tashiro, K., Segawa, T., Futami, T. et al. Establishment and characterization of a novel kidney cell line derived from the common bottlenose dolphin. In Vitro Cell.Dev.Biol.-Animal 59, 536–549 (2023). https://doi.org/10.1007/s11626-023-00786-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11626-023-00786-y