Abstract
Angiogenesis involves temporo-spatially coordinated endothelial cell (EC) proliferation, differentiation, migration, and sprouting. Notch signaling is essential in regulating EC behaviors during angiogenesis, but its downstream mechanisms remain incompletely defined. In the current study, we show that miR-223-3p is a downstream molecule of Notch signaling and mediates the role of Notch signaling in regulating EC migration and sprouting. In human umbilical vein endothelial cells (HUVECs), Notch activation by immobilized Dll4, a Notch ligand, upregulated miR-223-3p, and Notch activation–mediated miR-223-3p upregulation could be blocked by a γ-secretase inhibitor (DAPT). miR-223-3p overexpression apparently repressed HUVEC migration, leading to attenuated lumen formation and sprouting capacities. Transcriptome comparison and subsequent qRT-PCR validation further indicated that miR-223-3p downregulated the expression of multiple genes involved in EC migration, axon guidance, extracellular matrix remodeling, and angiogenesis. In addition, miR-223-3p antagonist transfection abolished Notch-mediated repression of EC migration and sprouting. By quantitative reverse transcription–polymerase chain reaction (qRT-PCR), western blotting, and reporter assay analysis, we confirmed that miR-223-3p directly targeted F-box and WD repeat domain–containing 7 (Fbxw7). Meanwhile, Fbxw7 overexpression could efficiently rescue the impaired migration capacity of ECs under miR-223-3p overexpression. In summary, these results identify that Notch activation–induced miR-223-3p suppresses EC migration and sprouting via Fbxw7.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Angiogenesis is defined as the process of forming organized and functional blood vessels from preexisting ones, and involves coordinated migration, proliferation, and differentiation of endothelial cells (ECs) (Greenspan and Weinstein 2021). During sprouting angiogenesis, ECs differentiate into tip and stalk cells. Tip cells as the leader of sprouts extend filopodia and migrate toward vascular endothelial growth factor (VEGF)–secreting cells, while stalk cells behind the tip cells proliferate to elongate sprouts (Chen et al. 2019; Greenspan and Weinstein 2021). EC migration is therefore an essential process of angiogenesis and can be divided into several sequential steps, namely sensing of the motile stimuli, degradation of the basement membrane (BM), extension along with the extracellular matrix (ECM), contraction of the cell body, and release of the traction forces (Fonseca et al. 2020; Muhleder et al. 2021). To accomplish these steps precisely, ECs need to integrate multiple molecular events involving extrinsic cell communications from pro-angiogenic stimuli, and intrinsic signaling networks that coordinate cell behaviors (Fonseca et al. 2020). Deregulated EC migration is involved in many human diseases (Fonseca et al. 2020), so it is important to uncover the molecular mechanisms regulating EC migration during angiogenesis.
Notch signaling, which mediates a direct cell–cell interaction employing a family of transmembrane receptors and ligands, is required for angiogenesis by regulating EC behaviors such as tip-stalk cell differentiation, proliferation, and migration (Tetzlaff and Fischer 2018). Genetic inactivation of Notch signaling increases EC migration and the number of tip cells (Dou et al. 2008). Conversely, Notch activation suppresses EC migration and leads to a compromised vascular network (Alabi et al. 2018), suggesting that Notch signaling is vital in regulating EC migration and sprouting (Tetzlaff and Fischer 2018). Although VEGFR2, Nrp1, and certain metabolic enzymes have been implicated in Notch-mediated EC behaviors (Phng and Gerhardt 2009; De Bock et al. 2013), the exact mechanism of Notch signaling in regulating EC migration has not been completely revealed.
microRNAs (miRNAs) have been reported to regulate multiple EC behaviors including migration and sprouting under physiological and pathological conditions (Tiwari et al. 2018). To identify potential Notch downstream miRNAs in ECs, we previously performed a small RNA sequencing and identified several miRNAs that are upregulated upon forced Notch activation in ECs (Yan et al. 2016). In the current study, we further validated one of them, miR-223-3p, in mediating the role of Notch signaling in regulating EC behaviors. miR-223-3p has been proved to be a potential antiangiogenic miRNA that prevents endothelial cell proliferation (Shi et al. 2013), in addition to other activities (Yuan et al. 2018). Our results show that miR-223-3p mediates Notch signaling in repressing EC migration and sprouting. Therefore, miR-223-3p is a novel Notch downstream molecule and mediates the role of Notch signaling in suppressing EC migration and sprouting.
Materials and methods
Cell culture and transfection
Primary human umbilical vein endothelial cells (HUVECs) were isolated using 1 mg/mL type I collagenase (Sigma-Aldrich, St Louis, MO) from human umbilical cord biopsies, which were obtained from the Department of Gynecology and Obstetrics of Xijing Hospital. HUVECs were cultured in an EC medium (ScienCell, San Diego, CA) containing 5% fetal calf serum (FCS), 1 × endothelial cell growth supplement (ECGS), 100 U/mL ampicillin, and 100 mg/mL streptomycin, and cells between passages 2 and 6 were used as previously described (Yan et al. 2016). The use of human samples was approved by the Ethics Committee of Xijing Hospital, Fourth Military Medical University.
HUVECs were transfected with 100 nmol/L nonsense control (NC), miR-223-3p antisense oligonucleotides (ASO), or Fbxw7 siRNA (RiboBio, Guangzhou, China) using Lipofectamine 2000™ (Invitrogen, Carlsbad, CA) according to the supplier’s protocol. Adenoviruses expressing miR-223-3p (AdmiR-223) and control adenovirus (AdCtrl) were purchased from Genechem (Shanghai, China) and used at multiplicity of infection (MOI) of 30 to overexpress miR-223-3p in HUVECs. Cell medium was changed 8 h after the addition of virus, and culture was continued for 48 h before harvest. Lentivirus expressing human Fbxw7 (NM_033632.3, 163 ~ 2286 bp from the first cDNA nucleotide) and control lentivirus were purchased from Hanbio Biotechnology (Shanghai, China) and used at MOI of 10 to overexpress Fbxw7 in HUVECs. To activate Notch signaling, culture plates were precoated with recombinant soluble Delta-like (Dll) 4 protein (Sino Biological Inc., Beijing, China) (0.5 μg/well) at 4 °C for 12 h, with PBS as a control. Solutions were then discarded and HUVECs (1 × 105/well) were seeded and cultured for the indicated period of time. The γ-secretase inhibitor DAPT (Selleck Chemicals, Houston, TX) was applied at 25 μmol/L, with dimethyl sulfoxide (DMSO) as a control.
Cell migration assay
For wound healing assay, HUVECs (1.5 × 105/well) were cultured in 12-well plates for 48 h to reach full confluence, and a scratch was made with a pipette tip. The medium was then replaced with an EC medium containing 1% fetal bovine serum (FBS). The scratch was photographed at 0 h and 16 h after the scratching, and the distance of cell migration was evaluated. For transwell assay, HUVECs (1 × 105) were seeded in Transwell chambers (Millipore, Burlington, MA) and cultured for 24 h. Cells were fixed with 4% paraformaldehyde and stained with 5% crystal violet. Cells migrating across the transwell were counted under a microscope.
Lumen formation assay
HUVECs (1 × 105/well) were seeded in 48-well plates precoated with matrigel basement membrane matrix (200 μL/per well, BD Bioscience, San Jose, CA), and incubated at 37 °C for 4 h. Cells were randomly photographed for five fields under a microscope, and cell cords forming lumen-like structure were quantified as mean number of branches per field and mean length per branch using the Image-Pro Plus 6.0 software.
Aortic ring sprouting assay
Male mice (6 ~ 8 wk old) were anesthetized and aortas were dissected. Aortic rings were prepared and incubated in a complete medium at 37 °C, and infected with adenovirus or transfected with ASO for 12 h as described (Yan et al. 2016). The aorta rings were then placed in a 96-well plate precoated with matrigel basement membrane matrix (100 μL per well). After solidification of the gel for 20 min at 37 °C, Opti-MEM (150 μL per well) including 2.5% FBS and VEGF (30 ng/mL, Sino Biological Inc.) was added and changed every other day for 4 d. To quantify the endothelial sprouts, each microvessel emerging from the main ring was considered as a sprout, and the microscope focus was adjusted to ensure that vessels in different planes were observed (Baker et al. 2011). The mean sprout numbers per ring and mean length per sprout were measured and compared using the Image-Pro Plus 6.0 software.
RNA sequencing (RNA-seq)
RNA-seq was conducted by commercial service (Gene Denovo Biotechnology Corporation, Guangzhou, China). In brief, HUVECs were infected with AdmiR-223 or AdCtrl for 48 h, and lysed with the TRIzol reagent (Invitrogen). Total RNA was extracted and evaluated for quality. Then, mRNA was enriched, fragmented, and reverse-transcribed into complementary DNA (cDNA). cDNA fragments were sequenced by using Illumina NovaSeq 6000. Original RNA-seq data have been deposited in the Genome Sequence Archive in BIG Data Center (Beijing) (accession #, PRJCA005424). The remaining data and material are available on reasonable request.
The online OmicShare tools (http://www.omicshare.com/tools) were used for the subsequent bioinformatic analysis.
Quantitative reverse transcription–polymerase chain reaction (qRT-PCR)
Total RNA was extracted from HUVECs with the TRIzol reagent, and cDNA was synthesized. qRT-PCR was conducted with the TBGreen kit (Takara, Dalian, China) and a QuantStudio 5 real-time PCR instrument (Life Technologies, Waltham, MA). Gene expression level was displayed as relative fold-change compared with β-actin. The miR-223-3p level was evaluated using a miRNA qRT-PCR kit (Takara), with U6 RNA as an internal reference. Primer sequences are exhibited in supplementary Table S1.
Western blotting
Total cellular proteins were extracted using the RIPA lysis buffer (Beyotime, Shanghai, China) containing phenylmethylsulfonyl fluoride (PMSF, 1 mM, Sigma-Aldrich), and quantified with a bicinchoninic acid (BCA) kit (Beyotime). Proteins were separated with sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and electro-blotted onto polyvinylidene fluoride membranes. Membranes were then probed with anti-Fbxw7 or anti-β-Actin as primary antibody, and HRP-anti-rabbit IgG or HRP-anti-mouse IgG as secondary antibody (Proteintech), respectively. Bands were detected using a chemoluminescence system (Tanon, Shanghai, China). Quantification was performed by ImageJ2x software (Rawak Software).
Reporter assay
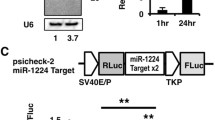
The 3’ untranslated region (3’UTR) of human Fbxw7 mRNA (NM_033632.3, region 2287–2786 bp from the first cDNA nucleotide) was synthesized and subcloned into pSI-Check2 plasmid to construct pSI-Fbxw7 wild type (Fbxw7-3’UTR-wt). A reporter, Fbxw7-3’UTR-mut, with two mutations in the 3’UTR complementary to the seed sequence of miR-223-3p was also generated by synthesis. HEK 293 T cells were infected with AdmiR-223-3p or AdCtrl adenovirus for 8 h, or transfected with miR-223-3p ASO or NC, and Fbxw7-3’UTR-wt (100 ng) or Fbxw7-3’UTR-mut (100 ng) plasmids was then transfected. Forty-eight hours later, cells were harvested, and the firefly and Renilla luciferase activities were analyzed with the dual-luciferase reporter assay system (Promega, Madison, WI), according to the instructions.
Statistical analysis
The Image-Pro Plus 6.0 software and Image J2x software were used for images analysis. Statistical analysis was conducted using the GraphPad Prism 8 software. Quantitative data were expressed as mean ± s.d. Student’s t-test was used for continuous variables between two groups. One-way ANOVA followed by Tukey’s post hoc test was used to compare the continuous variables more than two groups for one independent variable. p < 0.05 was considered as significant.
Results
miR-223-3p is a novel Notch signaling downstream molecule in ECs
To identify Notch downstream miRNAs in ECs, we have compared miRNA profiles of ECs with or without Notch activation, and verified that miR-342-5p functions downstream to Notch signaling to regulate EC proliferation and angiogenesis (Yan et al. 2016). miR-223-3p is another miRNA among the Notch activation–induced miRNAs in ECs. To validate miR-223-3p as a Notch downstream molecule, HUVECs were cultured in the presence of immobilized Notch ligand Dll4, which efficiently activates Notch signaling as shown by upregulated Hey1 (Fig. 1A). qRT-PCR confirmed that Dll4-mediated Notch activation upregulated miR-223-3p in HUVECs apparently (Fig. 1A). Blocking Notch signaling by DAPT, a γ-secretase inhibitor, canceled Dll4-stimulated Notch activation as well as miR-223-3p upregulation in HUVECs (Fig. 1B). These results indicate that Notch signaling positively regulates miR-223-3p expression in ECs.
Notch activation upregulates miR-223-3p expression. (A) HUVECs were stimulated with Dll4 or PBS. Forty-eight hours later, Hey1 and miR-223-3p expression were examined with qRT-PCR (n = 5). (B) HUVECs were stimulated as in (A) in the presence of DAPT or DMSO. Hey1 and miR-223-3p expression were observed with qRT-PCR (n = 5). Error bars = means ± s.d. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Statistical tests: two-tailed Student’s t-test for (A), and one-way ANOVA followed by Tukey’s post hoc test for (B)
miR-223-3p attenuates EC migration and sprouting
To access the function of miR-223-3p in ECs, we overexpressed miR-223-3p in HUVECs by adenovirus-mediated transfection (Fig. 2A). We examined cell migration using Transwell assay and wounding healing assay, and both assays showed that EC migration decreased apparently in miR-223-3p-overexpressing HUVECs as compared with the control (Fig. 2B). Consistent with cell behavior alterations, miR-223-3p overexpression inhibited the lumen formation capacity of HUVECs in vitro, as shown by lumen formation assay (Fig. 2C). Aortic ring sprouting assay further confirmed that miR-223-3p overexpression attenuated EC sprouting capacity (Fig. 2D). These results suggested that miR-223-3p overexpression inhibits EC migration and sprouting in angiogenesis.
miR-223-3p overexpression reduces EC motility and sprouting. (A) HUVECs were infected with Ctrl (AdCtrl) or miR-223-3p (AdmiR-223) adenovirus. Forty-eight hours later, miR-223-3p level was observed with qRT-PCR (n = 9). (B) EC migration was evaluated and compared using a Transwell assay (n = 3) and a wound healing assay (n = 3), respectively. (C) HUVECs were treated as in (A) and subjected to lumen formation assay. EC angiogenic capacity was determined by calculating the number and length of branches (n = 3). (D) Aortic rings were dissected and infected as in (A). EC sprouting capacity was determined and compared with an aortic ring sprouting assay (n = 5). Error bars = means ± s.d. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Statistical tests: two-tailed Student’s t-test for (A–D)
miR-223-3p suppresses a panel of migration and sprouting-related genes
In order to uncover the downstream mechanisms of miR-223-3p-mediated suppression of EC migration and sprouting, we compared transcriptomes between miR-223-3p overexpression and control HUVECs by RNA-seq (Fig. 3A). Gene set enrichment analysis (GSEA) indicated that transcripts involved in sprouting angiogenesis, EC migration, guidance, and ECM remodeling were positively enriched in the control group, indicating that miR-223-3p downregulates the expression of these genes (Fig. 3B). Heatmap comparisons also showed that sprouting-related and ECM-related genes were downregulated in miR-223-3p overexpression HUVECs (Fig. 3C) (Blanco and Gerhardt 2013; Feng et al. 2020). We further validated a part of the representative genes by qRT-PCR, and the result confirmed that the mRNA level of genes associated with angiogenic sprouting (Vegfr2, Vegfr3, Aplnr, Tie2, Jag1), axon guidance (Robo1, Nrp2), and ECM remodeling (Mmp2, Mmp16, Col1a2, Col3a1, Col4a1, Fn1) was remarkably downregulated under miR-223-3p overexpression (Fig. 3D). Therefore, miR-223-3p overexpression downregulates a panel of genes involved in angiogenic sprouting, axon guidance, and ECM remodeling, leading to compromised EC migration and sprouting.
miR-223-3p attenuates multiple migration and sprouting-related genes expression. (A) HUVECs were infected with AdCtrl or AdmiR-223, and subjected to RNA-seq 48 h later. FPKM data were analyzed by principal component analysis (PCA). (B) GSEA analysis of gene sets related to migration, sprouting, and ECM remodeling. (C) Sprouting- and ECM- related genes were shown by heatmap. (D) Sprouting- and ECM-related gene expression was validated using qRT-PCR (n = 3). Error bars = means ± s.d. **p < 0.01; ***p < 0.001; ****p < 0.0001. Statistical test: two-tailed Student’s t-test for (D)
miR-223-3p mediates the repressive effect of Notch activation on EC migration
We further evaluated the relationship between Notch activation and miR-223-3p upregulation in regulating EC migration and sprouting. HUVECs were transfected with an ASO targeting miR-223-3p and cultured with immobilized Dll4 or control. qRT-PCR showed that miR-223-3p ASO transfection efficiently decreased miR-223-3p level under Dll4 stimulation (Fig. 4A). Transwell assay and wound healing assay manifested that miR-223-3p inhibition rescued EC migration that was repressed by Notch activation (Fig. 4B and C). Consistently, lumen formation assay and aortic ring assay showed that while Notch activation attenuated lumen formation and sprouting, downregulating miR-223-3p rescued this phenotype (Fig. 4D and E). In addition, miR-223-3p inhibition partially restored the expression level of migration and sprouting-related genes downregulated by Dll4 stimulation (Fig. 4F). These analyses demonstrated that miR-223-3p mediates the inhibition of EC migration and sprouting under Notch activation.
Notch activation represses EC migration and sprouting through miR-223-3p. (A) HUVECs were treated with PBS, Dll4 plus NC, or Dll4 plus miR-223-3p ASO. miR-223-3p expression level was evaluated with qRT-PCR (n = 4). (B and C) HUVECs were treated as in (A). EC migration was evaluated by a Transwell assay (n = 3) (B) and a wound healing assay (n = 4) (C), respectively. (D) HUVECs were treated as in (A). EC angiogenic capacity was evaluated by a lumen formation assay (n = 3). (E) Aortic rings were dissected and treated as in (A). EC sprouting capacity was determined by an aortic ring sprouting assay (n = 5). (F) HUVECs were treated as in (A). The expression level of migration and sprouting-related genes was evaluated with qRT-PCR (n = 4). Error bars = means ± s.d. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; n.s, not significant. Statistical tests: one-way ANOVA followed by Tukey’s post hoc test for (A–F)
miR-223-3p directly targets Fbxw7 in ECs
We further accessed miR-223-3p target genes in ECs. Bioinformatic analyses suggested a panel of potential genes targeted by miR-223-3p, but qRT-PCR showed that only the expression of F-box and WD repeat domain–containing 7 (Fbxw7) was significantly downregulated in miR-223-3p overexpression HUVECs (Fig. 5A). Western blotting further validated that the protein level of Fbxw7 decreased significantly in miR-223-3p overexpression HUVECs (Fig. 5B). Meanwhile, the miR-223-3p blockade could rescue the inhibitory effect of Dll4 on the Fbxw7 protein level (Fig. 5C). In addition, reporter assay indicated that miR-223-3p significantly suppressed the expression of the luciferase gene containing 3’UTR of Fbxw7, which could be canceled when the miR-223-3p recognition site was mutated (Fig. 5D and Fig. S1A). We further evaluated the luciferase activity upon miR-223-3p blockade; the results showed that the luciferase activity significantly increased under miR-223-3p inhibition (Fig. 5D). These data suggest that Fbxw7 is a direct target of miR-223-3p in ECs.
miR-223-3p directly targets Fbxw7 in ECs. (A) Expression of the potential targeted genes was observed with qRT-PCR (n = 3). (B) Fbxw7 protein level was determined by western blotting (n = 3). (C) HUVECs were treated with PBS, Dll4 plus NC, or Dll4 plus miR-223-3p ASO. Fbxw7 protein level was evaluated with western blotting (n = 3). (D) Reporter assay. Luciferase activity was evaluated in HEK 293 T cells transfected with Fbxw7-3’UTR-wt (100 ng) or Fbxw7-3’UTR-mut (100 ng), together with AdmiR-223-3p, AdCtrl (n = 4), miR-223-3p ASO or negative control (NC) (n = 6). (E and F) HUVECs were transfected with Fbxw7 siRNAs for 48 h. EC migration was evaluated with a wound healing assay (n = 3) (E) and a Transwell assay (n = 3) (F). (G) HUVECs were transfected as in (E). EC angiogenic capacity was determined with a lumen formation assay (n = 3). Error bars = means ± s.d. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Statistical tests: two-tailed Student’s t-test for (A, B, and D), and one-way ANOVA followed by Tukey’s post hoc test for (C and E–G)
To further test the role of Fbxw7 in EC migration, we decreased the Fbxw7 expression level with siRNAs. qRT-PCR showed that the siRNAs efficiently reduced the expression level of Fbxw7 (Fig. S1B). Wound healing and Transwell assay demonstrated that Fbxw7 blockade significantly suppressed EC migration (Fig. 5E and F and Fig. S1C and D). Lumen formation assay further indicated that Fbxw7 blockade repressed EC angiogenic capacity (Fig. 5G). These results suggest that Fbxw7 acts as a pro-angiogenic factor and its blockade suppresses EC migration and angiogenic capacity.
Fbxw7 overexpression rescues miR-223-3p-induced migration arrest in ECs
To further confirm that Fbxw7 mediated the inhibition of EC migration under miR-223-3p, we overexpressed Fbxw7 in HUVECs using lentivirus before infecting with AdmiR-223-3p or AdCtrl. qRT-PCR and western blotting validated the increased expression level of Fbxw7 in the Fbxw7 overexpressing group (Fig. 6A and B). Wound healing assay showed that miR-223-3p-induced reduction in EC migration could be significantly rescued by Fbxw7 overexpression (Fig. 6C), which was further confirmed by a Transwell assay (Fig. 6C). Moreover, lumen formation and aortic ring assays showed that Fbxw7 overexpression efficiently rescued the deficiency of angiogenic capacities of ECs induced by miR-223-3p overexpression (Fig. 6D and E). Meanwhile, qRT-PCR demonstrated that Fbxw7 overexpression could partially upregulate some migration and sprouting-related gene expression under miR-223-3p overexpression (Fig. 6F). These results further demonstrate that miR-223-3p suppresses EC migration and sprouting ability by directly targeting Fbxw7.
Fbxw7 overexpression abolished miR-223-3p-mediated inhibition of EC migration. (A) HUVECs were infected with lentivirus expressing Fbxw7 and adenovirus expressing miR-223-3p, cells were harvested 48 h later. The Fbxw7 mRNA level was determined with qRT-PCR (n = 5). (B) HUVECs were treated as in (A). The Fbxw7 protein level was evaluated with western blotting (n = 5). (C) HUVECs were treated as in (A). The EC migration capacity was observed with a wound healing assay (n = 5) and a Transwell assay (n = 3). (D and E) ECs were treated as in (A). EC angiogenic and sprouting abilities were evaluated with lumen formation and aortic ring assays (n = 3 for lumen formation assay, n = 5 for aortic ring assay), respectively. (F) HUVECs were treated as in (A). The expression level of migration and sprouting-related genes was determined with qRT-PCR (n = 4). Error bars = means ± s.d. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; n.s, not significant. Statistical tests: one-way ANOVA followed by Tukey’s post hoc test for (A–C, E, and F)
Fbxw7 overexpression rescues the repressive effects of Notch activation on EC migration
To further evaluate the relationship between Notch and Fbxw7, we overexpressed Fbxw7 with lentivirus in HUVECs under Dll4 stimulation. Western blotting showed that Dll4 stimulation reduced the protein level of Fbxw7, which could be restored by Fbxw7 overexpression (Fig. 7A). Wound healing and Transwell assays demonstrated that Fbxw7 overexpression partially rescued the repressive effects of Notch activation on EC migration (Fig. 7B and C). In addition, lumen formation and aortic ring assays indicated that Fbxw7 overexpression could further restore the defects of the angiogenic ability of ECs under Dll4 stimulation (Fig. 7D and E). These results suggest that Notch activation represses EC migration and sprouting ability by targeting Fbxw7.
Fbxw7 overexpression restored EC migration and sprouting abilities under Notch activation. (A) HUVECs were infected with lentivirus expressing Fbxw7 under Dll4 stimulation, cells were harvested 24 h later. The Fbxw7 protein level was determined with western blotting (n = 3). (B) HUVECs were treated as in (A). The EC migration capacity was observed with a wound healing assay (n = 3). (C) HUVECs were treated as in (A). The EC migration ability was determined with a Transwell assay (n = 3). (D) HUVECs were treated as in (A). EC angiogenic ability was observed with a lumen formation assay (n = 5). (E) Aortic rings were treated as in (A). An aortic ring assay was used to evaluate EC sprouting ability (n = 5). Error bars = means ± s.d. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Statistical tests: one-way ANOVA followed by Tukey’s post hoc test for (A–E)
Discussion
Notch signaling plays critical roles in angiogenesis by regulating EC behaviors in multiple aspects (Tetzlaff and Fischer 2018). During sprouting, Dll4 is highly expressed in tip cells, which can activate Notch signaling in adjacent stalk cells, and Jagged family Notch ligands expressed by stalk cells reversely inhibit Notch activation in tip cells, promoting tip/stalk differentiation (Blanco and Gerhardt 2013). Notch activation inhibits stalk ECs competing for the tip cell fate (Blanco and Gerhardt 2013), and conversely, Notch inhibition with genetic ablation or pharmacologic inhibitors leads to increased EC migration and excessive formation of tip cells (Dou et al. 2008; Tetzlaff and Fischer 2018), a process called hyper-sprouting. Previous studies have reported several downstream effectors likely mediating Notch signaling in ECs (Phng and Gerhardt 2009; Tetzlaff and Fischer 2018). For instance, Notch activation typically represses VEGFR signaling and certain metabolism enzymes, such as PFKFB3 (Blanco and Gerhardt 2013; De Bock et al. 2013). However, it has been unclear how Notch inhibition or activation endows ECs with tip or stalk phenotypes, which are characterized by, for simplicity, high migration/low division or high division/low migration, respectively.
Our group has identified a group of miRNAs that likely mediate the effect of Notch signaling on EC phenotypes. Previously, we have shown that miR-342-5p downstream to Notch receptors inhibits EC proliferation and promotes migration by targeting endoglin and modulating several angiogenic pathways, and regulates neural stem cells as well (Yan et al. 2016; Gao et al. 2017). In the current study, we further demonstrate that miR-223-3p is a Notch downstream molecule in ECs and is upregulated upon Notch activation. Our results indicate that miR-223-3p can at least partly mediate the effects of Notch signaling on repressing EC migration and sprouting (Kumar et al. 2014). The mechanism of Notch regulating miR-223-3p is currently not clear. In T-cell acute lymphoblastic leukemia (T-ALL), previous studies have suggested that Notch signaling, in collaboration with NF-κB, could upregulate miR-223-3p through trans-activating its promoter (Kumar et al. 2014). More in-depth studies are required to elucidate the mechanism of Notch signaling in regulating miR-223-3p in ECs.
Our transcriptomic analysis shows that miR-223-3p overexpression downregulated a considerable large panel of genes related with cell migration in ECs. These include several receptor tyrosine kinases (RTKs), such as Vegfr2, Vegfr3, Tie2, and Aplnr, which play critical roles in promoting EC migration and sprouting (Blanco and Gerhardt 2013; Joussen et al. 2021). This is consistent with recent reports showing that miR-223-3p restrains angiogenesis by preventing growth factor signaling (Shi et al. 2013). Upon binding to these receptors by their ligands, such as VEGF and ANGPT2, RTKs mediate EC activation and promote several sequential processes of cell migration, including polarization, filopodia and lamellipodia formation, and ECM degradation and remodeling, leading to sprouting and new vessel formation (Fonseca et al. 2020). Indeed, our transcriptomic analysis and qRT-PCR validation showed that genes involved in ECM degradation and synthesis are also downregulated by miR-223-3p overexpression. During angiogenesis, ECs secrete a series of proteolytic enzymes, such as MMPs, to degrade ECM and basement membrane (BM), and then remodel ECM by synthesizing new ECM molecules including fibronectin and collagens (Karamanos et al. 2021). Our data show that Mmp2 and Mmp16, as well as Fn1, Col1a2, Col3a1, and Col4a1, were all downregulated in miR-223-3p overexpression EC (Karamanos et al. 2021). Another group of miR-223-3p-downregulated genes is axon guidance molecules. At least three pairs of guidance molecules, including semaphorin-neuropillin, Netrin-Unc5/DCC, and Slit-Robo, are involved in angiogenesis (Autiero et al. 2005). miR-223-3p downregulated Semaphorins, Slit2, and Robo1, suggesting that miR-223-3p can also inhibit the pathway-finding ability of ECs in sprouting. In summary, our data imply that miR-223-3p overexpression in ECs mainly downregulates multiple genes in terms of cytokine signaling, ECM degradation and remodeling, and axon guidance. These changes disabled ECs to be activated by cytokine stimulation, invade through ECM, and deposit new BM.
miR-223-3p is a multi-functional miRNA that is involved in regulating gene expression under various physiological and pathogenic conditions, such as rheumatoid arthritis and cancer (Yuan et al. 2018). Different genes have been previously identified as miR-223-3p targets under specific conditions (Zhang et al. 2020). In ECs, miR-223-3p could suppress the expression of β1 integrin and RhoB (Shi et al. 2013). However, in our transcriptomic data, the level of β1 integrin mRNA showed no apparent difference between miR-223-3p-overexpressing ECs and the Ctrl group, and the RhoB mRNA level even increased after miR-223-3p overexpression. Among a panel of other potential miR-223-3p targets predicted by bioinformatic analysis, Fbxw7 is likely regulated by miR-223-3p in ECs (Kurashige et al. 2012; Kumar et al. 2014; Zhou et al. 2015; Liu et al. 2017; Jiang et al. 2019; Shao et al. 2019). qRT-PCR, western blotting, and reporter assay further validated that Fbxw7 is a direct target of miR-223-3p in ECs. Fbxw7 is a ubiquitin E3 ligase and has been previously shown to participate in regulating EC migration and angiogenesis through degradation of Notch receptor and hypoxia-inducible factor-1α (HIF-1α) (Ji et al. 2018). Depletion of Fbxw7 markedly impairs EC motility and angiogenesis (Yeh et al. 2018; Luo et al. 2021). Our results demonstrated that Fbxw7 mediated the inhibitory effects of miR-223-3p on EC migration, and Fbxw7 overexpression could efficiently rescue the weakened EC motility by miR-223-3p overexpression and Notch activation.
In addition, the influence of miR-223-3p on EC proliferation might be complicated. Our preliminary data suggested that miR-223-3p overexpression could increase EC proliferation. These results are in contrast with some other reports (Shi et al. 2013). These inconsistencies could be attributed to the complex functions of Fbxw7, which is a E3 ligase–mediating degradation of many molecules involved in EC proliferation. Thus, as two examples, Fbxw7 could target Notch1 to promote proliferation (Hoeck et al. 2010), but could also target MYC to repress proliferation (Hu et al. 2021; Sun et al. 2021), likely depending on the context of ECs and the microenvironment. Other targets of miR-223-3p and Fbxw7, such as Erk and Klf2 (Shi et al. 2013; Wang et al. 2013), respectively, could further increase the complexity. Our data in the current study demonstrate that Notch signaling upregulates miR-223-3p to attenuate EC migration by targeting Fbxw7. The precise role and detailed mechanisms of miR-223-3p in regulating EC proliferation require further explorations in the future.
Conclusion
miR-223-3p is a novel downstream effector of Notch signaling in ECs. Notch signaling upregulates miR-223-3p to attenuate EC migration, contributing to compromised sprouting through directly targeting Fbxw7.
References
Alabi RO, Farber G, Blobel CP (2018) Intriguing roles for endothelial ADAM10/Notch signaling in the development of organ-specific vascular beds. Physiol Rev 98:2025–2061
Autiero M, De Smet F, Claes F, Carmeliet P (2005) Role of neural guidance signals in blood vessel navigation. Cardiovasc Res 65:629–638
Bajbouj K, Ramakrishnan RK, Hamid Q (2021) Role of matrix metalloproteinases in angiogenesis and its implications in asthma. J Immunol Res 2021:6645072
Baker M, Robinson DS, Lechertier T, Barber PR, Tavora B, D’Amico G, Jones DT, Vojnovic B, Hodivala-Dilke K (2011) Use of the mouse aortic ring assay to study angiogenesis. Nat Protoc 7:89–104
Blanco R, Gerhardt H (2013) VEGF and Notch in tip and stalk cell selection. Cold Spring Harb Perspect Med 3:a006569
Chen W, Xia P, Wang H, Tu J, Liang X, Zhang X, Li L (2019) The endothelial tip-stalk cell selection and shuffling during angiogenesis. J Cell Commun Signal 13:291–301
De Bock K, Georgiadou M, Schoors S, Kuchnio A, Wong BW, Cantelmo AR, Quaegebeur A, Ghesquiere B, Cauwenberghs S, Eelen G, Phng LK, Betz I, Tembuyser B, Brepoels K, Welti J, Geudens I, Segura I, Cruys B, Bifari F, Decimo I, Blanco R, Wyns S, Vangindertael J, Rocha S, Collins RT, Munck S, Daelemans D, Imamura H, Devlieger R, Rider M, Van Veldhoven PP, Schuit F, Bartrons R, Hofkens J, Fraisl P, Telang S, Deberardinis RJ, Schoonjans L, Vinckier S, Chesney J, Gerhardt H, Dewerchin M, Carmeliet P (2013) Role of PFKFB3-driven glycolysis in vessel sprouting. Cell 154:651–663
Dou GR, Wang YC, Hu XB, Hou LH, Wang CM, Xu JF, Wang YS, Liang YM, Yao LB, Yang AG, Han H (2008) RBP-J, the transcription factor downstream of Notch receptors, is essential for the maintenance of vascular homeostasis in adult mice. FASEB J 22:1606–1617
Feng J, Chen L, Jiang Y, Tao Y (2020) The role of Apelin/APJ in a mouse model of oxygen-induced retinopathy. Invest Ophthalmol vis Sci 61:47
Fonseca CG, Barbacena P, Franco CA (2020) Endothelial cells on the move: dynamics in vascular morphogenesis and disease. Vasc Biol 2:H29–H43
Gao F, Zhang YF, Zhang ZP, Fu LA, Cao XL, Zhang YZ, Guo CJ, Yan XC, Yang QC, Hu YY, Zhao XH, Wang YZ, Wu SX, Ju G, Zheng MH, Han H (2017) miR-342-5p regulates neural stem cell proliferation and differentiation downstream to Notch signaling in mice. Stem Cell Rep 8:1032–1045
Greenspan LJ, Weinstein BM (2021) To be or not to be: endothelial cell plasticity in development, repair, and disease. Angiogenesis 24:251–269
Hoeck JD, Jandke A, Blake SM, Nye E, Spencer-Dene B, Brandner S, Behrens A (2010) Fbw7 controls neural stem cell differentiation and progenitor apoptosis via Notch and c-Jun. Nat Neurosci 13:1365–1372
Hu L, Lv X, Li D, Zhang W, Ran G, Li Q, Hu J (2021) The anti-angiogenesis role of FBXW7 in diabetic retinopathy by facilitating the ubiquitination degradation of c-Myc to orchestrate the HDAC2. J Cell Mol Med 25:2190–2202
Ji Q, Xu X, Song Q, Xu Y, Tai Y, Goodman SB, Bi W, Xu M, Jiao S, Maloney WJ, Wang Y (2018) miR-223-3p inhibits human osteosarcoma metastasis and progression by directly targeting CDH6. Mol Ther 26:1299–1312
Jiang L, Lv L, Liu X, Jiang X, Yin Q, Hao Y, Xiao L (2019) miR-223 promotes oral squamous cell carcinoma proliferation and migration by regulating FBXW7. Cancer Biomark 24:325–334
Joussen AM, Ricci F, Paris LP, Korn C, Quezada-Ruiz C, Zarbin M (2021) Angiopoietin/Tie2 signalling and its role in retinal and choroidal vascular diseases: a review of preclinical data. Eye (lond) 35:1305–1316
Karamanos NK, Theocharis AD, Piperigkou Z, Manou D, Passi A, Skandalis SS, Vynios DH, Orian-Rousseau V, Ricard-Blum S, Schmelzer CEH, Duca L, Durbee M, Afratis NA, Troeberg L, Franchi M, Masola V, Onisto M (2021) A guide to the composition and functions of the extracellular matrix. FEBS J. https://doi.org/10.1111/febs.15776
Kumar V, Palermo R, Talora C, Campese AF, Checquolo S, Bellavia D, Tottone L, Testa G, Miele E, Indraccolo S, Amadori A, Ferretti E, Gulino A, Vacca A, Screpanti I (2014) Notch and NF-kB signaling pathways regulate miR-223/FBXW7 axis in T-cell acute lymphoblastic leukemia. Leukemia 28:2324–2335
Kurashige J, Wantanabe M, Iwatsuki M, Kinoshita K, Saito S, Hiyoshi Y, Kamohara H, Baba Y, Mimori K, Baba H (2012) Overexpression of microRNA-223 regulates the ubiquitin ligase FBXW7 in oesophageal squamous cell carcinoma. Br J Cancer 106:182–188
Luo W, Garcia-Gonzalez I, Fernandez-Chacon M, Casquero-Garcia V, Sanchez-Munoz MS, Muhleder S, Garcia-Ortega L, Andrade J, Potente M, Benedito R (2021) Arterialization requires the timely suppression of cell growth. Nature 589:437–441
Muhleder S, Fernandez-Chacon M, Garcia-Gonzalez I, Benedito R (2021) Endothelial sprouting, proliferation, or senescence: tipping the balance from physiology to pathology. Cell Mol Life Sci 78:1329–1354
Phng LK, Gerhardt H (2009) Angiogenesis: a team effort coordinated by notch. Dev Cell 16:196–208
Shao J, Fan G, Yin X, Gu Y, Wang X, Xin Y, Yao Y (2019) A novel transthyretin/STAT4/miR223–3p/FBXW7 signaling pathway affects neovascularization in diabetic retinopathy. Mol Cell Endocrinol 498:110541
Shi L, Fisslthaler B, Zippel N, Fromel T, Hu J, Elgheznawy A, Heide H, Popp R, Fleming I (2013) MicroRNA-223 antagonizes angiogenesis by targeting beta1 integrin and preventing growth factor signaling in endothelial cells. Circ Res 113:1320–1330
Sun JX, Chang TF, Li MH, Sun LJ, Yan XC, Yang ZY, Liu Y, Xu WQ, Lv Y, Su JB, Liang L, Han H, Dou GR, Wang YS (2018) SNAI1, an endothelial-mesenchymal transition transcription factor, promotes the early phase of ocular neovascularization. Angiogenesis 21:635–652
Sun JX, Dou GR, Yang ZY, Liang L, Duan JL, Ruan B, Li MH, Chang TF, Xu XY, Chen JJ, Wang YS, Yan XC, Han H (2021) Notch activation promotes endothelial quiescence by repressing MYC expression via miR-218. Mol Ther Nucleic Acids 25:554–566
Taha M, Shaker OG, Abdelsalam E, Taha N (2020) Serum a proliferation-inducing ligand and MicroRNA-223 are associated with rheumatoid arthritis: diagnostic and prognostic implications. Mol Med 26:92
Tetzlaff F, Fischer A (2018) Control of blood vessel formation by Notch signaling. Adv Exp Med Biol 1066:319–338
Tiwari A, Mukherjee B, Dixit M (2018) MicroRNA key to angiogenesis regulation: miRNA biology and therapy. Curr Cancer Drug Targets 18:266–277
Wang H, Chen J, Zhang S, Zheng X, Xie S, Mao J, Cai Y, Lu X, Hu L, Shen J, Chai K, Chen W (2020) miR-223 regulates autophagy associated with cisplatin resistance by targeting FBXW7 in human non-small cell lung cancer. Cancer Cell Int 20:258
Wang R, Wang Y, Liu N, Ren C, Jiang C, Zhang K, Yu S, Chen Y, Tang H, Deng Q, Fu C, Wang Y, Li R, Liu M, Pan W, Wang P (2013) FBW7 regulates endothelial functions by targeting KLF2 for ubiquitination and degradation. Cell Res 23:803–819
Yan XC, Cao J, Liang L, Wang L, Gao F, Yang ZY, Duan JL, Chang TF, Deng SM, Liu Y, Dou GR, Zhang J, Zheng QJ, Zhang P, Han H (2016) miR-342–5p is a Notch downstream molecule and regulates multiple angiogenic pathways including Notch, vascular endothelial growth factor and transforming growth factor beta Signaling. J Am Heart Assoc 5:e003042
Yeh CH, Bellon M, Nicot C (2018) FBXW7: a critical tumor suppressor of human cancers. Mol Ther 17:115
Yuan X, Berg N, Lee JW, Le TT, Neudecker V, Jing N, Eltzschig H (2018) MicroRNA miR-223 as regulator of innate immunity. J Leukoc Biol 104:515–524
Yumimoto K, Nakayama KI (2020) Recent insight into the role of FBXW7 as a tumor suppressor. Semin Cancer Biol 67:1–15
Zhang MW, Shen YJ, Shi J, Yu JG (2020) MiR-223-3p in cardiovascular diseases: a biomarker and potential therapeutic target. Front Cardiovasc Med 7:61056
Zhou X, Jin W, Jia H, Yan J, Zhang G (2015) miR-223 promotes the cisplatin resistance of human gastric cancer cells via regulating cell cycle by targeting FBXW7. J Exp Clin Cancer Res 34:28
Funding
This study was supported by the National Natural Science Foundation of China (31671523, 31730041, 82003110) and Natural Science Foundation of Shaanxi Province (2020JQ-441).
Author information
Authors and Affiliations
Contributions
R.N.W. and Z.Y.Y. performed experiments and collected data. L.L., X.X.F., B.C., and X.Y.Z. assisted with experiments and data collection. H.H., X.C.Y., and Q.J.Z. designed the experiments and prepared the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The use of human samples was approved by the Ethics Committee of Xijing Hospital, Fourth Military Medical University. All human participants signed informed consent for the use of their umbilical cord biopsies. The animal experiments were permitted by the Animal Experiment Administration Committee of the Fourth Military Medical University.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ruonan Wang and Ziyan Yang are contributed equally to this study
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, R., Yang, Z., Liang, L. et al. Notch activation suppresses endothelial cell migration and sprouting via miR-223-3p targeting Fbxw7. In Vitro Cell.Dev.Biol.-Animal 58, 124–135 (2022). https://doi.org/10.1007/s11626-022-00649-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11626-022-00649-y